Linking changes in the soil microbial community to C and N dynamics during crop residue decomposition
2021-09-10CyrineREZGUllsabelleTRlNSOUTROTGATTlNMarieBENOlTKarineLAVALWassilaRlAHANGLET
Cyrine REZGUl,lsabelle TRlNSOUTROT-GATTlN,Marie BENOlT,Karine LAVAL,Wassila RlAHANGLET
1 AGHYLE Research Unit,UniLaSalle,Rouen-Team,Mont-Saint Aignan 76134,France
2 Agroecology and Environment Research Unit,Isara,AgroSchool for Life,Lyon 69007,France
Abstract Crop residues are among the main inputs that allow the organic carbon (C) and nutrients to be maintained in agricultural soil.It is an important management strategy that can improve soil fertility and enhance agricultural productivity.This work aims to evaluate the extent of the changes that may occur in the soil heterotrophic microbial communities involved in organic matter decomposition and C and nitrogen (N) mineralization after the addition of crop residues.Soil microcosm experiments were performed at 28°C for 90 days with the addition of three crop residues with contrasting biochemical qualities:pea (Pisum sativum L.),rapeseed (Brassica napus L.),and wheat (Triticum aestivum L.).Enzyme activities,C and N mineralization,and bacterial and fungal biomasses were monitored,along with the bacterial and fungal community composition,by the highthroughput sequencing of 16S rRNA and ITS genes.The addition of crop residues caused decreases in β-glucosidase and arylamidase activities and simultaneous enhancement of the C mineralization and net N immobilization,which were linked to changes in the soil microbial communities.The addition of crop residues decreased the bacterial and fungal biomasses 90 days after treatment and there were shifts in bacterial and fungal diversity at the phyla,order,and genera levels.Some specific orders and genera were dependent on crop residue type.For example,Chloroflexales,Inquilinus,Rubricoccus,Clitocybe,and Verticillium were identified in soils with pea residues;whereas Thermoanaerobacterales,Thermacetogenum,and Hypoxylon were enriched in soils with rapeseed residues,and Halanaerobiales,Rubrobacter,and Volutella were only present in soils with wheat residues.The findings of this study suggest that soil C and N dynamics in the presence of the crop residues were driven by the selection of specific bacterial and fungal decomposers linked to the biochemical qualities of the crop residues.If crop residue decomposition processes showed specific bacterial and fungal operational taxonomic unit (OTU) signatures,this study also suggests a strong functional redundancy that exists among soil microbial communities.
Keywords:crop residues,C and N mineralization,enzyme activities,bacterial and fungal diversity
1.lntroduction
C and N transformations from soil organic matter (OM)are mainly (micro) biologically based.Indeed,soil organic materials are the trophic resources of heterotrophic soil organisms (Patersonet al.2009).OM plays an important role in many ecosystem services provided by soils (Dominatiet al.2010).Its degradation by soil organisms allows the turnover of nutrients (mainly C,N,phosphorus (P),and sulfur (S)),which is essential for plant growth and the durability of ecosystems.Crop residue inputs have been proposed as a sustainable and cost-efficient management tool in agricultural practices to preserve soil organic matter(Lehtinenet al.2014;Arcandet al.2016;Shahbazet al.2017).The long-term balance between soil C inputs through crop residues and lossesviamineralization and oxidation determines the soil organic C content and nutrient cycling in the agrosystem (Ghimireet al.2017).
Various crop residues often exhibit different physicochemical properties,thereby affecting the soil in different ways.Their biochemical compositions and physical structures affect mineralization (Prescott 2010).Some studies have demonstrated,under controlled or field conditions,that hemicellulose,lignin,and the physical size of crop residues,affect the C and N decomposition rates (Hadaset al.2004;Abivenet al.2005;Cayuelaet al.2009;Moreno-Cornejoet al.2014).The soluble C content determines the initial rate of crop residue decomposition(Hadaset al.2004),whereas the lignin content controls the medium-to long-term fate of added C (Trinsoutrotet al.2000;Nicolardotet al.2007).According to Ghimireet al.(2017),the addition of crop residues at a higher rate potentially increases the C mineralization.Moreover,a study conducted by Redinet al.(2014) showed that the application of 25 species of crop residues with a broad range of chemical compositions affected C mineralization.
In addition,some researchers have investigated the effect of incorporating the residues of different plant organs (roots,leaves,stems,etc.) on C and N mineralization (Trinsoutrotet al.2000;Fanget al.2007;Aberaet al.2012;Redinet al.2014).As expected,these plant organs also affected soil N mineralization.Indeed,roots displayed significantly lower N mineralization compared to the shoots and leaves(Abbasiet al.2014).The biochemical composition or quality parameters,such as total N concentration,lignin,cellulose,and hemicellulose contents,exert a significant influence on crop residue decomposition.In parallel,some ratios and indexes (such as C:N ratio,lignin:N ratio,and lignocellulosic index (LCI)) are considered useful indicators for estimating both decomposition and the N release of added residues(Vahdatet al.2011;Aberaet al.2012;Abbasiet al.2014).
The heterotrophic microbial communities control the coupling of C and N cycles in soils (Recouset al.2017).Indeed,the diversity and the activity of these microbial communities,as well as the proportions of the major elements (C,N,P,S,etc.) in the crop residues,determine the partition between the C and the N that is mineralized or assimilated in soils (Chenet al.2014).In this case,the relationship between the addition of crop residues and the microbial communities has attracted increasing attention(Pascaultet al.2010a;Kriaučiūnienėet al.2012;Leeet al.2017;Fanget al.2018;Sauvadetet al.2018).Suleimanet al.(2018) indicated that the addition of wheat straw residues causes a change in the diversity and function of microbial communities.Moreover,the addition of different quantities of crop residues promotes the abundance of some bacterial groups (BacteroidetesandProteobacteria),while low-quality crop residues favor fungal abundance(Fanget al.2018).Lianet al.(2019) showed that soybean residues significantly impact the bacterial community,resulting inBacteroidetes,Actinobacteria,Firmicutes,andAcidobacteriabeing enriched at the initial stage of crop residue input.
Microbial communities in soils produce several enzyme activities which are involved in the decomposition of crop residues (Frasieret al.2016;Lashermeset al.2016;Hünninghauset al.2017;Luoet al.2017;Zhenget al.2018).Fanget al.(2019) showed a significant increase in several enzyme activities (β-glucosidase,cellobiohydrolase,xylosidase,N-acetyl-glucosaminidase,and aminopeptidase)with a marked increase at 10 days after the addition of wheat residues.These authors suggested that these increases in enzyme activities were linked to the stimulation of microbial growth and the abundance of some functional genes.In parallel,Sauvadetet al.(2016) showed that the relationship between the evolution of microbial communities and the production of enzymes during the decomposition of crop residues is related to residue qualities.Indeed,the study conducted by Aminet al.(2014) found that microorganisms exhibited an initial rapid growth in the presence of a highquality litter and produced enzymes that are not efficient in degrading recalcitrant components.Meanwhile,in the presence of the more recalcitrant maize roots,microbial biomass grew more slowly but produced enzymes of higher efficiency.This high enzyme efficiency could be explained by the synergistic action of hydrolytic and oxidative enzymes,even in the early stages of decomposition.
This study is part of a larger program that aims to evaluate the impact of innovative crop rotations (pea-wheat-barleyvs.rapeseed-wheat-barley) in the Normandy region of France.As different crops produce various residues,an understanding of residue degradation kinetics and its influence on soil health is of major interest to the evaluation of the crop systems.The objective of this paper is to evaluate whether the application of crop residues will affect the soil heterotrophic microbial community involved in OM decomposition and C and N mineralization.Although the degradation of plants of various quantities and qualities is well-known,its consequences on soil communities and their functions have received less attention.To better understand these effects,we have followed the evolution of microbial communities in relation to C-N fluxes and enzyme activities at key stages in the decomposition of different qualities of crop residues,in the pedoclimatic context of Normandy(France).The focus was on monitoring the abundance and diversity of bacterial and fungal communities,enzyme activities,and C and N mineralization as a result of incorporating contrasting crop residues (legume and nonlegume crop residues) harvested in the field.
2.Materials and methods
2.1.Soil sampling
The soil was collected from an arable field located in the Normandy region (France).This region has a temperate oceanic climate,with a mean annual rainfall ranging from 80-110 mm.The soil is sandy loam,classified as luvisol.This soil was chosen because of its characteristics,which are representative of the common pedological types of soil in Normandy.Soil samples were collected randomly from a depth of 0-20 cm from three points in each plot,using a soil auger.The soil analysis indicated that the soil texture was characterized by 6.56% clay,68.10% silt,and 25.34% sand,with a mean pH (soil H2O) of 7.6 in the topsoil.The average soil C and N contents were (17.14±3.82) and (1.27±0.07) g kg-1of dry soil,respectively.Before drying,20 g of fresh sieved soil was used to determine water content by drying the soil at 105°C for 24 h.
2.2.Collection of crop residues
Three predominant plant species available on-farm were selected as part of the crop rotation.These were pea (Pisum sativumL.),rapeseed (BrassicanapusL.),and wheat(TriticumaestivumL.).Crop residues (stem and leaf) were dried at room temperature and were then analyzed,milled,and passed through a 1-mm sieve.Triplicate samples of crop residues were taken and analyzed for their C,N,soluble(SOL) compounds,hemicellulose (HEM),cellulose (CEL),and lignin (LIG) concentrations.Total C and N contents of the residues were determined using an elemental analyzer(NA,2000;Fisons Instruments,Milan,Italy).This technique is based on the quantitative“dynamic flash combustion”method.
To characterize the biochemical composition of decomposing residues,the SOL compounds of the dried residues were determined by hot water extraction (100°C)for 30 min,followed by extraction with a neutral detergent(100°C) for 60 min using the method described by Linères and Djakovitch (1993).The resulting non-extracted material was then successively subjected to selective extraction for HEM and CEL,as described by Van Soest (1963).The mass of the residues was weighed after each step in the serial extraction,and the difference between the mass of the material subjected to a specific extraction and the mass of the residues accounted for that specific component.The mass of the final residual non-extractable fraction of the plant material was considered as LIG mass.From biochemical fractionation,the LCI was calculated as described by Melilloet al.(1989).It corresponds to the ratio between the content of crop residues in LIG and the sum of the contents of LIG,CEL,and HEM.
2.3.Laboratory incubations
Fresh soil,equivalent to 25 g of dried soil,was weighed and transferred into glass jars.Distilled water was added to the soil to achieve a soil moisture content of 80% of field capacity.The treatments comprised a control without the addition of crop residues and three crop-residue sources,i.e.,pea (P.sativumL.),rapeseed (B.napusL.) and wheat(T.aestivumL.).Crop residues were weighed and added to the jars at a rate equivalent to 2‰ of organic C of the crop residue weight,which corresponded to 0.114,0.109,and 0.106 g for pea,rapeseed,and wheat,respectively.Incubation experiments were conducted under constant C and non-limiting N conditions,according to the AFNOR Standard (AFNOR XP U44-163) (AFNOR 2009).Then,25 g of dry soil mixed with crop residues was incubated in 2-L glass jars for CO2monitoring,and in 50-mL plastic containers for the determination of mineral N and microbiological parameters.These containers were placed in large plastic trays with a few small holes in the covers to allow for gas exchange.
A total of 81 jars corresponded to soils with added pea,rapeseed,and wheat residues (three treatments) and soils without crop residues (control)×9 incubation times (1,3,7,14,21,28,49,70,and 90 days)×3 replicates for CO2monitoring.A total of 45 jars for mineral N monitoring (3 treatments+controls×5 incubation times (1,7,14,49,and 90 days)×3 replicates) were used,and some of them were taken for the determination of microbiological parameters (3 treatments+controls×2 incubation times (1 and 90 days)×3 replicates).All jars were placed in a completely randomized design.The soil was then incubated under controlled conditions at 28°C.Soil moisture was checked/adjusted every two days by weighing the glass jars and adding the required amount of distilled water.
2.4.C and N mineralization measurements
The CO2produced by the soil was trapped in 10 mL of 0.5 mol L-1NaOH in a 40-mL flat-bottom flask.The traps were changed periodically to renew the atmosphere in the jars and prevent saturation of the NaOH.The CO2produced by the soil and trapped by NaOH was determined by conductimetry using a conductivity meter (Metler Toledo Seven Multi,Darmstadt,Germany)
The initial concentration of total mineral N on day 1 was determined by extracting soil samples with 100 mL of 1 mol L-1KCl added directly to the flask immediately after incorporation of the crop residues.Thereafter,mineral N was extracted by shaking for 1 h with 100 mL of 1 mol L-1KCl followed by filtration (Whatman No.40).Extracts were stored at -20°C until analysis.The total mineral N of the extract was determined by using the Gallery Automated Photometric Analyser-Thermo (Thermo Fisher Scientific,Vantaa,Finland).
2.5.Total DNA extraction and quantification
Nucleic acids were extracted from 0.5 g of soil using a FastDNA SPIN Kit for soil (MP-Biomedicals,Santa Ana,CA,USA),according to the instructions of the manufacturer.Three replicates were performed for each treatment.DNA was quantified by a fluorimetric measurement using Hoeschst 33258 fluorochrome at 360/460 nm excitation/emission wavelengths,with a Fluorescent DNA Quantitation Kit (Biorad,Hercules,CA,USA) of dsDNA stored at -20°C(Gangneuxet al.2011).The DNA extracted from the soil corresponded to the total microbial biomass,and it was expressed as µg of extracted DNA per g of dry soil (µg g-1dry soil).
2.6.Real-time PCR amplification for bacterial and fungal biomass quantification
The 18S rRNA amplifications for fungal biomass estimation by 18S rDNA real-time qPCR were carried out with a total volume of 25 µL.The qPCR mix was prepared as follows:5 ng of soil microbial DNA,0.5 µmol L-1of each primer (FU18S1 5´-GGAAACTCACCAGGTCCAGA-3´and Nu-SSU-1536 5´-ATTGCAATGCYCTATCCCCA-3´)(Borneman and Hartin 2000),25 µL of absolute qPCR SYBR Green Mix (Roche,Basel,Switzerland),and 0.25 mg mL-1BSA (GeneOn Bioscience,Germany).Standard curves were obtained using serial dilutions of linearized plasmids containing the cloned 18S rRNA gene ofFusarium graminearum,and the results are expressed as 18S rRNA gene copy number per gram of dry soil.The amplification protocol (40 cycles of PCR,20 s at 95°C,30 s at 62°C,and 30 s at 72°C) was performed using the LightCycler 480 real-time PCR system (Roche,Basel,Switzerland).The efficiency of the qPCR ranged from 95-99%.
The 16S rRNA amplifications for bacterial biomass estimation by 16S rRNA real-time PCR were carried out under the same conditions as the 18S rDNA PCR,except for the primers (63f 5´-CAGGCCTAACACATGCAAGTC-3´ (Marchesiet al.1998) and BU16S4 5´-CTGCTGCCTCCCGTAGG-3´)derived from 341F (Muyzeret al.1993).The efficiency of the qPCR ranged from 98-100%.Standard curves were obtained using serial dilutions of linearized plasmids containing the cloned 16S rRNA genes fromPseudomonas aeruginosa,and the results are expressed as 16S rRNA gene copy number per gram of dry soil.Two independent qPCR assays were performed for the bacterial and fungal dsDNA estimations.Three replicates were performed for each treatment.
2.7.High-throughput sequencing of the bacterial 16S rRNA and fungal lTS genes
Sixteen samples of DNA extracted from soil (three soils with added residues and the control soil without residues×2 replicates×2 sampling times:T1 and T90)were analyzed for the bacterial 16S rRNA gene and for the ITS region of fungal gene amplicons by high-throughput sequencing using the Illumina MiSeq.For each sample,DNA extracted from soils was used for amplification of the V4-V5 hypervariable region of bacterial 16S rRNA gene(Claessonet al.2010) and ITS2 of fungal genes using an optimized and standardized amplicon-library preparation protocol (Metabiote®,GenoScreen,Lille,France).An internal bacterial (artificial bacterial community comprising 17 different bacteria (ABCv2)) and fungal (artificial fungal community comprising 11 different fungi (AFC)) positive control and a negative (sterile water) control were also performed.Briefly,PCR reactions were performed using 5 ng of genomic DNA and 192 fusion barcoded primers(at 0.2 µmol L-1final concentrations) with an annealing temperature of 50°C for 30 cycles.PCR products were purified using Agencourt AMPure XP magnetic beads(Beckman Coulter,Brea,CA,United States),quantified according to GenoScreen’s protocol,and mixed in an equimolar amount.Sequencing was performed using 250-bp paired-end for bacteria and 300-bp paired-end for fungi-sequencing chemistry on the Illumina MiSeq platform(Illumina,San Diego,CA,United States) at GenoScreen(Lille,France).
The raw paired-end reads (495 447 for bacteria and 1 465 138 for fungi) were demultiplexed per sample and subjected to the following steps:(1) search and removal of both forward and reverse primers using CutAdapt with no mismatches allowed in the primer sequences;(2) qualityfiltering using the PRINSEQ-lite PERL script (Schmieder and Edwards 2011) by truncating bases at the 3´ end with Phred quality score<30;(3) paired-end read assembly using FLASH (Magoč and Salzberg 2011) with a minimum overlap of 30 bases and >97% overlap identity.Taxonomic and diversity analyses were performed with the Metabiote Online ver.2.0 pipeline (GenoScreen,Lille,France),which is based in part on QIIME Software ver.1.9.1 (Caporasoet al.2010).Following pre-processing,full-length 16S rRNA gene sequences were checked for chimera sequences (inhouse method based on Usearch 6.1).For both bacteria and fungi,similar sequences with a nucleic identity-defined threshold (97% identity for an affiliation at the genus level on the V4-V5 regions of the 16S rRNA gene and the ITS regions,respectively) were clustered with Uclust ver.1.2.22q(Edgar 2010) through an open-reference OTU-picking process and complete-linkage method,finally generating groups of sequences,or“Operational Taxonomic Units.”An OTU cleaning step,involving elimination of singletons,was performed.The most abundant sequence of each OTU was considered the reference sequence of its OTU and taxonomically compared to the Greengenes database.The taxonomy of each 16S rRNA and ITS gene sequence was analysed by the RDP classifier algorithm ver.2.2 (Coleet al.2014) against the database of Greengenes (version 13_8;www.greengenes.gov) for the 16S rRNA gene and UNITE7.0 (alpha version;https://unite.ut.ee) for the ITS.Alpha-diversity metrics (Chao1 index) within samples were computed using QIIME ver.1.9.1.
2.8.Enzyme activities
Soil enzymes have been reported as useful soil-state bioindicators because they provide information on the soil’s ability to perform biogeochemical reactions (Sinsabaugh and Shah 2012;Almeidaet al.2015).We have chosen to measure β-D-glucosidase (β-GLU;EC:3.2.1.21),N-acetylβ-glucosaminidase (NAG;EC:3.2.1.30),and arylamidase(ARYLN;EC:3.5.1.5) activities,which decompose different substrates with varying complexity (Sinsabaugh and Follsatd Shah 2012).The GLU enzyme is related to the C cycle,acting in the cleavage of cellobiose into glucose molecules(Almeidaet al.2015).The ARYLN enzyme catalyses the hydrolysis of N-terminal amino acids from arylamides(Dodor and Tabatabai 2007).NAG enzymes catalyse the hydrolysis of chitin,which is important in C and N cycling in soils.Both ARYLN and NAG activities are associated with microbial N acquisition,and they play a major role in N mineralization in soils (Ekenler and Tabatabai 2004;Jianet al.2016).Enzyme activity measurements were performed in 96-well microplates in triplicate according to the AFNOR Standard (AFNOR 20130:2018).Briefly,for β-GLU and NAG activities,4 g soil samples were mixed for 10 min at 250 r min-1with 25 mL water.Soil suspensions were incubated respectively with 0.05 mol L-14-nitrophenyl β-Dglucopyranoside (Sigma-Aldrich,Merck KGaA,Darmstadt,Germany) and 0.01 mol L-14-N-acetyl-β-D-glucosaminide(Sigma-Aldrich,Merck KGaA,Darmstadt,Germany).The reaction was stopped (with 0.5 mol L-1CaCl2and 0.1 mol L-1Tris at pH 12) after 1 h,each plate was centrifuged (5 min at 1 500× g),and absorbance was measured on a microplate reader (Varioskan Flash-Thermo Scientific,Vantaa,Finland).The amounts ofp-nitrophenol were obtained by measuring the absorbance at λ=405 nm,with comparison to calibration curves.
For ARYLN activity,4 g soil samples were mixed for 10 min at 250 r min-1with 25 mL Trizma base (50 mmol L-1,pH 7.5).Soil solutions were incubated with 0.008 mol L-1L-leucine β-naphthylamide hydrochloride (Sigma-Aldrich,Merck KGaA,Darmstadt,Germany).The β-naphthylamine produced was extracted with acidified ethanol and converted to an azo compound by reacting withp-dimethylaminocinnamaldehyde (DMCA).The amount of β-naphthylamine was obtained by measuring the absorbance at λ=540 nm,with comparison to calibration curves.
Three replicates were performed for each soil (with and without residues) at two sampling times corresponding to one day after incubation (T1) and 90 days after incubation(T90),for a total of 24 samples.Enzyme activities are expressed as nmol min-1g-1dry soil.
2.9.Statistical analysis
Statistical tests were carried out to compare the HEM,CEL,LIG,and SOL fractions between the three crop residues.Statistical tests were also carried out to evaluate the effects of the addition of crop residues on the total microbial biomass (dsDNA),the total bacterial and fungal biomasses (16S and 18S rRNA gene copy numbers),bacterial and fungal diversity (high-throughput sequencing),enzyme activities (β-GLU,NAG,and ARYLN),and C and N dynamics.The measured data were compared by Tukey’s test with significant differences at 5% (P<0.05).The diversity of bacterial and fungal communities was analyzed by a Venn diagram.Principal component analysis(PCA) was performed to compare crop residue effects on microbial parameters.The calculations of the correlations between the different parameters were carried out using a Spearman’s rank correlation procedure,and the significance level of these correlations was tested using a pairwiset-test.All analyses were performed using R (The R Development Core Team 2019).
3.Results
3.1.Composition of crop residues
The C content of the residues ranged from 438 to 467 g kg-1,while the N content ranged from 6.1 to 15.5 g kg-1,respectively.Pea residues exhibited the highest N content,while wheat residues exhibited the highest C content.The biochemical characterization of crop residues showed significant differences among their compositions (Table 1).Pea residues exhibited the highest SOL fraction (37%) and the lowest content of HEM (2%).The LIG content varied between 6% in wheat residues and 12% in rapeseed residues,whereas the CEL content was equivalent in the three crop residues and corresponded to the most abundant component.For the three crop residues,the C:N ratio ranged from 28.1±3.06 to 76.6±4.60 (pea residues<rapeseed residues<wheat residues),while the LIG:N ratio varied from 1.24±0.17 to 0.50±0.50.LIG:N was the highest in rapeseed residues,while the lowest values were recorded in the pea residues.Regarding the LCI ratio,rapeseed residues were the most recalcitrant compared to pea and wheat residues (Table 1).
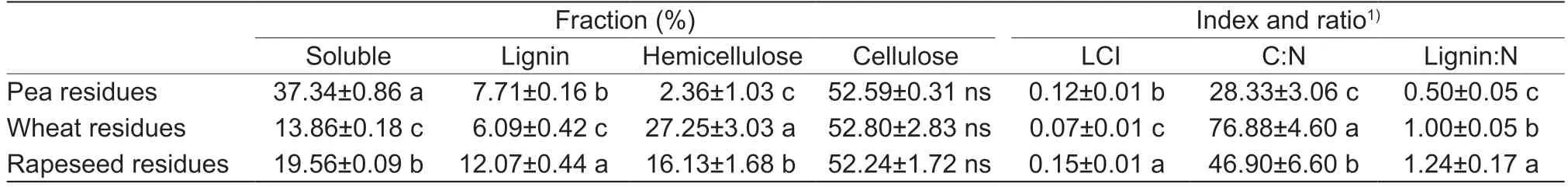
Table 1 Main characteristics of the three crop residues
3.2.C and N mineralization after the addition of crop residues
Soils without residues presented the lowest C mineralization(943 mg C-CO2kg-1dry soil) compared to soils with added crop residues.In each treatment,net C mineralization was calculated by subtracting mineralized C in the control soil from mineralized C in the soils with residues (Fig.1-A).During the beginning of the incubation (until day 9),soils with pea residues exhibited the highest C mineralization(Fig.1-B).Then (until day 90),net C mineralization was similar for soils amended with pea and rapeseed residues(2 200 mg C-CO2kg-1of dry soil) and significantly higher for soil with wheat residues (2 400 mg C-CO2kg-1of dry soil;Fig.1-A).
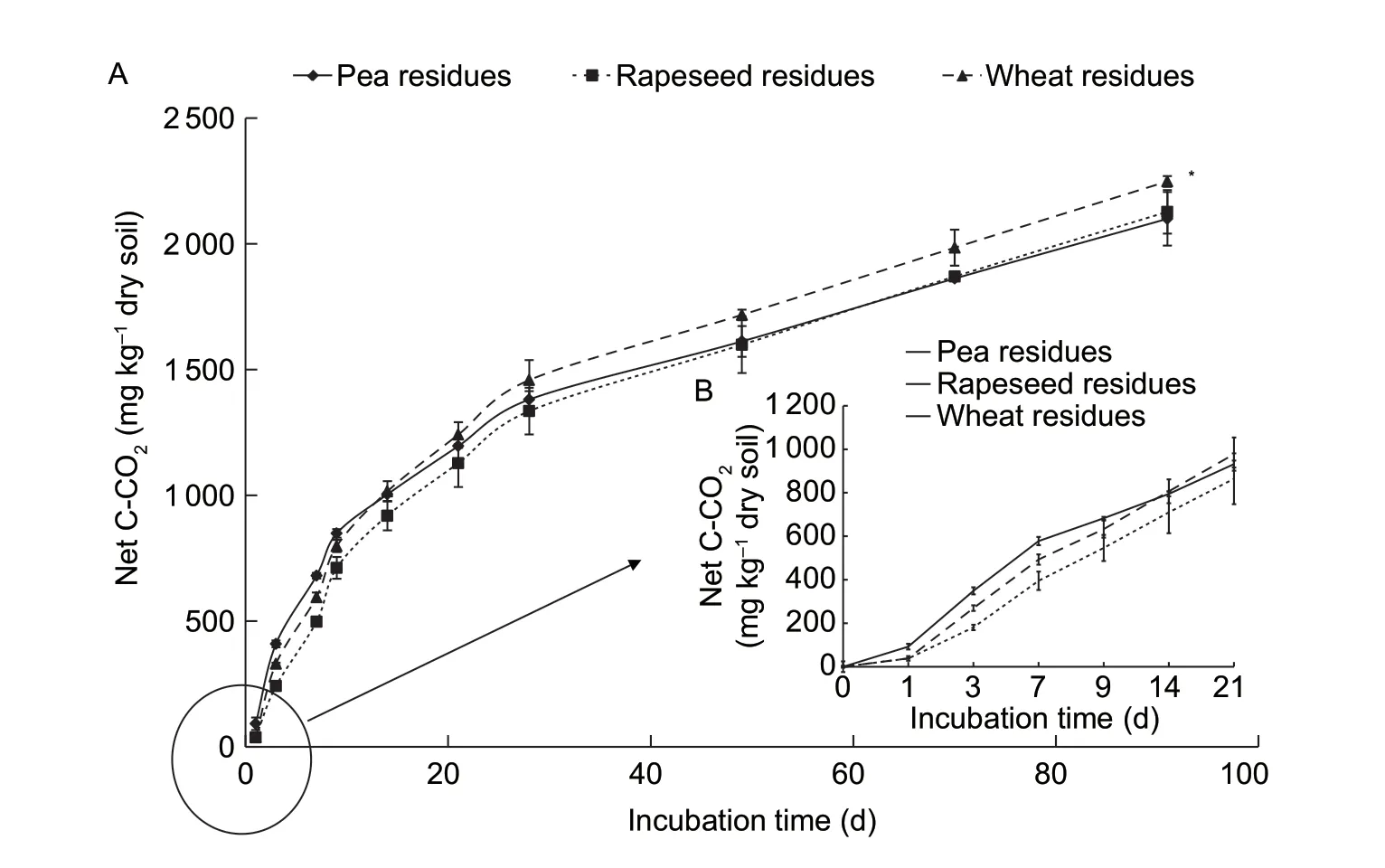
Fig.1 Net C mineralization in soil after the addition of crop residues after 90 days of incubation (A) and after 21 days of incubation(B).Data are mean±SD (n=3).* indicates statistical significance between soils with different crop residues at P<0.05 according to Tukey’s test.
Regarding the N dynamic,it was expressed as net cumulative mineralized N by subtracting the amount of mineral N recovered in control soils (Fig.2).No matterwhich crop residue is added,the amount of N in soil without residues is higher than the amount found in the soil with crop residues.Therefore,all the residues drove a net N immobilization during the 90 days of the experiment.Wheat residues showed the highest N immobilization (309 mg N kg-1of dry soil) compared to rapeseed (263 mg kg-1of dry soil) and pea residues (182 mg N kg-1of dry soil).
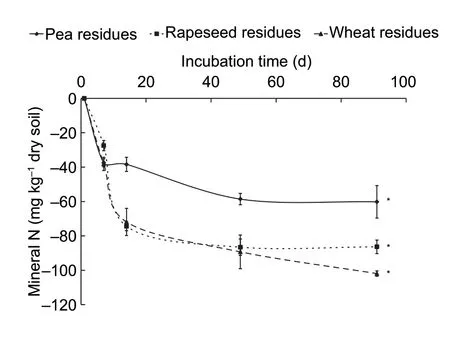
Fig.2 Net N immobilization in soils under three crop residue additions at different incubation times.Data are mean±SD(n=3).* indicates statistical significance between soil with different crop residues at P<0.05 according to Tukey’s test.
3.3.Enzyme activities after the addition of crop residues
The three tested enzymes did not respond in a similar way to the presence of crop residues (Fig.3).The β-GLU activity was 1.4 times significantly higher in amended soils compared to control soils at the beginning of the experiment.Whereas NAG activities were 1.3 and 1.2 times significantly higher after pea and wheat residue addition,respectively,compared to the control.For ARYLN activity,no significant effect was observed after the addition of crop residues.At the end of the incubation,β-GLU activities decreased by about 6.71 and 7.28% following the addition of pea and rapeseed residues.The most marked decrease was observed for ARYLN activity,which decreased by about 38 and 35% after the addition of wheat and rapeseed residues.NAG activity was negatively correlated with LIG and LCI;however,no significant effect was observed on its activity after the addition of crop residues.Other measured enzyme activities were not correlated with the biochemical qualities of the crop residues.
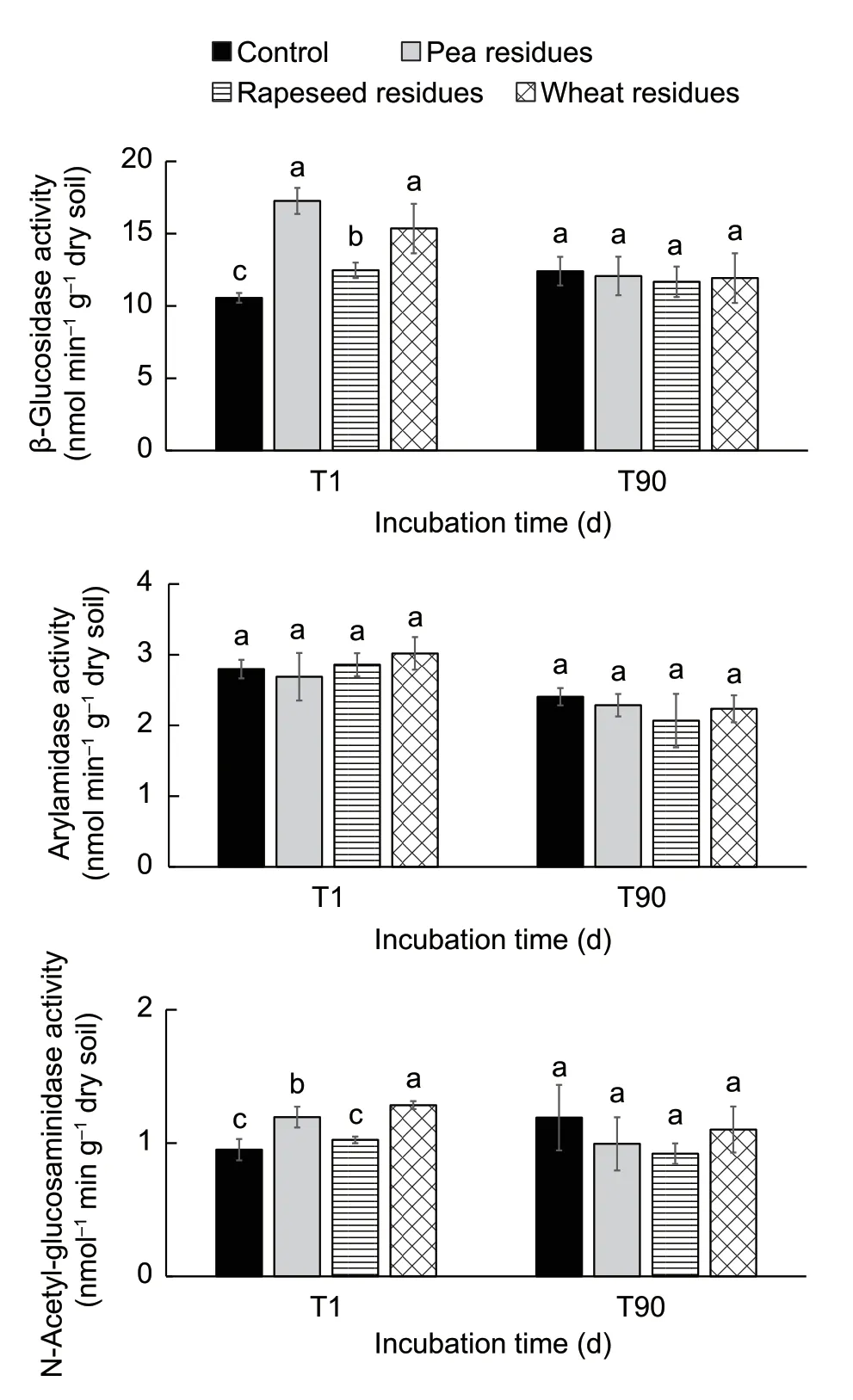
Fig.3 β-Glucosidase,arylamidase,and N-acetylglucosaminidase enzyme activities in soils with and without crop residues at two incubation times.T1,one day after incubation;T90,90 days after incubation.Data are mean±SD (n=3).Letters (a,b and c) indicate statistical significance between soils with crop residues and soil without crop residues (control)for each sampling time at P<0.05 according to Tukey’s test.
3.4.Abundance of the total bacterial and fungal biomasses after the addition of crop residues
At the beginning of the incubation,the addition of crop residues slightly increased total microbial biomass(assessed by the quantification of DNA extracted from soil)compared to the control (about 20%;Table 2).At the end of the incubation (T90),except for soils with pea residues that showed a slight decrease in microbial biomass,no difference was observed between soils amended with rapeseed or wheat residues and the control soil.A significant effect was observed between T1 and T90 on total microbial biomass.
Regarding bacterial biomass (Table 2),no crop residue effect was observed at the end of the incubation.However,a significant increase was observed between soils with added residues and the control at the beginning of the incubation(T1).For fungal biomass,various effects were observed.A significant decrease in soils with added pea and wheat residues and an increase in soils with rapeseed residues was observed at the end of the incubation.
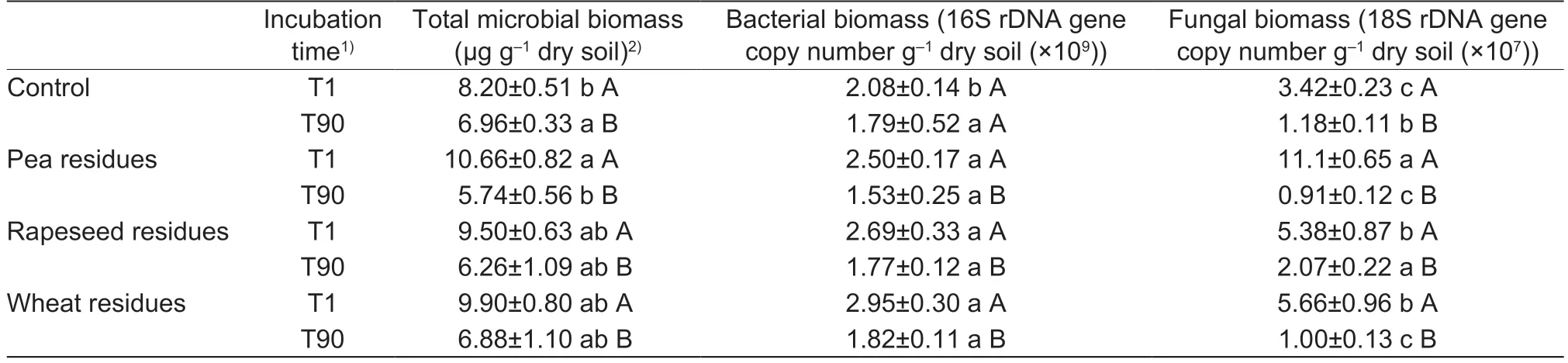
Table 2 The total microbial,bacterial and fungal biomasses from soil without residues (control) and soils with crop residues at two incubation times
3.5.High-throughput sequencing analysis of the microbial communities after the addition of crop residues
For bacterial community composition,Miseq sequencing yielded a total of 427 701 single reads,and their distribution ranged from a minimum of 24 622 to a maximum of 35 845 reads per sample of 16S rDNA.The rarefaction curves of the soil samples reached the plateau for all samples (Appendix A).The OTUs were assigned to 27 phyla,70 orders,and 200 genera.When the abundance exceeds 1% (>1%),we have considered the phylum as dominant in the bacterial community,as reported by several authors (Lópezet al.2017;Wanget al.2018;Denget al.2019).The abundant phyla are shown in Fig.4-A.Among them,the most dominant phylum corresponded to Proteobacteria (48%),which included α-Proteobacteria (25%),δ-Proteobacteria(9%),β-Proteobacteria (7%),and γ-Proteobacteria (6%),followed by Actinobacteria (19%),Acidobacteria (8%),Bacteroidetes (8%),Chloreflexi (4%),Nitropirae (3%),Gemmatimonadetes (2%),and Fimicutes (2%).
At the phylum level,Proteobacteria was the most abundant group regardless of the sampling time or the considered residues.Proteobacteria increased significantly between T1 and T90 in all treatments.Proteobacteria varied significantly between the amended soil with wheat residues (48%) and soils with pea and rapeseed residues(≈51%).At the end of the incubation,the most significant decrease of β-Proteobacteria was observed in the soil with wheat residues,while only a slight decrease for γ-Proteobacteria was observed in soil with pea residues.No significant difference was observed for δ-Proteobacteria after the addition of residues,whereas the abundance of α-Proteobacteria was significantly higher after the addition of rapeseed residues.Actinobacteria and Bacteroidetes decreased significantly at T90,especially after the addition of pea and rapeseed residues,respectively.The relative abundances of Chloroflexi,Gemmatinomonadetes,and Firmicutes increased about 1.5,2,and 1.6 times in soils with wheat,pea,and rapeseed residues,respectively,in comparison to the control soil at the end of incubation (T90).The less abundant phyla (<1%) are shown in Fig.4-B,and they are represented by Armatimonadetes,Elusimicrobia,Chlorobi,Fibrobacteres,GN02,OD1,WPS-2,BHI80-139,GAL15,OP11,WS2,and WS3.Among these phyla,Armatimonadetes,Chlorobi,GN02,GAL 15,WPS-2,and OD1 were,in most cases,significantly different between the control and the amended soils at the end of the incubation(T90).Indeed,the relative abundances of WPS-2,WS3,Armatimonadetes,and GN02 increased significantly compared to the control,while the abundances of Chlorobi,OP3,GAL 15,Fibrobacteres,and OD1 decreased.Overall,the diversity of the bacterial profile based on minor phyla showed similar profiles in soil amended with rapeseed residues and the control soil,and these profiles were different from those observed in soils amended with wheat or pea residues.
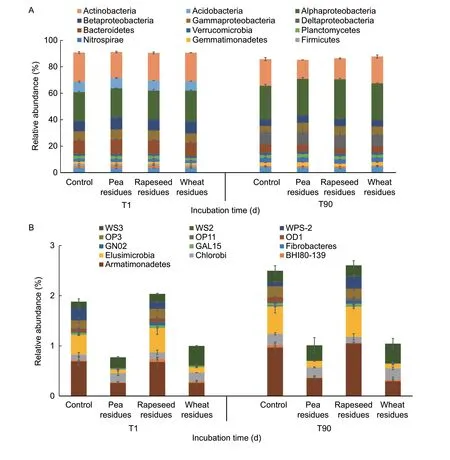
Fig.4 The relative abundances of major bacterial phyla exceeding 1% (A) and minor bacterial phyla at less than 1% (B) in the soils with and without crop residues (control) at two incubation times.T1,one day after incubation;T90,90 days after incubation.Data are mean±SD (n=2).
At the order level,five orders (Chloroflexales,Rhodothermales,Nitrososphaerales,Rickettsiales,and Ktedonobacterales) were identified after the addition of pea residues,mainly dominated by Rickettsiales (Fig.5).Under rapeseed residues,four orders were identified,with the highest abundance in Thermoanaerobacterales OTUs.In soils with wheat residues,Halanaerobiales,Rubrobacterales,Planctomycetales,and Chthoniobacterales were identified.The bacterial orders were represented by a relatively equal number of OTUs.The number of genera-specific OTUs was higher than those identified for the order level after the addition of the three crop residues (Fig.5).Indeed,12 genera were identified under pea residues,with the highest abundance in Inquilinus.Under rapeseed residues,eight genera were observed with an average of equal abundance of OTUs.Seven genera were specifically identified after the addition of wheat residues,mostly represented by OTUs assigned to Nocardioides,Aquicella,and Rubrobacter.
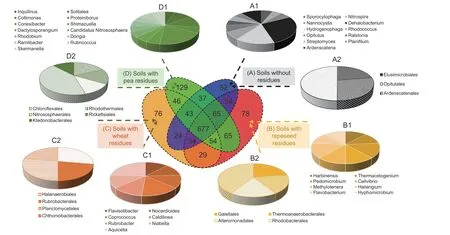
Fig.5 Venn diagram for bacterial diversity showing the unique and shared OTUs between soils with and without crop residues.OTUs were defined at 97% sequence similarity.Pie charts showing the abundances of unique OTUs at genera (A1,B1,C1 and D1) and order levels (A2,B2,C2 and D2) in soils without residues (A),with rapeseed residues (B),with wheat residues (C),and with pea residues (D).
For fungal community composition,Miseq sequencing yielded a total of 91 571 single reads,and the rarefaction curves of the soil samples reach the plateau for all samples(Appendix A).The OTUs were assigned to 13 fungal phyla,including Ascomycota,Basidiomycota,Mortierellomycota,Chytridiomycota,etc.The most dominant fungal phyla included Basidomycota,Ascomycota,and Mortierellomycota (Fig.6).These phyla are differentially impacted by the addition of crop residues.The abundance of Basidomycota and Ascomycota increased after the addition of crop residues.Basidomycota was greater after the addition of rapeseed residues.In contrast,the relative abundances of Ascomycota identified in the study were significantly higher after the addition of wheat(18.08±2.24% in control and 32.89±0.78% after the addition of wheat residues) and pea residues (18.08±2.24% in control and 6.98±1.22% after the addition of pea residues).Mortierellomycota phyla decreased after the addition of the three crop residues and was significantly more important after wheat residues.Entomophthoromycota and Mucoromycota phyla were present only after the addition of pea and wheat residues,respectively.The addition of crop residues markedly enriched soils with Cercozoa;the abundance of this fungal group increased three times after the addition of pea residues and about two times after the addition of wheat and rapeseed residues.

Fig.6 The relative abundances of fungal phyla in the soils with and without crop residues (control) at two incubation times.T1,one day after incubation;T90,90 days after incubation.Data are mean±SD (n=2).
At the order level,two orders (Chaetosphaeriales and Basidiobolales) were identified after the addition of pea residues,mainly dominated by Basidiobolales (Fig.7).Under rapeseed residues,three orders were selected and were represented by a relatively equal number of OTUs.In soils with wheat residues,six orders were selected,with the highest abundance in Malasseziales OTUs.
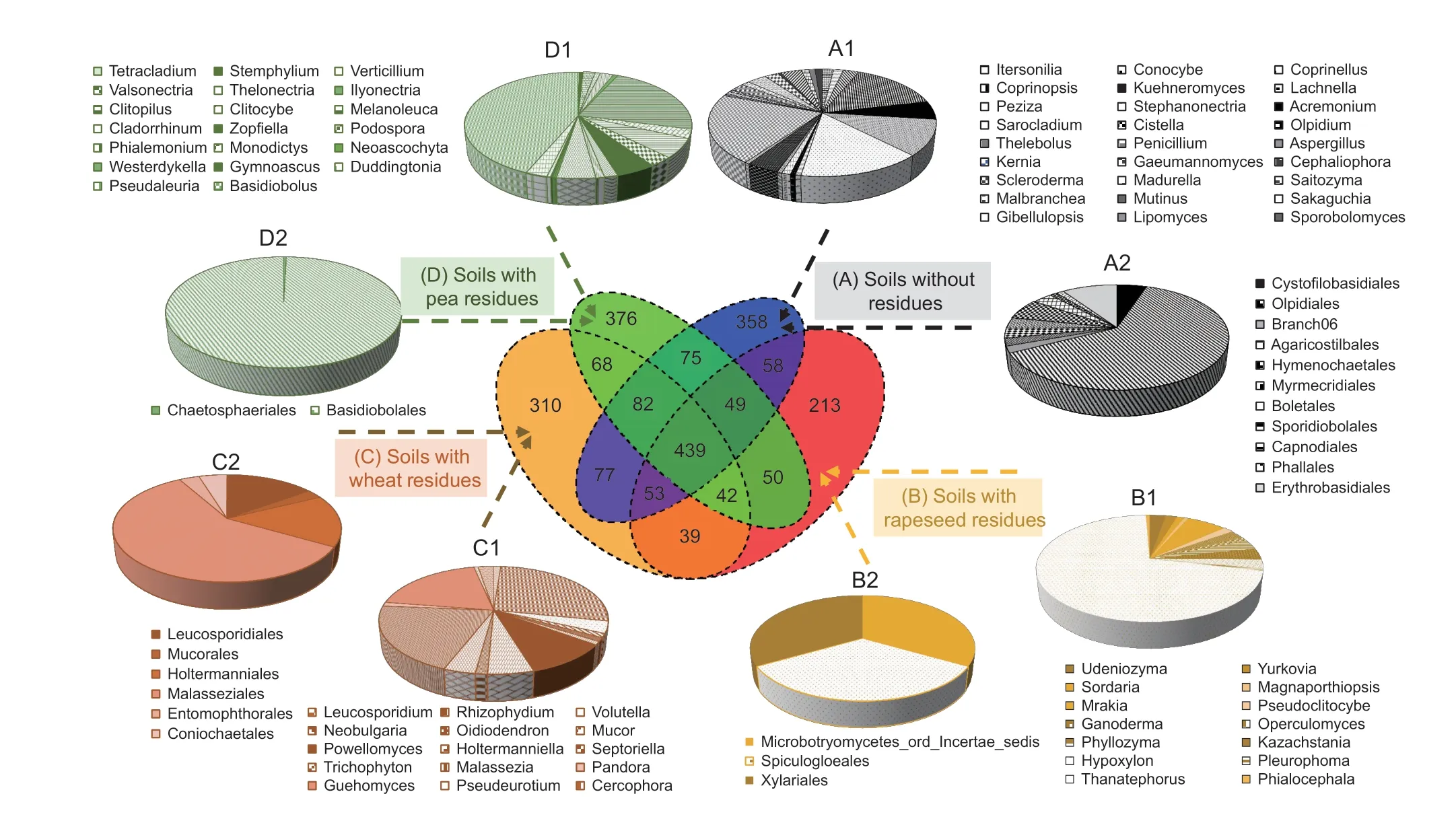
Fig.7 Venn diagram for fungal diversity showing the unique and shared OTUs between soils with and without crop residues.OTUs are defined at 97% sequence similarity.Pie charts showing the abundances of unique OTUs at genera (A1,B1,C1 and D1) and order levels (A2,B2,C2 and D2) in soils without residues (A),with rapeseed residues (B),with wheat residues (C),and with pea residues (D).
Among the 275 identified fungal genera,the number of genera-specific OTUs was higher than those identified for the order level under the addition of the three crop residues (Fig.8).In fact,20 genera were identified under pea residues,with the highest abundance in Basidiobolus.Fourteen and 15 genera were specifically identified after the addition of rapeseed and wheat residues,respectively.
3.6.The effects of the biochemical qualities of crop residues on the diversity of bacterial and fungal communities
The effects of crop residue biochemical qualities on the diversity of bacterial and fungal communities have been studied using a PCA (Fig.8).The results showed that the diversity of soil bacterial and fungal communities is significantly different according to both the addition of crop residues (controlvs.amended soil) and their biochemical characteristics (pea,wheat,and rapeseed).Based on the diversity of bacterial and fungal communities,the PCA showed three clusters,including control soils,soils with rapeseed or wheat residues and soil with pea residues(Fig.8).For bacterial and fungal diversities,the two axes of the PCA explain 65.8% of the total variance.The PCA clearly illustrates the link between the biochemical composition of crop residues and the microbial diversity at the genera and order levels.The PCA score plot revealed that the changes in bacterial and fungal diversity were more pronounced in soils with pea residues in comparison to soils with rapeseed and wheat residues.The specific bacterial genera (Dongia,Collimonas,Candidatus Nitrososphaera,Inquilinus) and fungal orders and genera (Basidiobolales,Chaetosphaeriales,Verticillium,Clitocybe,Neoascochyta)identified under soils amended with pea residues were linked to the soluble fraction and total N content of crop residues.While soils amended with rapeseed and wheat residues are not discriminated based on bacterial and fungal genera,some genera seem to be dependent on plant species.The addition of rapeseed residues affected Niabella,Cellvibrio,and Hypomicrobium bacterial genera and Spiculogloeales,Microbotryomycetes_ord_Incertae_sedis,Xylariales Udeniozyma,Yurkovia,and Sordaria fungal orders and genera,which were strongly linked to the C:N ratio,and to a lesser extent,to lignin and cellulose contents.For soil amended with wheat residues,Flavobacterium,Rubrobacter,and Caldilinea bacterial genera and Leucosporidiales,Mucorales,Leucosporidium,and Rhizophydiumgenera fungal orders and genera were closely correlated with hemicellulose content.The circular arrow in Fig.8 represents the gradual change in bacterial and fungal diversity,which should be linked to the initial biochemical composition of the crop residues.The distribution of bacterial and fungal diversity at genus and order levels highlighted by the PCAs are consistent with those described by the Venn diagram (Figs.5 and 7).
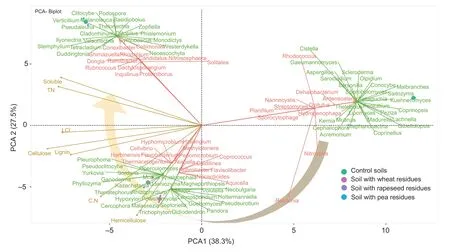
Fig.8 Principal component analysis (PCA) of crop residue effect on bacterial and fungal diversity at the end of incubation(90 days).The circular arrow indicates the gradual changes in the bacterial and fungal diversity in relation to the initial biochemical composition of the crop residues.Bacterial diversity is designated in red color,fungal diversity designated in green color,and biochemical composition of crop residues is designated in yellow color.
4.Discussion
4.1.Relationship between composition of the crop residues and C mineralization
In this study,a higher rate of C mineralization at the beginning of the incubation (until day 9) was observed after the addition of pea residues,which presented the highest content of SOL compounds (37%) (Fig.1-B).Bertrandet al.(2007) reported that labile compounds (SOL fraction)determine the decomposition rate of crop residues in the early stages.These results agree with those of our study since SOL fraction and net C mineralization (day 1) were significantly correlated (r=0.74,P<0.05;Appendix B).Concentrations of total sugars as soluble compounds have been considered to exert dominant control in the earlier stages of crop residue decomposition (Johanssonet al.2011).Some studies show that legume residues have the highest content of soluble sugars compared to wheat and rapeseed residues,which explains the highest level of C mineralization observed after the addition of pea residues(Hashemet al.2013;Stewartet al.2015).At the end of the incubation,a decline in mineralization rate was observed.
This observation had been previously explained by the increase in crop residue recalcitrance over time due to the greater proportion of remaining recalcitrant compounds at the end of decomposition (Šnajdret al.2011;Wickingset al.2012).Some studies reported that at late stages,CEL,HEM,and LIG compounds,which are more recalcitrant,defined the decomposition of crop residues (Machinetet al.2011;Šnajdret al.2011).In our study,however,no significant correlation was found between these recalcitrant fractions and the C mineralization rate (Appendix B).
Some researchers have shown a negative correlation between LIG fraction and C mineralization,and they explain that this is due to the resistant nature of LIG to the rapid microbial decomposition that promotes the formation of a complex phenyl-propanol structure,which often covers the cellulose-hemicellulose matrix (Stewartet al.2015).Due to the complexity of this matrix,microbial substrate use efficiency for LIG is low because this efficiency decreases with increasing LIG content (Bahriet al.2008;Cotrufoet al.2013).The results of this study showed the highest C mineralization rate in soils with wheat residues,which have the lowest amount of LIG (3%).Crop residues vary in their structural and chemical compositions (Adairet al.2008),and their decomposition rates are generally negatively related to the number of recalcitrant compounds present in their biomass,such as lignin,phenols,and tannins (Bertrandet al.2007;Castellanoet al.2015).In this case,crop residues with the lowest LIG content (less SGH-lignin content) had a higher C mineralization rate (Stewartet al.2015).
4.2.Relationship between crop residue composition and N dynamics
Crop residue incorporation in soils is subject to biological degradation (Berg and McClaugherty 2008).During biological degradation,crop residue C is used in respiration by decomposers,which releases CO2and provides energy(Chenet al.2009).With this energy source,microbial communities absorb N from the soil to promote their propagation.Therefore,the availability of mineral N in the soil can play an important role in the decomposition of crop residues (Chenet al.2014).Its influence depends mainly on the N demand of the microorganisms.In this study,the monitoring of the mineralization kinetics was carried out under non-limiting N conditions so as to avoid a confounding effect of N availability (Recouset al.1995;Trinsoutrotet al.2000).In this case,it is expected that the N residue content will not significantly affect the decomposition rate of crop residues.This study’s results showeda decrease in the rate of mineral N in the soil after the addition of crop residues.For each residue,a net N immobilization is observed for the entire duration of the incubation.It has been previously demonstrated that crop residues with low N (or with a high C:N ratio>20) lead to N immobilization in the early stages of decomposition (Jensen 1994;Johnsonet al.2007).
The results showed an important N immobilization in the soils,with wheat residues presenting the highest proportion between the immobilized N and the N residue content (1.68%).Moreover,a negative correlation was also observed between the rate of mineral N in soils with added residues and the C:N ratio of the crop residues(r=-0.92,P<0.05;Appendix B).Therefore,microbial activity and N immobilization are favored for residues with a higher rate of mineralized C,such as wheat residues (Hadaset al.2004;Kriaučiūnienėet al.2012),due to the degradation of crop residues and the subsequent consumption of mineral N by the soil microbial communities.
4.3.Effect of crop residue addition on enzyme activities
In this study,the addition of crop residues induces an immediate increase in hydrolytic enzyme activities (β-GLU and NAG) at the beginning of the experiment.This is consistent with results shown by Aminet al.(2014) who demonstrated that the addition of residues increases enzyme (cellulase,xylanase,and laccase) activities after only 10 min of incubation.Other authors have shown stimulation of some enzyme activities (β-glucosidase,cellobiohydrolase,xylosidase,phosphatase,N-acetylglucosaminidase) after the addition of wheat residues(Fanget al.2018).This contribution stimulates the growth of starved microbial populations,and therefore,the rate of production of enzymes (Piotrowska and Wilczewsk 2012;Aminet al.2014;Sauvadetet al.2017).ARYLN,which is considered as an N acquiring enzyme (Dodor and Tabatabai 2007),did not change at the beginning of the incubation because of the non-limiting N conditions in the soils.At the end of the incubation,decreases of β-GLU and ARYLN activities were noted,and these two enzymes were strongly correlated with each other (r=0.78,P<0.05;Appendix B).Hydrolase enzymes are associated with the degradation of parietal polysaccharides (Cusacket al.2010;Bellet al.2014) and peaked at earlier stages of degradation after the addition of crop residues (Rinkeset al.2011;Šnadjret al.2011).The decrease in these enzyme activities at the end of the experiment could be explained by several factors,including the depletion of less complex organic compounds(Geisseleret al.2009;Piotrowska and Wilczewsk 2012) and the contribution of oxidative enzymes in the degradation of recalcitrant compounds (Piotrowska and Wilczewsk 2012)as demonstrated by the strong negative correlation between NAG activity and LCI (r=-0.69,P<0.05;Appendix B) and NAG activity and LIG (r=-0.83,P<0.05;Appendix B).In addition,the negative correlation between NAG activity and fungal biomass (r=-0.80,P<0.05;Appendix B) reinforces the previous findings (Geisseleret al.2009;Kriaučiūnienėet al.2012;Piotrowska and Wilczewsk 2012) showing the large contribution of fungal oxidative enzymes in the degradation of crop residues at the end of incubation.
4.4.Effect of the addition of crop residues on the abundance of microbial communities
The addition of crop residues enhances total,bacterial,and fungal biomasses only at the beginning of the incubation.Several researchers have argued that the addition of crop residues induces the stimulation of total biomass by enhancing its growth activity,resulting in a significant increase in total biomass (Geisseleret al.2009).Some researchers have shown the same findings as our study using different methods to analyze microbial biomass under the addition of wheat,pea,and rapeseed residues(Kriaučiūnienėet al.2012;Nguyen and Marschner 2017;Sauvadetet al.2017).The late stage of the residue’s decomposition is marked by the colonization of crop residues by a population that slowly degrades the most recalcitrant fraction,such as LIG (Fontaineet al.2003;Sauvadetet al.2017).The correlations observed between fungal biomass and both LIG and LCI (r=0.57,P<0.05 andr=0.72,P<0.05;Appendix B) confirmed the findings in the literature showing the involvement of fungi in the degradation of recalcitrant compounds by using LIG as a C source (Boeret al.2005;Kriaučiūnienėet al.2012).As suggested by the literature and as demonstrated by the significant correlations between LIG and some bacterial strains (Cellvibrio,Pedomicrobium,Haliangium,Harbinensis,Flavobacterium,etc.) (Appendix C),bacteria can also degrade LIG,sometimes more effectively than the fungal population (Peresteloet al.1996;Vargas-Garcíaet al.2007).The observed increase in fungal biomass at the end of incubation after the addition of rapeseed residues,could be linked,as suggested by Berget al.(2015),to the higher Mn content of rapeseed residues(Isikhuemhenet al.2014),compared to pea and wheat residues.Indeed,Berget al.(2015) showed a relationship between the crop residue decomposition rate and Mn concentrations,suggesting the stimulation of fungal Mn peroxidase enzymes (Hatakka and Hammel 2011).
4.5.Effect of the addition of crop residues on the taxonomic composition of bacterial communities
The addition of crop residues impacted the soil bacterial community by inducing a significant change in the abundance of some bacterial groups.The rapid change in the community observed at the early stages of residue degradation is explained by the modification of simple compounds (sugars,starch,etc.).The addition of crop residues enhanced copiotrophs (R-strategists) bacterial phyla,which are stimulated in C-rich environments (Fiereret al.2007;Pascaultet al.2010b;Sauvadetet al.2017;Suleimanet al.2018;Zhenget al.2018) and are mostly represented by β-and γ-Proteobacteria sub-classes(Acosta-Martínezet al.2008;Verzeauxet al.2016;Suleimanet al.2018).In this study,these phyla increased because of substrate availability and their fast growth capacity (Anejaet al.2006;Bernardet al.2007;Fiereret al.2007;Pascaultet al.2013;Trivediet al.2013;Taoet al.2019).After 90 days,α-and δ-Proteobacteria,and Firmicutes increased after the addition of crop residues as shown in previous studies(Ramirezet al.2012;Verzeauxet al.2016;Leeet al.2017;Suleimanet al.2018).Because of the depletion of labile C compounds,these oligotrophs (K-strategists) phyla grow slowly in nutrient-poor environments (Pascaultet al.2010a;Sauvadetet al.2017;Suleimanet al.2018) and can use a wide range of recalcitrant C compounds (Gaillardet al.2003).In addition,most of the bacteria affiliated with α-and δ-Proteobacteria,and Firmicutes can create cellulosomes,which are involved in the digestion of recalcitrant compounds(Ziganshinet al.2013;Saccoet al.2016;Leeet al.2017).Actinobacteria and Bacteroidetes abundances decreased 90 days after the crop residue input into the soil.These results are in agreement with those of Suleimanet al.(2018) but in discrepancy with several studies describing Actinobacteria and Bacteroidetes as a bacterial group able to degrade complex substrates (Pascaultet al.2010b;Leeet al.2017;Lianet al.2019).However,the reduction in Actinobacteria may be related to the decrease of the major order Actinomycetales,which includesSporichthaandNocardiagenera (Taoet al.2019).
Minor phyla (Armatimonadetes,GN02,WPS-2,and OD1)increased in this study after the addition of crop residues,while Chlorobi decreased.Armatimonadetes phyla are present in most soils at relatively low abundances (below 1%) as shown in this study (Denget al.2019).Based on these limited phenotypic data,one can only reasonably note that all the currently available Armatimonadetes groups are oligotrophs (Leeet al.2014).Regarding GN02 and OD1 bacteria,they are not culturable;this could explain the limited available information about their trophic state.Brown and Jumpponen (2015) highlighted that many of these rare bacteria are digestion-resistant and have a symbiotic nature with their environment.WPS-2 bacteria were described as dominant and positively correlated with the C:N ratio(Hermanset al.2017),which disagrees with the results of this study (WPS-2 bacteria were negatively correlated with the C:N ratio,r=-0.62,P<0.05;Appendix C).Other OTUs that belonged toChlorobiwere abundant in the earlier stage of decomposition,but they were always found in very low proportions (Ruiet al.2009;Liuet al.2016).Overall,our findings highlight the changes in bacterial community composition based on the turnover between copiotrophic and oligotrophic bacterial phyla.
At the order level,correlational analyses provided insight into the crop residue characteristics responsible for the observed selection in bacterial community composition.Among the orders that were most affected and appeared after the addition of each crop residue,the Halanaerobiales,Thermoanaerobacterales,Halanaerobiales,Actinomycetales,etc.(Appendix C) exhibited significant correlations with some crop residue compositions.These orders are poorly described in the literature.Thus,there remains a large amount of unexplained variation in the observed bacterial order composition as linked to their biochemical characteristics (Liet al.2018).Various bacterial genera also showed significant correlations with crop residue composition (Appendix C).Rubrobacter has also been shown to degrade HEM (r=0.73,P<0.05;Appendix C) and xylan and could,therefore,play a significant role in the degradation of these components (De Mandalet al.2017).
The relative abundance of oligotrophic taxa,such as Nocardiodes and Niabella,increased during the residue degradation (present at the end of the incubation at T90),indicating the abilities of these microorganisms to degrade polymers with different complexities,such as HEM and CEL (Appendix C) (Reddyet al.2013;Kerdraonet al.2019).The positive relationship between these genera and HEM suggested their direct or indirect involvement in the degradation of the HEM fraction,which may be related to their abundance after the addition of wheat residues,which presents the highest HEM content.Soils with pea residues are rich in genera likeInquilinus,Dongia,Conexibacter,Collimonas,andDactylosporangium,which are involved in nutrient recycling and the degradation of crop residues.Among them,Conexibacteris highly correlated to SOL,TN,and the C:N ratio (Appendix C) and is described in literature as mainly associated with the degradation of litter with low C:N (Kocejaet al.2019).Except forConexibacterandCollimonas,which are highly correlated with the SOL fraction (P<0.01;Appendix C),the other specific genera are weakly correlated with the SOL fraction (Appendix C),but they are highly correlated among themselves and toCollimonasgenus (P<0.01;Appendix C).This may explain their presence after the addition of pea residues,which presents the highest SOL content.Some bacterial genera have been identified as CEL and LIG consumers,andCellvibrioandFlavobacteriumare included among them.These bacteria were positively correlated with LIG (r=0.71,P<0.03;Appendix C) and highly abundant under rapeseed residues,which presented the highest LIG content,confirming the previous findings (Zhang and Lueders 2017;Moreno-Espíndolaet al.2018).
4.6.Effect of the addition of crop residues on the taxonomic composition of fungal communities
In the present study,Ascomycota,Basidiomycota,and Mortierellomycota were the dominant phyla in all treatments,accounting for >18% of the fungi,which was much greater than the other phyla.This is consistent with results shown by Srouret al.(2020) and Zhanget al.(2020).Ascomycota are the main fungal decomposers in many soil ecosystems(Maet al.2013;Zhouet al.2016).Ascomycota play a major role in recycling plant residues,and they might form symbioses with mycorrhizae or endophytes (Ivancevic and Karadelev 2013;Choudharyet al.2018;Zhanget al.2020).Ascomycota are the most diverse group of saprotrophic fungi(Xionget al.2014) and the key decomposers in agricultural soils,which expand in response to organic inputs (Maet al.2013;Wanget al.2018).Basidiomycota,rather than Ascomycota,dominates terrestrial ecosystems,probably due to relatively less data available for agricultural soils(Tedersooet al.2014).Basidiomycota plays a particularly important role in degrading plant litter with high lignin content(Blackwoodet al.2007;Entwistleet al.2018;Zhanget al.2020).Our results showed that the relative abundance of Basidiomycota after the addition of rapeseed residues was significantly higher,indicating that rapeseed residues could provide a higher lignin content for the growth of Basidiomycota.
To further detail the effects of the addition of crop residues with contrasting biochemical characteristics on specific groups of soil fungi,important orders and genera were detected and appeared only after the addition of each crop residue.However,a variety of fungal genera showed significant correlations with crop residue compositions.For instance,ClitocybeandVerticilliumpresent only after added pea residues had been shown to degrade carbohydrates(Fernandoet al.2016;Geethanjali and Jayashankar 2016).These specific genera are negatively correlated (r=-0.78,P<0.05;Appendix D) to the C:N residue ratio.Hypoxylongenus,known for its ability to degrade LIG,was detected only after the addition of rapeseed (which presented the highest LIG content) (Lópezet al.2017).These specific genera are positively correlated with LIG content (r=0.82,P<0.05;Appendix D).Volutellagenera play an important role in HEM degradation (Masigolet al.2019).The results of this study showed a positive correlation (r=0.76,P<0.05)between the abundance of these genera and HEM fraction(mostly present in wheat residue).This may explain their abundance after the addition of wheat residues.Finally,this study confirms that the residue biochemical qualities are the main drivers of the shift in bacterial and fungal communities during crop residue decomposition in soil.
The decomposition of crop residues is an essential process in cropping systems as they are the main exogenous source of C.Our results showed that the residue qualities have distinct effects on soil microbial communities and C and N dynamics.The literature reported that the crop residues with the highest labile fraction affect the bacterial pathway at the beginning of decomposition,and crop residues with the highest recalcitrant fraction affect the fungal pathway at the end of decomposition,which suggests that a specific function is linked to specific taxa.Our findings highlighted the concomitant presence of some bacterial and fungal groups throughout the decomposition of crop residues.Indeed,many bacterial and fungal genera and orders were co-correlated among them and with the same biochemical fraction of the crop residues.This finding highlights the strong functional redundancy of soil microbial communities and underlines the complex relationships between structure and function of soil microbial communities.Finally,the correlations observed among fungal groups are more abundant and much stronger than those among bacterial groups,supporting the idea that potential interactions in fungal communities are stronger than those of bacterial communities in soils during crop residue decomposition.
5.Conclusion
This study complements previous knowledge about the effects of crop residue biochemical composition on microbial communities,C and N dynamics,and the soil microbiological state.It is well-known that crop residue inputs increase C mineralization and induce N immobilization in a range that is influenced by the crop residue’s initial biochemical composition.The main hypothesis,which stated that these changes might be related to changes in the composition and biomass of soil heterotrophic microbial communities and enzyme activities,was confirmed.The findings of this study showed that the soil C and N dynamics in the presence of the crop residues were driven by the selection of specific bacterial and fungal decomposers linked to the initial biochemical qualities of crop residues.
These findings are of major concerns from the perspective of agroecology development.Indeed,crop residue management is a recognized practice that may help to maintain OM in soils and contribute to N management during the intercrop period.Moreover,it seems that crop residues drive the microbial community composition in soils.This is of great interest when agricultural practices need a decrease in chemical inputs and an improvement in soil biodiversity and its related functions,such as nutrient cycling.
Acknowledgements
The authors would like to thank François D’Hubert of the Chambre d’Agriculture of Normandy and the farmers of the association of Economic and Environmental Interest Group(EEIG).This research was funded by the Normandy region and was supported by the Vivepois Project (D16-12746),France.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Errata regarding previously published articles
- Viricidal activity of several disinfectants against African swine fever virus
- Application of methyl jasmonate postharvest maintains the quality of Nanguo pears by regulating mitochondrial energy metabolism
- Melatonin treatment induces chilling tolerance by regulating the contents of polyamine,γ-aminobutyric acid,and proline in cucumber fruit
- Modification of total and phosphorus mineralizing bacterial communities associated with Zea mays L.through plant development and fertilization regimes
- Yield performance and optimal nitrogen and phosphorus application rates in wheat and faba bean intercropping
