Inter-regulatory role of microRNAs in interaction between viruses and stem cells
2021-09-03AfsoonAfshariRaminYaghobiGhazalRezaei
Afsoon Afshari,Ramin Yaghobi,Ghazal Rezaei
Afsoon Afshari,Shiraz Nephro-Urology Research Center,Shiraz University of Medical Sciences,Shiraz 7193711351,Iran
Ramin Yaghobi,Ghazal Rezaei,Shiraz Transplant Research Center,Shiraz University of Medical Sciences,Shiraz 7193711351,Iran
Abstract MicroRNAs(miRNAs)are well known for post-transcriptional regulatory ability over specific mRNA targets.miRNAs exhibit temporal or tissue-specific expression patterns and regulate the cell and tissue developmental pathways.They also have determinative roles in production and differentiation of multiple lineages of stem cells and might have therapeutic advantages.miRNAs are a part of some viruses’ regulatory machinery,not a byproduct.The trace of miRNAs was detected in the genomes of viruses and regulation of cell reprograming and viral pathogenesis.Combination of inter-regulatory systems has been detected for miRNAs during viral infections in stem cells.Contraction between viruses and stem cells may be helpful in therapeutic tactics,pathogenesis,controlling viral infections and defining stem cell developmental strategies that is programmed by miRNAs as a tool.Therefore,in this review we intended to study the interregulatory role of miRNAs in the interaction between viruses and stem cells and tried to explain the advantages of miRNA regulatory potentials,which make a new landscape for future studies.
Key Words:MicroRNAs;Viruses;Stem cells;Regulatory
INTRODUCTION
More than 27 years have passed since the discovery of the first microRNA(miRNA)named miRNA-lin-4 inCaenorhabditis elegans(C.elegans)[1].The function of this molecule was not explainable at that time.By discovering the second miRNA inC.elegans[2]and then in humans and fruit flies,these types of molecules became a real challenge in biology.
miRNAs are small non-coding RNAs(typically 22 nucleotides in length)that are derived from hairpin-shaped precursors with 70 to 100 nt.They are known as posttranscriptional regulatory tools over specific mRNA targetsviadirect base-pairing interactions[3,4].In most species,miRNAs are phylogenetically conserved in a wide variety of key biological processes including embryogenesis and maintenance of“stemness”[5].Also,miRNAs exhibit temporal or tissue-specific expression patterns and play an important role in development timing[6].Traces of miRNAs were detected in viruses,too.While studying the role of RNAi in B lymphocytes infected with Epstein-Barr virus(EBV),some small RNAs were found that not only could be cloned from the cells but were encoded by the viral genome itself.These small RNAs were proved to be miRNAs and named miR-BHRF1-1,miR-BHRF1-2,miR-BHRF1-3,miR-BART1 and miR-BART2[7].Finally,this fact was accepted that miRNAs are a part of some viruses’ regulatory systems,not a byproduct.Moreover,it is not easy to discriminate between cellular miRNAs actively induced or repressed by viral factors and those miRNAs altered by host responses[8].
The pluripotency of embryonic stem cells(ESCs)is an important feature that helps the researchers to study different roles of miRNAs[9,10].The therapeutic potential of human embryonic stem cells(hESC)provides exciting new opportunities for cellbased therapies.However,it is required to understand the molecular regulatory networks that control the properties of the cells such as self-renewal and differentiation potential[4,11].Genetic inactivation of the molecular machinery essential for proper maturation of miRNAs has been the cause of aberrant stem cell self-renewal and/or differentiation[12],indicating that the regulation of transcriptional network by miRNAs might control stem cell functions[13].A combined inter-regulatory relation has been detected for miRNAs during infection of the stem cells with viruses,which might be helpful in therapeutic tactics,viral pathogenesis and control.Therefore,in this review,we aimed to study the inter-regulatory role of miRNAs in the interaction between the viruses and stem cells.
BIOGENESIS OF MIRNAS AND FUNCTIONS
miRNAs have been detected to be ubiquitous molecular regulators for controlling the quality of gene expression in different species.Important and critical processes in cells are happening under regulatory conditions made by miRNAs such as cellular development,proliferation,differentiation,apoptosis and metabolism[14-16].Molecular biogenesis of miRNAs starts from nuclear transcription conducted by RNA polymerase II in the nucleus.The result of transcription of miRNA genes by RNA polymerase II is making a long molecule called primary miRNA[16,17].This long primary miRNA molecule has features of normal mRNA molecules such as 5’ cap and 3’ polyadenylation that makes one or more 70-80 nucleotide hairpin structures by folding.Later,these stem loop(hairpin)structures are recognized by an enzyme called Drosha,which acts as RNase III enzymes and works collectively with its cofactor DiGeorge syndrome critical region 8.The result of the function of Drosha and DiGeorge syndrome critical region 8 of primary miRNA is cleavage in approximately 22 bp down the stem which yields ~60 nucleotide precursor miRNA that contains two nucleotide overhangs in its 3’ end[18,19].
Precursor miRNA molecules are detected by a transporting vehicle called exportin-5 and transferred from the nucleus to the cytoplasm.In the cytoplasm another RNase III enzyme,Dicer,starts processing precursor miRNA into a ~22 nucleotide miRNA duplex[20,21].One of the strands of the miRNA duplex,which is called “guide”strand,enters into a functional complex called RNA-induced silencing complex(RISC).The second strand,which is known as “passenger” strand(also known as miRNA),degrades frequently.The degree of base pairing at the 5’ ends of miRNA duplexes determines the strand selection for entering into the RISC.This means the less stable strand in base pairing at the 5’ end preferentially incorporates into RISC[22].The miRNA within the RISC serves as target recognition of miRNA in the cytoplasm through base complementary commonly with sequences in the 3’untranslated region(UTR)of target mRNAs.The nucleotides in miRNAs that attach to the 3’UTR of target miRNAs are composed of 6-8 nucleotides in the 5’ of miRNAs,which is called “seed” sequence[23,24].
Furthermore,miRNAs might be fully complementary or imperfectly matched to their targets.The former results in the induction of nucleolytic cleavage of target miRNA in the region of base pairing and causes a rapid decay of the entire transcript[25],and the latter causes translational repression[23](Figure 1).One of the plausible mechanisms that is proposed for translational inhibition of miRNA within the RISC is through translocating targets of miRNA into P bodies,which are cytoplasmic structures without ribosomes.This translocation is attributed to a P body component(GW182)that attach to Argonaute proteins in the miRNA within the RISC complex.In P bodies,the targets of miRNA might be deadenylated,decapped,degraded or held in stasis[24].
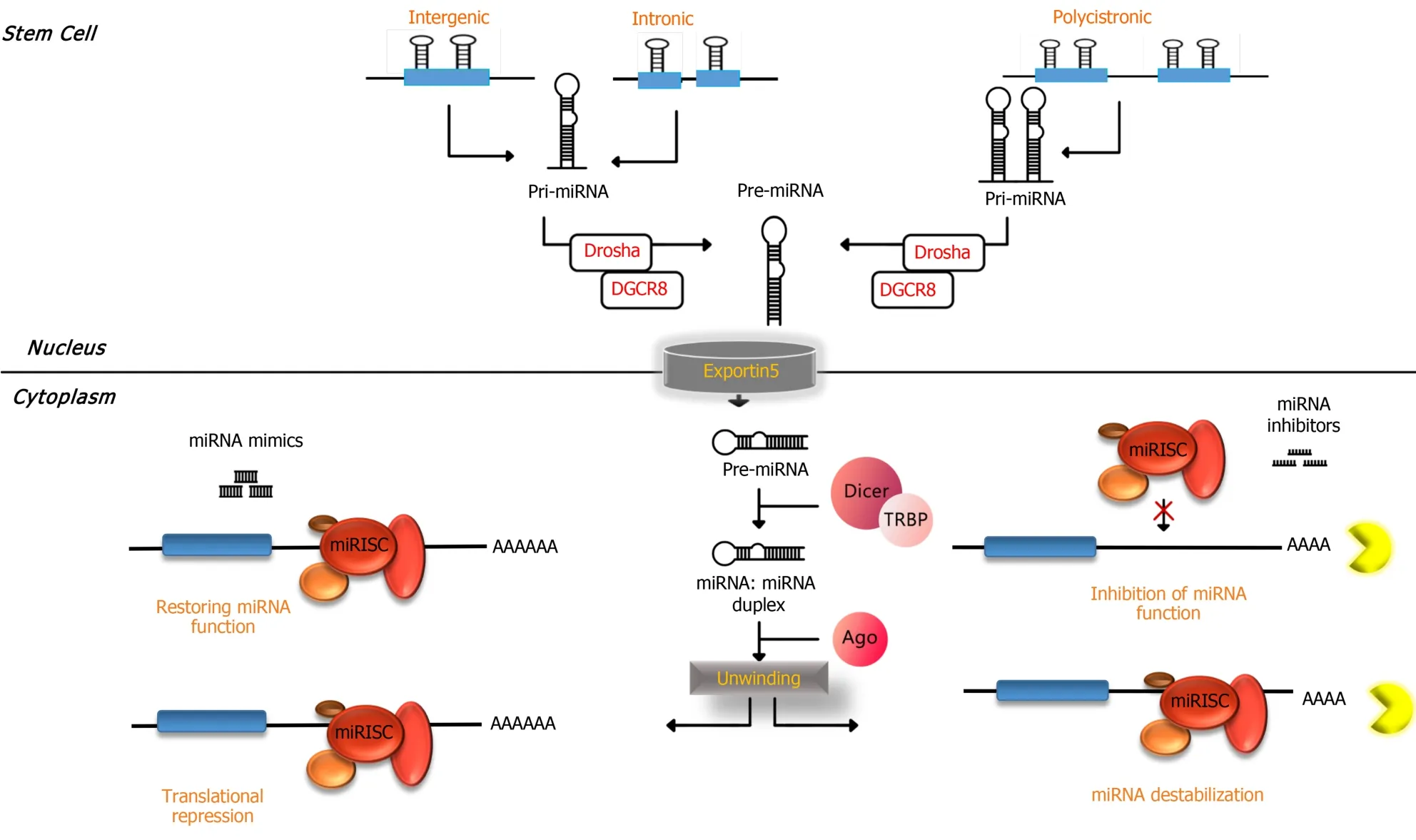
Figure 1 MicroRNA biogenesis begins with the generation of the primary microRNA transcript.The microprocessor complex,comprised of Drosha and DiGeorge syndrome critical region 8,cleaves the primary microRNA(miRNA)to produce the precursor miRNA.The precursor miRNA is exported to the cytoplasm in an exportin-5 dependent manner and processed to produce the mature miRNA duplex.Finally,mature miRNA duplex is loaded into the Argonaute family of proteins to form a miRNA-induced silencing complex.miRNA:MicroRNA;Pri-miRNA:Primary microRNA;Pre-miRNA:Precursor microRNA;miRISC:MicroRNA within the RNA-induced silencing complex;DGCR8:DiGeorge syndrome critical region 8;Ago:Argonaute;TRBP:Transactivation response element RNA-binding protein.
MIRNAS AND VIRUSES
Identification of viral miRNAs and their targets
Discovery of viral miRNAs was a new attractive area for researchers in order to investigate the mechanisms used by these tiny molecules for gene expression regulation in their host cells.These studies were the basis of substantial progress in understanding the life cycle of the virus and their interactions with their host cells[7].The fundamental method for identification and study of viral miRNAs is isolation of small RNAs after infection of cells,copy DNA cloning and sequencing[17,26,27].
Human cytomegalovirus(HCMV)is known to be the prototype of β-herpesviruses that has the largest genome size and can persist lifelong in hematopoietic cells such as granulocytes.Several miRNAs of HCMV have been detected and cloned,especially during the lytic phase of infection[28].Murine CMV(MCMV),which is a close relative of HCMV,makes a well-designed animal model of CMV and does not replicate in mice.Most miRNAs detected in MCMV are expressed during lytic replication of the virus.However,none of the MCMV miRNAs have a significant homology with miRNAs in HCMV[29,30].Through bioinformatic methods,several miRNAs have been predicted for herpes simplex virus(HSV)-1 and HSV-2.However,only one of them has been verified[28,31].
EBV is capable of immortalizing normal B cells,and this ability is related to many malignancies in human.The ENBA transcript is the source of three miRNAs known as BHRF1 miRNAs[26,32,33].Additionally,during latent infection the virus can produce more than 14 BART miRNAs[33,34],7 of which are similar to miRNAs detected in EBV-related virus of monkey and provides the first example of miRNA conservation within the families of herpesviruses[35].Kaposi’s sarcoma-associated herpesvirus(KSHV/HHV8)is a γ-herpesvirus family that is related to human malignancies,such as Kaposi’s sarcoma,primary effusion lymphoma and Castleman’s disease[36].This virus encodes a transforming protein called Kaposin[37];also,miRNAs are derived from primary effusion lymphoma derived cells.
In adenoviruses,the miRNAs are derived from a non-coding transcript,called virus-associated RNA.This product induces resistance during interferon-related defenses and facilitates viral replication.It is detected that virus-associated RNA is processed by six Dicer into miRNA that facilitates the adenovirus infection[38].
As a typical example of polyomaviruses,SV40 is characterized as an excellent model of oncogenesis in the simian cells such as monkeys that leads to verruca and fibrosarcoma.One miRNA has been detected for SV40,the target of which is found inin vivoexperiments[33,35,39,40].
Five miRNAs have been detected bioinformatically in human immunodeficiency virus(HIV-1),but only one of them has been cloned successfully[27,28,41-43],which might be related to its low abundance.
Diverse functions provided by viral miRNAs
Viruses are capable of regulating the expression of viral proteins during lytic or latent phases of infection[44].This model of regulation has been detected in EBV infection.One of transforming proteins named LMP-1,is controlled through miRNAs.In this process,a cluster of BRAT miRNAs attaches to the 3’UTR of LMP-1 mRNA and causes repression in its protein expression.This phenomenon finally renders to resistance in apoptosis of the infected cells[45].This regulatory process has also been detected in the beginning of HCMV expression.In this virus,miRUL112-1 regulates immediate early IE1(UL123,IE72)genes,which is a transcription factor that is essential for expression of many viral genes in infected cells[46,47].In SV40,a miRNA that is expressed late during infection reduces the expression of T antigen by targeting the 3’UTR of early transcripts.This T antigen repression helps the infected cells not to sensitize cytotoxic T cells[48].Additionally,some overexpressed small non-coding RNAs encoded by HIV-1 reduce the level of viral transcripts and facilitate the maintenance or formation of latency after viral infection[49,50].
Based on the fact that miRNAs do not activate the host’s immune response,they are ideal means for viruses in establishing stable latency in their hosts through regulating some host miRNAs.For instance,there are 12 miRNAs encoded by KSHV that are related to its latency process and finally cause transformation of the cells during production of Kaposi’s sarcoma[51].KSHV-miR-K12-11 is directly related to viral induced malignancy.This viral miRNA contains 100% homology in seed sequence with has-miR-155 that acts as an oncogene.miR-155 is upregulated in the lymphomas and is also a critical factor for B cell development[52,53].It seems that these two miRNAs regulate the same set of cellular targets such as a transcription repressor called BACH-1[39,54].
Interaction between virus miRNA and target genes
miRNAs are determinative elements in regulating the gene expression puzzle,and they control gene expression in diverse processes.Therefore,viral miRNAs are involved in regulation of not only their own gene,but also many host genes in order to subvert many cellular defense mechanisms[7,55].In this process,many miRNAs target predictors such as MiRanda[56],which is one of the earliest target predictors.The algorithm used in MiRanda could predict the targets of a microRNA through matching the 3’UTR binding sites among mRNAs of the virus and/or host[33,56-58](Figure 2).
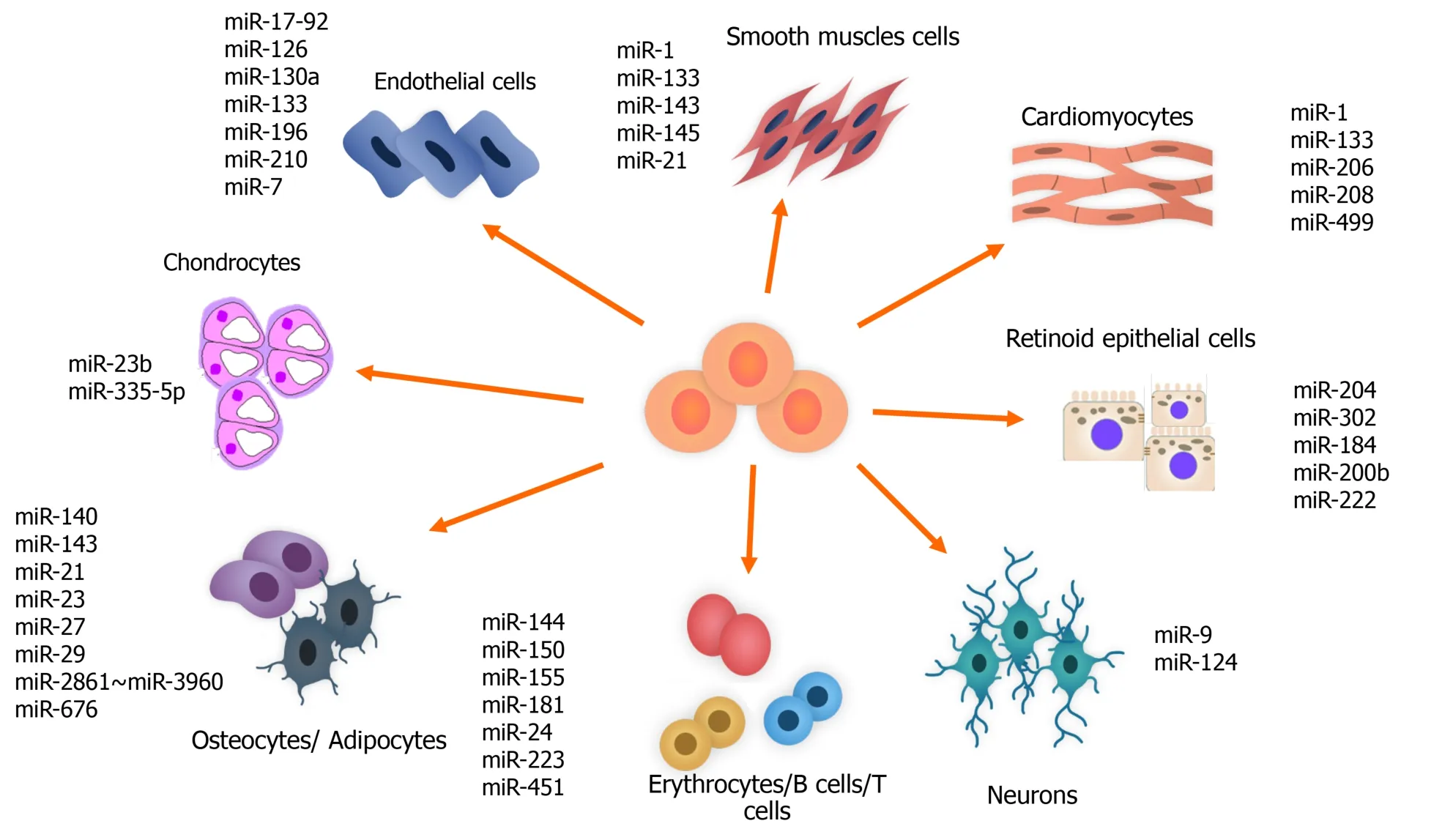
Figure 2 Interplay between viral infection and microRNA expression.miR:MicroRNA.
Action of virus miRNAs on mRNAs of host cells:HCMV miRNAs(miR-UL23 and miR-UL24)[28,58]are able to recognize some host mRNAs such as synaptonemal complex protein 1,cathepsin S precursor and IL-1 receptor related protein that are important in the biological function in the cells[56-58].
Another example is miR-BART1 in EBV that can regulate the function of many host cell mRNAs.This miRNA binds to the 3’-UTR of Bcl-2 mRNA and regulates the process of apoptosis and proliferation.Other targets of miR-BART1 are Zinc Finger protein 177 and stromal cell derived factor 1 mRNAs that regulate them through binding to the 3’-UTR[28,59].EBV miR-BART5 target PUMA that is a p53-regulated pro-apoptotic Bcl2-family member in infected neural precursor cells and inhibition of miR-BART5 leads to an increase in PUMA-mediated apoptosis[60].Additionally,EBV contains miRNAs that directly target antiviral molecules like miR-BHRF1-3 that downregulate CXCL11,which is a major player of host defenses against EBV[61].
miR-LAT is encoded by the latency-associated transcript gene of HSV-1[62].By its anti-apoptotic effectviadownregulation of TGF-β1 and SMAD-3 expression,both of them are linked in the TGF-β signaling pathway,cause the survival of the infected neurons and contribute to the persistence of HSV in a latent form[31].
Thrombospondin 1 is one of the molecules targeted by multiple miRNAs of KSHV,and the function of thrombospondin 1 in normal conditions inhibits angiogenesis and cell growth by activating TGF-β.Therefore,Kaposi sarcoma tumors exhibit a reduction in thrombospondin 1 activity[51].Additionally,there are more regulators of cell survival and growth that are the targets of viral miRNAs such as BCLAF1.BCLAF1 is detected to be a target of miR-K5 of KSHV in endothelial and B cells and can increase the reversibility of latent infection[63].KSHV miR-K11 and miR-K6 in the endothelial cells can target MAF(a transcription factor involved in final phases of many cell types),and it seems that KSHV miRNAs regulate the endothelial cells during infection that results in oncogenesis[64].miR-K11 of KSHV is an ortholog of cellular miR-155,and one of their shared targets is an oxidative stress repressor of transcription named BACH1 that coordinates with MAF proteins to repress heme oxygenase 1[39,54].
Some key cellular products such as major histocompatibility complex class I-related chain B(MICB)are targeted by different viral miRNAs(miR-UL112-1 of HCMV,miRK7 of KSHV and miR-BART2 of EBV),and the important fact is that their target sites in 3’UTR of MICB mRNA do not have overlapping sites for the three miRNAs.The importance of MICB is due to its ability to activate natural killer cells and CD8+ T cells in response to viral infections[65,66].
During viral infections,many alterations happen in the expression rate of cellular miRNAs in dealing with antiviral strategies of host defense and/or alterations in host cellular environment.As an illustration,both miR-155 and miR-146a expression is induced during EBV infection of B cells[67].In the latency phase of EBV infection,promoters of miR-146a and miR-29b are activated in the host cells,and the former causes reduction in interferon-responsive genes[67]and the latter results in downregulation of TCL1,which has a role in the host cell survival and proliferation[68].
Two viral proteins are encoded by oncogenic human papillomaviruses(E6 and E7)that inhibit the p53 and Rb pathways,respectively.Subsequently,the cellular miRNAs that are controlled through these two pathways are profoundly influenced,and miR-34a is downregulated by E6 that leads to an increase in cell growth[69].Also,E6 causes a reduction in miR-218,which is the reason of increase in LAMB3 in HPV-16 infected cells,and their final outcome is enhancement in cell migration and tumorigenicity[70].
HCMV infection makes changes in the function of the mTOR signaling pathway that has vital regulatory effects over cellular processes dealing with metabolism,growth and survival.It is reported that during HCMV infection,miR-100 and miR-101 are reduced,which is essential for regulating the mTOR signaling,and through this HCMV captures the control of cell critical processes[71].
It is documented that two miRNAs(miR-17-5p and miR-20a)of the cells harboring HIV-1 infection are suppressed.These miRNAs are responsible for targeting and controlling a cellular histone acetylase and proposed cofactor of the HIV-1 Tat transactivator called PCAF[8].In MCMV infection the function of specific miRNA specifically triggered miR-27a activity,which is rapidly decreased following infection[72].
Action of virus miRNAs on mRNA in the virus itself:Finding the complementary sequences,especially in the 3’UTR part of viral genes,sheds light into the fact that viral miRNAs regulate their own gene products as well.Antisense strand of viral DNA polymerase transcript BALF5 has a complementary site for miR-BART2 in its 3’UTR part,and it is suggested that expression of theBALF5gene should be partially controlled through miR-BART2[28].Additionally,miR-BART2 can bind to the 3′UTR of BMLF1(EB2),and miR-ART1 can adhere to the 3′UTR ofBBLF4mRNA andLMP2AmRNA,which contribute to regulating the expression of EBV genes[73].The other example is miR-UL112-1 of HCMV that is known to be the first viral miRNA that has targets in both virus and host.In the virus,it targets the viral IE72 transcript and in the host servs as an agent for reduction of the MICB protein,and the results are induction of viral latency[46,47].
Cellular transformations made by viral miRNAs:Viral miRNAs can attach and regulate both viral and cellular mRNAs.The ultimate goal of viral miRNAs for reprogramming the cellular processes include controlling lytic-latent switch,promoting cell proliferation,survival,and differentiation and finally controlling the host immune responses,for preparing a cellular microenvironment in favor of facilitating viral life cycle[74].
Functions of viral miRNAs in the infection and replication process:Considering the ability of viral miRNAs in employment of host cell components of gene expression machinery,it is expected to observe its consequences in the host gene expression.The study of Sullivanet al[48]showed that SV40 can regulate the expression rate of evading the immune system of the host.Sequence of miRNA is located on the late strand of the SV40 circular genome and overlaps the early mRNAs produced from the opposite strand of the viral genome.Early mRNA expression of viral large T-antigen is controlled by production of miRNA at the late stage of the replicative cycle.SV40 Tantigen is the foremost target for activation of cytotoxic T lymphocytes of the host,and the miRNA mediated decrease in T-antigen lowers the susceptibility of the infected cellsin vitro.This suggests that one of the functions of the SV40 miRNA may be related to its ability in evading the immune surveillance during the latency in the host[48,75-78].
In EBV infection,miR-BART2 binds to the mRNA ofBALF5,which results in latent infection.Hence,miR-BART2 may regulate the latent-lytic switch by preventing premature BALF5 expression[28].LMP1,which is targeted by miR-BART1-5p,miRBART16 and miR-BART17-5p,acts as a viral mimic for tumor necrosis factor receptor and induces cell proliferation during latency[45].
In HIV infection,miR-N367 binds to the U3 negative response element and reduces the activity of its promoter.The outcome of this process is inhibition in the HIV replication and viral persistent infection[43,49,50,79,80].HIV probably produces some miRNAs that can inhibit the expression of molecules and cytokines related to restriction of immunologic function of the host such as CD28 and CD4[41,43,80].
MiR-UL112-1 is located in the antisense sequence of theUL114(a viral DNA glycosylase)gene in HCMV[81].HCMV miR-UL148D and miR-US33 might be involved in their expression regulation and are located antisense of US29 and UL150,respectively[28,82].
HSV-1 and HSV-2 viruses use viral miRNAs in order to downregulate ICP0 and ICP34.5 that are products of immediate early genes[83-85].Additionally,miR-H3 and miR-H4 are located in antisense sequence of theICP34.5gene of HSV-1 and HSV-2 viruses[83].Also,miR-H2 in both of the mentioned viruses is transcribed antisense of ICP0 and diminishes the production of ICP0 protein[84-86].miR-H6 of HSV-1 targets ICP4,while it is not located antisense of theICP4gene[85].In KSHV infection,miR-K9* targetsORF50,which is a critical gene in activating the lytic phase of viral life cycle[87].
Functions of viral miRNAs in inducing tumorigenesis:It has been reported that some of viral miRNAs have a close relationship with tumors.This theory proposes that the viral miRNAs target the tumor suppressor genes of the host and facilitate tumor appearance.An example for this process is miR-BHRF1-1 in EBV that targetsP53(tumor suppressor gene)andBCL-2(apoptosis regulatory factor)mRNAs of the host and pave the way for the formation of tumors[26].
During EBV infection,two miRNA families,let-7 and miR-200,as tumor suppressors were downregulated.Viral products such as BARF0,EBNA1 and LMP2A are responsible for downregulation of the miR-200 family and also ZEB1,ZEB2 and Ecadherin[88].Moreover,in the EBV infected B cells and epithelial cells,miR-200b and miR-429 induces lytic replication[89].
Furthermore,microarray analysis results showed that some miRNAs such as miR-34b,-34c,-18a,-200a/b,-449a,-31 and let-7 were dysregulated in nasopharyngeal carcinoma,and the abnormality produced in the production rate of their target was the reason of proliferation of nasopharyngeal carcinoma[90].EBNA1 upregulation has been related with the latent infection of EBV in epithelial cell tumors[91,92].
Identification of cellular miRNAs and their viral targets:Beside the fact that a key function of viral miRNAs is to regulate the cell’s milieu,evidence also exists that highlights the role of cellular miRNAs in reshaping the course of viral infections.In this regard,several experiments detected cellular miRNAs that act in regulation of viral infections[93-96].
The role of small interfering RNAs[93]and another class of small noncoding RNAs(called piRNAs)has been detected to deal with suppressing the replication of mammalian retroviruses[94,95].In addition,experiments have validated the bioinformatically proposed miRNAs and revealed that some cellular miRNAs could indeed target various infecting viruses such as HIV-1[97-101].Commonly,cellular miRNAs act as restrictors of viral replication.miR-32 is reported to confine the replication of primate foamy virus type 1[100].During HIV-1 infection,viral mRNAs have been targeted by a set of human miRNAs including miR-28,miR-125b,miR-150,miR-223,and miR-382 in order to repress viral replication[99].
Considering the fact that cellular miRNAs can target mRNA products of the viral or cellular genome,it might be a question that RNA viruses can escape from this kind of regulation.However,interaction of cellular miRNA viral RNA has been reported.miR-32 in 293T cells can target primate foamy virus type 1,and in mice miR-24 and miR-93 target vesicular stomatitis virus RNAs[100,102].Additionally,in CD4+ T cell cultures,several cellular miRNAs were detected to target HIV-1 RNAs that facilitate maintenance of the viral latency[99].
MicroRNAs in stem cells
Stem cells are known as undifferentiated cells that are capable to differentiate into various cell lineages.Commonly,stem cells are classified as ESCs,induced pluripotent stem cells(iPSCs)and adult stem cells.They are named according to where they have originated from,for example,mesenchymal stem cells(MSCs),hematopoietic stem cells,cardiac stem cells,neural stem cells(NSCs),endothelial stem cells,etc.ESCs are pluripotent stem cells derived from the inner cell mass of a blastocyst or earlier morula stage embryos in the epiblast tissue[103],whereas iPSCs are directly generated through somatic cell reprogramming[104].The ability of stem cells in pluripotency and self-renewal makes them virtuous candidates for clinical therapies.In preclinical animal experiments,different cell types have been originated from stem cells,such as cardiovascular cells[105],neural cells[106]and osteoblasts[107],which are used for transplantation in order to repair the damaged organs[108,109].
miRNAs have been detected to be key regulators of the stem cells and in ESCs with ablated Dicer or DiGeorge syndrome critical region 8(Dicer-/- or Dgcr8-/-).Abnormal differentiation has been reported[110,111].Additionally,miRNAs fulfill this task by targeting the factors related to pluripotency at the 3’UTR.In human ESCs,miR-145 would repress Oct4,Sox2 and Klf4 mRNAs in order to subside the pluripotency potential of ESCs[112].
miR-296,miR-470 and miR-134 play roles in mouse ESC differentiation through targeting the coding region of transcription factors such as Nanog,Oct4 and Sox2 in mouse ESC differentiation[113].Classifications of miRNAs modulating the fate of stem cells that are recognized include c-Myc-induced miRNAs,miRNAs targeting P53 and early embryonic miRNA cluster and finally embryonic stem cell specific miRNAs,which also are known as ESC-specific cell cycle-regulating miRNAs[114-117].
Cell reprogramming
The process of reprogramming the differentiated somatic cells into a pluripotent state is referred to as cell reprogramming.For cell reprogramming,some technologies such as nuclear transplantation and iPSC reprogramming are needed.Nuclear transplantation is executed by transferring a nucleus from a donor individual into an oocyte,which is enucleated previously.But iPSC technology involves the reprogramming of somatic stem cells into a pluripotent state by repressing the expression of pluripotency related genes or proteins[104,118].Human iPSC is generated through transduction of combinations of Oct3/4,Sox2,Nanog and Lin28[119].
Although using viral mediated transduction of cell reprogramming is risky due to random integration of the virus into the host cell genome and causes tumorigenicity,the efficiency of this method is higher(0.02%-0.08%)than using other methods such as virus-free methods[120].Moreover,another method that was using synthetically modified mRNA is used to generate more efficient(1.4%)human iPSCs with lower tumorigenicity potential[121].
Another example of miRNA reprogramming potential is miR-302 of human ESCs that can activate the critical genes for cell cycle progression and reprogramming of the somatic cells[122].Studies show that somatic and cancer cells might be reprogrammed by miR-302 cluster into a less/trans differentiated state through alterations in the epigenetic programming,the same as iPSCs[123,124].
Regulating stem cells during reprogramming via miRNAs
miRNAs not only can reprogram cells,but also might have mechanisms for regulating this process and regulate the efficiency of iPSC reprogramming.There are reports of over-expression of embryonic stem cell specific miRNAs such as miR-290,or the miR-302 family enhances the efficiency of reprogramming[125].Other human miRNA clusters such as miR0372(that is an ortholog of miR-290 and miR-302 clusters in mouse),miR-17-92,miR-106b-25 and miR-106a-363 clusters(which is very similar to miR-302 cluster sequence)are documented as enhancers of reprogramming efficiency[126,127].The ability of miRNAs in reprogramming the somatic cells into iPSCs happens in a direct manner.miR-302 cluster is reported to reprogram human skin cancer cells into a pluripotent condition[128].Mouse and human somatic cells can be directly transfected into a pluripotent stateviadirect transfection by the miR-200c,miR-302 and miR-369 family[129].The important benefit of this method is reaching efficiency above 10% and the lowest tumorigenicity[108].
The other way for miRNA participation in the process of reprogramming is regulating the cell cycle factors.In this regard,two miRNA families(miR-25 and miR-130/301/721)can target p21,which is a cell cycle inhibitor,and this phenomenon leads to a promotion in the efficiency of reprogramming[127,130].Furthermore,in a somatic cell reprogramming process,a reduction in miR-34a can promote the efficiency of the process significantlyviatargeting p53[131].
Mechanism of miRNA-mediated stem cell reprogramming
miRNAs regulate stem cell reprogrammingviaa process that consists of three main steps:initiation,maturation and stabilization[132].Some miRNAs that are activated through OSK(Oct4,Sox2,Klf4)or OSKM(Oct4,Sox2,Klf4,c-myc),such as miR-200b,miR-200c,miR-106a-363 cluster,miR-302-367 cluster and miR-93/106b,are detected to be involved in the initiation phase that is mesenchymal to epithelial transition,during iPSC initiation[127,132,133].Upregulation of some miRNAs such as miR-19,miR-17,miR-290 and miR-8 family and downregulation of miR-30/Let-7 family are critical for activation and maintenance of pluripotency[132].
Role of miRNAs in stem cells pluripotency
As to regulation of pluripotency,miRNAs control this processviadirect targeting of 3’UTRs of pluripotency factors.miR-145 subsides OSK(Oct4,Sox2,Klf4)genes that deal with pluripotency by repressing them,and three miRNAs consisting of miR-134,miR-296 and miR-470 regulate pluripotency in ESCs through targeting the coding sequences of Oct4,Sox2 and Klf4[112,113].A study reported that the miR-290 family had epigenetic effects on DNA molecules such as methylation in order to regulate differentiation and pluripotency of ESCs[134].An interesting study also showed that Oct4,Sox2,Nanog and Tcf3 had binding sites in the promoter region of most miRNAs that are preferentially or exclusively expressed in ESCs.These transcription factors also regulate the expression of miRNAs[135].
Self-renewal in stem cells by miRNAs
The strategy of genetic reprogramming by miRNAs in the stem cells can be used for their potential of survival,proliferation and tissue repair post-transplantation[136].Different miRNAs have various capabilities,like being apoptotic,anti-apoptotic and neutral.This is also dependent on the kind of cell line in which they are expressed and the range of their targets in each kind of cells[137-140].Therefore,activated miR-290 and miR-143 in ESCs contribute to the proliferation and cell cycle progression[114,136].Also,miR-143 is abundant in embryonic development,especially during myocardial proliferation and cardiogenesis.Furthermore,increased expression of miR-143 is detected in some carcinomas as well[141].
The expression level of some miRNAs(miR-378,miR-689,miR-21,miR-574-5P,miR-696 and miR-370)was significantly increased during liver regeneration[142,143],but these miRNAs had no expression alteration during hepatic differentiation of human umbilical cord MSCs.This research confirms the ability of these miRNAs in selfrenewalvsdifferentiation[144].
Differentiation in stem cells by miRNAs
One of the important regulatory roles of miRNAs is their modulating role in stem cell differentiation.This ability is used for differentiating the cells originating from the stem cells into various adult cells for treatment of different diseases.The miR-302 family,which is located on ch.4,and the miR-200,miR-372 and miR-520 families,which are located on ch.19,are highly expressed in hESCs and are downregulated at the time of differentiation in adult cells[145,146](Figure 3).
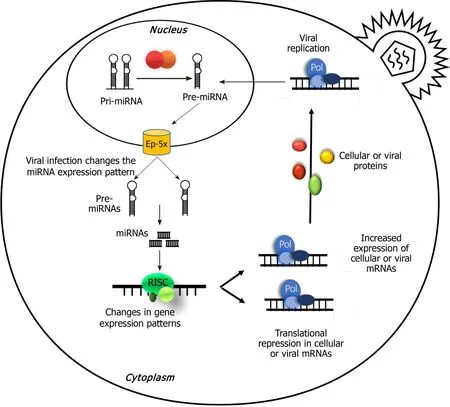
Figure 3 Importance and specificity of microRNAs in stem cell differentiation.miRNA:MicroRNA;Pri-miRNA:Primary microRNA;Pre-miRNA:Precursor microRNA;RISC:RNA-induced silencing complex;Pol:Polymerase;Exp-5:Exportin 5.
The miR-204 and miR-302 families are also known to be related to differentiation and maturation of retinal pigment epithelium cells from hESCs.Increase in the miR-204 family and decrease in the miR-302 family is detected too.During retinal pigment epithelium differentiation,miR-184,miR-200b and miR-222,which are known as retinal pigment epithelium-specific miRNA signatures,increase[147,148].
Significant increase of miR-145 during hESC differentiation results in a repression in pluripotency by direct targeting the genes related to self-renewal and paves the way for differentiation[149].The same process for differentiation is detected in murine ESCs through repression of Sox2 and Klf4 by miR-200c,miR-203 and miR-183[150],and miR-134,miR-296 and miR-470 target Nanog,Oct4 and Sox2[113].For facilitating ESC differentiation,silencing the self-renewal genes is necessary as well.Let-7 is detected to be a critical miRNA for controlling the level of stem cell factors[110].
It is hard for her to sit comfortably while wearing the body brace and so she sits, still and unnatural5, staring out the window. Her face is tense and tired and older somehow, much older than her seventeen years. She doesn’t even remember the world of a seventeen-year-old girl -- it’s as if that world never was. And she thinks she knows what Dorothy must have meant when she said, “Oh, Toto, I don’t think we’re in Kansas anymore.” It is more than an issue of geography, she is quite certain.
Furthermore,miRNAs accompany the cells during their differentiation process until they reach their final fate,which could be differentiated into various specialized cells such as cardiovascular,neural,osteogenic,chondrogenic and hematopoietic.The following parts briefly explain each process.
In the process of cardiovascular differentiation of cardiomyocyte progenitor cells and stem cells,miRNAs have regulatory roles.miR-499,viatargeting Sox 6,facilitates the differentiation of human-derived cardiomyocyte progenitor cells into cardiovascular cells[151].miRNAs conduct the cardiovascular differentiation of ESCs and iPSCs.For instance,miR-1 regulates the cardiac differentiation of ESCs and iPSCs in the infracted heart[139].This miRNA also,by targeting Klf4,promotes smooth muscle cell differentiation of retinoid acid-induced ESCs[152].
The modulatory roles of miRNAs have been detected in neurogenesis.miR-21 is related to neural differentiation of the subventricular zone in the adult mammalian brain[153].Nuclear receptor TLX is targeted by miR-9 that results in NSC differentiation and inhibition of the expression of pri-miR-9.A negative regulatory loop that finally results in a balance between proliferation and differentiation of the NSCs is created through the action of TLX and miR-9[154].Another example of making a loop in regulating adult NSC differentiation is methyl-CpG binding protein 1-miR-184-Numbl loop.In this case,acute deficiency of methyl-CpG binding protein 1 results in an increase in miR-184 that directly targets Numbl(Numb-like),which is the regulator of brain development[155].
The role of miRNAs in neurogenesis of ESCs and iPSCs is detected by targeting some neural differentiation,relatively.Suppression of the miR-371-3 family that is highly expressed in human iPSCs and ESCs is a classic example of this process[156].Other examples are downregulation of miR-132 by suppressing Nurr1 during differentiation of the tyrosine hydroxylase positive neurons[157],inhibition of activin and BMP-dependent pathways activate miR-125 that results in the suppression of Smad4 and finally differentiation of hESCs into the neural lineage[158],and creation of a regulation loop by Oct4 and miR-302 during differentiation of hESCs through NR2F2[159].
Any progress in expanding the ability of generating osteogenic and chondrogenic cells from other sources of cells is of great therapeutic value,and miRNAs are able to regulate these processes through targeting specific transcriptional factors and pathways,such as extracellular signal-regulated kinase-dependent pathway that has a critical role in osteoblast differentiation.Activation of RUNX2 through phosphorylation promotes expression of Osterix that results in the activity of alkaline phosphatase.Focal adhesion kinase is activated by extracellular proteins after activation of extracellular signal-regulated kinase 1/2.miR-138 has the ability to suppress differentiation of hMSCs into osteoblasts by targeting focal adhesion kinase[160].
miR-23b and miR-335-5p are related to induction of chondrogenic differentiation in human and mice MSCs,respectively.The former suppresses protein kinase A signaling in humans,and the latter targets Daam1 and ROCK1 in mice[161].
VIRUSES,MIRNAS AND STEM CELLS:LANDSCAPE OF VIEW
Considering the capabilities of stem cells,viruses and miRNAs provides insights into many potential sources for different aspects of molecular medicine,such as finding anti-viral therapies,pathogenesis and control of viral diseases and many more.The following parts briefly point to the few but valuable steps toward the mentioned goals by preparing the studies that have been done in this regard.
Therapeutic approaches of miRNAs in virally infected stem cells
In their study,Qianet al[166]reported the potential of umbilical cord blood derived mesenchymal stem cells-derived(uMSC)exosomes as effective anti-HCV agents through transporting a series of antiviral exosomal miRNAs to the target cells.Among the exosomes derived from different cell types,uMSC exosomes were the best candidate for repressing HCV infection while showing lower cytotoxicity compared with other antiviral agents.They claim that their study is the first in introducing new functional and therapeutical role for uMSC exosomes and providing new insights and prospects for the development of optimal antiviral agents in the future.Also,specific exosomal miRNAs derived from uMSCs result in the augmentation of the original effect of the host cell miRNAs[167].
Pathogenesis abilities of miRNAs in virally infected stem cells
Japanese encephalitis virus infection is a central nervous system neuroinflammation disease that is commonly more detected in children and old-age people.Human microglial cells were infected with Japanese encephalitis virus,and the miRNAmicroarray profiling reported the expression level of different miRNAs.miRNAs are involved in molecular pathogenesis of Japanese encephalitis virus and might be helpful in developing antiviral strategies against this infection[168].
Another study that detected the value of miRNAs in pathogenesis is a study that was done for elucidating the role of miRNAs in pathogenesis of Zika virus(ZIKV;a mosquito-borne virus resulting in newborn brain abnormalities such as microcephaly).This study focused on intracellular and extracellular vesicle-derived miRNAs and the host mRNA transcriptome of the neural stem cells during ZIKV infection.It was shown that some miRNAs,especially miR-4792,dysregulated at the intracellular level and had altered levels in extracellular vesicles during ZIKV infection[169].
Regulatory effects of miRNAs in virally infected stem cells
ZIKV envelope protein had the ability to alter the miRNome profile of human fetal neural stem cells,leading to alterations in proliferation and differentiation of fetal neural stem cells.Furthermore,they detected 14 upregulated and 11 downregulated miRNAs,among which miR-204-3p and miR-1273g-3p were directly responsible for regulating the expression level of NOTCH2 and PAX3,respectively.They also revealed through GO analysis that altered miRNAs in their study were in close relationship with the cell cycle and development processes[170].
Furthermore,for understanding the underlying mechanisms of mother-to-fetus transmission during ZIKV infection,mRNA and miRNA expression profiles were studied in human umbilical cord mesenchymal stem cells infected with two lineages of ZIKV,African(MR766)and Asian(PRVABC59).The results indicated that,during viral infection,miR-142-5p and its cellular targets(IL6ST and ITGAV)were decreased in a significant manner.The results of this study certify the importance of miRNAs in modulation of viral replication,especially during ZIKV infection[171].
During coxsackievirus B3 infection in Hela cells,miRNA alteration profiling specified 34 miRNAs whose predicted targets were mainly associated with cellular differentiation and transcriptional regulation.This study might be the first step in detecting the regulatory ability of miRNAs during viral infections like coxsackievirus B 3 infection[172].
Controlling capacities of miRNAs in virally infected stem cells
Coronavirus disease in 2019 is the cause of severe acute respiratory syndrome coronavirus 2 infection that,unfortunately,has no specific treatment and is still spreading among the world population.Hyunet al[173]studied the regulatory ability of the miRNAs derived from MSC extracellular vesicles as a potential novel therapeutic factor.Their study could introduce some therapeutic miRNAs by critical roles in the viral biology of the infected cells.Among them,miR-92a-3p,miR-103a-3p,miR-181a-5p,miR-26a-5p and miR-23a-3p are able to block RNA replication in severe acute respiratory syndrome coronavirus 2 and suppress virus-mediated proinflammatory responses by human bronchial epithelial cells and lung fibroblasts,all of which express angiotensin-converting enzyme 2 receptors.
CONCLUSION
Overall,this review provides a comprehensive view on the changes in the host miRNAs induced by viral infection and highlights the importance of miRNAs in the discovery and characterization of cellular factors involved in the modulation and regulation of viral replication and pathogenesis.Using the ability of stem cells in producing miRNAs against viruses might also be a giant step forward in the path of control and therapy of persistent viral infections.Programing of cross talk between viruses and stem cells by miRNAs may be helpful in therapeutic tactics,pathogenesis and controlling viral infections,and stem cell development strategies need to be evaluated in future studies.
杂志排行
World Journal of Stem Cells的其它文章
- Differences and similarities between mesenchymal stem cell and endothelial progenitor cell immunoregulatory properties against T cells
- Mesenchymal stem cells for enhancing biological healing after meniscal injuries
- Modulating poststroke inflammatory mechanisms:Novel aspects of mesenchymal stem cells,extracellular vesicles and microglia
- Antler stem cells and their potential in wound healing and bone regeneration
- Therapeutic prospects of mesenchymal stem/stromal cells in COVID-19 associated pulmonary diseases:From bench to bedside
- Mesenchymal stem cells as a potential therapeutic tool to cure cognitive impairment caused by neuroinflammation
