Genome-wide analysis of the invertase genes in strawberry(Fragaria×ananassa)
2021-08-12YUANHuazhaoPANGFuhuaCAIWeijianCHENXiaodongZHAOMizhenYUHongmei
YUAN Hua-zhao,PANG Fu-hua,CAI Wei-jian,CHEN Xiao-dong,ZHAO Mi-zhen,YU Hong-mei
Institute of Pomology,Jiangsu Academy of Agricultural Sciences/Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement,Nanjing 210014,P.R.China
Abstract Sugar is an important material basis in fruit development,and strawberry fruit flavour and sweetness largely depend on the sugar content and variety.Invertases (INVs) play an important role in the regulation of sugar accumulation because they irreversibly catalyse the hydrolysis of sucrose into the corresponding nucleoside diphosphate-glucose,glucose or fructose in fruit.In this work,we provided a comprehensive analysis of the INV gene family in octoploid strawberry(Fragaria×ananassa),including the gene structure,chromosomal locations,conserved domains,and gene evolution and expression profiles during strawberry fruit development.Our study revealed that polyploid events resulted in the abundant amplification (almost three-or four-fold) of the INV gene in the F.×ananassa genome,and these amplified INV genes showed dominant expression in strawberry fruit.More than half of the FaINVs transcripts with low expression had incomplete coding sequences by alternative splicing.Previous studies have shown that cell wall invertases (CWINV) are involved in the regulation of phloem unloading and sink strength establishment.The expression of FaCWINV1 was markedly upregulated during fruit development and strongly expressed in ripe fruit.Moreover,a significant correlation was observed between the total sugar content and the FaCWINV1 expression level.These findings suggest that FaCWINV1 may be involved in sugar accumulation in strawberry fruit.Taken together,the results of our study will be beneficial for further research into the functions of INVs in the regulation of fruit ripening.
Keywords:strawberry,sugar,invertases,fruit ripening
1.Introduction
The cultivated strawberry (Fragaria×ananassa) is the most economically important soft fruit species globally and always ranks first in cultivation area and yield harvest among small berries (Hummer and Hancock 2009).Strawberry has been described as the“queen of fruit”because it has high levels of nutritious elements,such as vitamin C,folate,and phenolic constituents (Proteggenteet al.2009).The quality of strawberry is mainly determined by the content and variety of soluble sugars,and their accumulation during the maturation process largely determines the sweetness at harvest.In addition,sugars also participate in various processes of plant growth and development,such as seed development,vascular tissue differentiation,and flower induction (Iraqi and Tremblay 2001;Ruan 2014).The three major soluble sugars in strawberry are sucrose,glucose,and fructose (Jiaet al.2016).The sugar content is generally notably high and increases significantly during growth and development in fleshy fruits (Jiaet al.2013).High sugar levels accumulate in the fruit depending on the expression of various genes associated with sugar biosynthesis,metabolism,and transportation (Sturm 1999).
Invertases (EC 3.2.1.26) play an important role in the regulation of sugar accumulation because they irreversibly catalyse the hydrolysis of sucrose into the corresponding nucleoside diphosphate-glucose,glucose or fructose in fruit (Vargas and Salerno 2010).Based on the optimum pH,subcellular localization and solubility,invertases in higher plants can be divided into three subfamilies:vacuolar invertase (VINV),cell wall invertase (CWINV) and neutral/alkaline invertase (NINV) (Roitsch and Gonzalez 2004).VINVs and CWINVs are enzymes localized to the cell wall and vacuole with acidic optimum pH,respectively (Shersonet al.2003).NINVs are soluble enzymes localized to the cytosol (Jiet al.2005).CWINVs are insoluble and proposed as core enzymes in the sucrose unloading pathway (Foyer 1987).By hydrolysis of sucrose to fructose and glucose in sink organs,CWINVs are involved in the concentration gradient of the sucrose between source and sink organs,which ensures continuous photosynthesis assimilation product transportation from source leaves into sink organs(Lemoineet al.2013).Therefore,the enzymatic activity of CWINVs regulates phloem unloading and sink strength establishment.In addition,the importance of CWINVs in plant growth and development was also demonstrated in previous studies.For example,antisense repression ofNin88andLin7using anther-specific RNA interference suppressed the supply of carbohydrates to pollen,resulting in male sterility (Goetzet al.2001;Goelet al.2006).In tomato,silencing LIN5 using an RNA interference(RNAi)-based approach produced an altered flower and fruit morphology (Zanoret al.2009).VINVsare mainly responsible for coordinating osmotic pressure,sucrose accumulation,and the sucrose-to-hexose ratio in vacuoles(Koch 2004).For example,silencingSlVIN1generated smaller fruits with higher sucrose content in tomato (Klannet al.1996).However,impaired expression of potatoVInvprevented sucrose accumulation in cold-stored tubers(Bhaskaret al.2010).Because of the lack of N-terminal signal peptide and protein glycosylation,NINV proteins are unstable and strongly affected by the external environment(Pelleschiet al.1997).To date,the gene function of NINVs has not been thoroughly elucidated.NINVs were considered to hydrolyse sucrose in tissues with low activities of CWINVs and VINVs,thereby providing substrates for the tricarboxylic acid cycle and energy for cell metabolism (Rossouwet al.2010).
Cultivated strawberry (F.×ananassa) is an allo-octoploid species exhibiting heterozygosity and complexity of the polyploid genome and hybridization between two octoploid strawberries,F.virginianaandF.chiloensis(Edgeret al.2019).To date,research on the function of invertase genes has been especially limited in strawberry.In this study,FaINVswere identified and characterized from theF.×ananassagenome.The gene structure,chromosomal locations,conserved domains,and gene evolution ofFaINVswere analysed.Based on theF.×ananassatranscriptome database,the dominant expression patterns ofFaINVsin the polyploid strawberry genome were examined.Finally,the temporal and spatial expression patterns ofFaINVsin strawberry fruit were examined by qRT-PCR.Our results will be beneficial for further investigations into the functions of INVs in the regulation of fruit ripening.
2.Materials and methods
2.1.Plant materials
Strawberry variety ‘Benihoppe’ is widely used all over the world,and permits are not required for the collection of plant samples.Strawberry plants (F.×ananassa,‘Benihoppe’)were obtained from the China national strawberry germplasm resource nursery,and grown in the plastic greenhouse at Jiangsu Academy of Agricultural Sciences,China.We divided the whole process of strawberry fruit development into seven stages according to the fruit weight and colour;small green (G1),big green (G2),degreening(G3),white (G4),initial red (G5),partial red (G6),and full red(G7),which were minor adjustments of the periods reported in previous studies (Jiaet al.2013;Yuanet al.2019).The receptacle fruits were immediately frozen in liquid nitrogen and stored at–80°C after being cut into small pieces.
2.2.ldentification and correction of invertase genes
To predict the invertase (INV) genes inF.×ananassa,the sequence of INVs (EC 3.2.1.26,β-fructosidase,β-fructofuranosidase) from other species in the NCBI Database (http://www.ncbi.nlm.nih.gov) was used as a query to search in theF.×ananassaCamarosa Genome Database(Fragaria×ananassaCamarosa Genome Assembly v1.0.a1)by BLASTp program with a cutoffE-value of 10–5(Edgeret al.2019).Then,the candidate genes were BLASTn(Altschul 1990) searched against the NR Database to confirm whether they wereINVgenes (higher bit score).Similarity searches were also performed using the BLASTn program in our previous high-quality reference transcriptome(SMLR) for octoploid strawberry by single-molecule long-read sequencing to correct the mispredicted gene structure in the gene annotation ofF.×ananassaCamarosa Genome.SMLR data can be acquired from the BioProject at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under the accession number PRJNA510223.
2.3.Sequence analysis of invertase genes
The deduced protein lengths,molecular weights,and isoelectric points were predicted by the ProtParam tool(Gasteigeret al.2005).Multiple sequence alignments were performed using ClustalW (Li 2003).A phylogenetic analysis was carried out by the neighbour-joining method using MEGA-X Software (Kumaret al.2018).A neighbourjoining analysis with pairwise deletion and bootstrap analysis with 1 000 replicates was performed using the p-distance model.The exon–intron structure information was acquired by comparing the gene coding frame with the corresponding genomic DNA sequence,and the exon–intron structure information was graphed using the GSDS2.0 tool (Huet al.2015).
2.4.Chromosomal mapping of invertase genes
The chromosomal location information of theINVgenes was retrieved from the position of the genes stored in the GFF file of theF.×ananassaCamarosa Genome.Based on this information,we manually marked the position of each gene on the chromosome to obtain the chromosomal distribution of each gene.
2.5.Illumina data analysis
The RNA-Seq datasets (SRP111905) were downloaded from the Sequence Read Archive (SRA) at the NCBI (http://www.ncbi.nlm.nih.gov/sra).The RNA-Seq dataset samples were collected from four successive developmental stages(green fruit,GF;white fruit,WF;turning stage,TS and red fruit,RF) of strawberry (F.×ananassa) fruit with two biological replicates.The clean reads were mapped to our previous high-quality reference transcriptome (SMLR) for octoploid strawberry by Bowtie2 (ver.2.2.4) (Langmead and Salzberg 2012),and the unmapped reads were extracted.Transcript expression levels were measured in the RNA-Seq analysis as reads per kilobase per million mapped reads (RPKM).
2.6.qRT-PCR analysis
Total RNA was extracted from fruits using the polysaccharide and polyphenolics-rich RNAprep Pure Kit (Tiangen,Beijing,China);and genomic DNA contamination was eliminated using DNase I (TaKaRa Bio,Shiga,Japan).cDNA was synthesized using an M-MLV Reverse Transcriptase Kit (Promega,Madison,WI,USA) according to the manufacturer’s protocol.qRT-PCR was carried out in a CFX Connect Real-Time PCR System (Bio-Rad,CA,USA) using the UltraSYBR Mixture (Low ROX) (CWBIO,Beijing,China)according to the manufacturer’s protocol.The strawberry ubiquitin gene was selected as an internal standard for the normalization of gene expression.Each fruit sample was quantified in triplicate independent biological replicates and three technical replicates.The relative RNA level of each gene was calculated using the comparative Ct (2−ΔCT) method and normalized using log10.The data were visualized with HemI Software (Heatmap Illustrator,ver.1.0) (Denget al.2014).
2.7.Soluble sugar measurement
All fruit samples were ground into a fine powder with liquid nitrogen.One gram was extracted in 10 mL of ethanol-0.2%metaphosphoric acid (4:1 by vol) for 24 h at 4°C and then centrifuged at 10 000×g for 10 min.The supernatant was collected in fresh tubes,and the residues were resuspended and incubated in 10 mL of ethanol-0.2% metaphosphoric acid (4:1 by vol) for 24 h at 4°C and then centrifuged at 10 000×g for 10 min.The supernatant was combined for further HPLC analysis.HPLC was performed as previously described (Jiaet al.2011).
2.8.Data availability statement
The samples of RNA-Seq datasets were collected from the Sequence Read Archive (SRA) at the NCBI (http://www.ncbi.nlm.nih.gov/sra),and the accession number was SRP111905.
3.Results
3.1.Genome-wide identification and characterization of the invertase gene family in strawberry
To predict theINV(EC 3.2.1.26,β-fructosidase,β-fructofuranosidase) genes inF.×ananassa,the sequence ofINVsfrom other species was used as a query to search theF.×ananassaCamarosa Genome Database.From the BLASTn output,44 protein-coding loci in theF.×ananassagenome were likely putativeINVs,including nineVINVs,sixCWINVsand 29NINVs.The cultivated strawberry(F.×ananassa) is allo-octoploid (2n=8x=56).The majority ofINVsshowed four copies,which were located on the four subgenomes.We named theINVsbased on the gene function and subgenome donor,such asFaVINV1-1.Due to the complexity of the allo-octoploid genome,errors occurred in the gene annotation of theF.×ananassaCamarosa Genome.Furthermore,we employed our previous highquality reference transcriptome (established by singlemolecule long-read sequencing (SMLR)) for octoploid strawberry to confirm and correct the candidates (Yuanet al.2019).Two candidates,FaCWINV2-1andFaCWINV2-2,were considered to be incomplete because of the incomplete genome assembly.FaVINV2-2.3 and FaNINV8-4were suspected to be pseudogenes with imperfect or shorter open reading frames,and all the remaining candidates were adjusted and confirmed to be complete.The characteristics of the FaINVs are shown in Table 1,including the deduced protein length,molecular weight,and isoelectric point.The maximal deduced proteins of theseF.×ananassaINVs contained 549–691 amino acid residues except for the pseudogenes,which is similar to INVs in other plant species,such as cassava (Yaoet al.2014),Populus(Chenet al.2015) and pepper (Shenet al.2018).The FaVINVs have lower isoelectric points of 4.66 and 6.41,which is a typical feature of soluble vacuole-localized enzymes,and except for FaNINV4,all the remaining FaINVs contained slightly higher isoelectric points,which are generally localized to the cytosol,mitochondria,plastids,and nucleus,with FaCWINVs possessing the highest isoelectric point (7.60–9.15).

Table 1 Characteristics of strawberry INV genes
3.2.Conserved domains and gene structure analyses
To further investigate the conserved patterns of strawberry INV proteins,a multiple sequence alignment was performed using the protein sequences ofFaINVsby the ClustalW program.Previous studies suggested that acid INVs (AIs)contain13 typical conserved functional regions,including two essential motifs,β-fructosidase motif and catalytic site,while NINVs possess 12 conserved regions (Chenet al.2015).The alignment analysis showed that all the strawberry FaNINVs contained 12 intact conserved regions except FaNINV8-4 (Fig.1),and all the FaCWINVs and FaVINVs contained 13 well-conserved regions except FaVINV2-2.3,FaCWINV2-1,and FaCWINV2-2,which were short in the N-terminal domain (Fig.2).We also searched the conserved domains of FaINV proteins using the PFAM Database.All of the FaCWINVs and FaVINVs contained an unknown function domain (DUF3357),a glycosyl hydrolases family 32 N-terminal domain (Glyco_hydro_32N) and a C-terminal domain (Glyco_hydro_32C);on the contrary,all of the FaNINVs contained an alkaline and neutral invertase domain(Glyco_hydro_100).
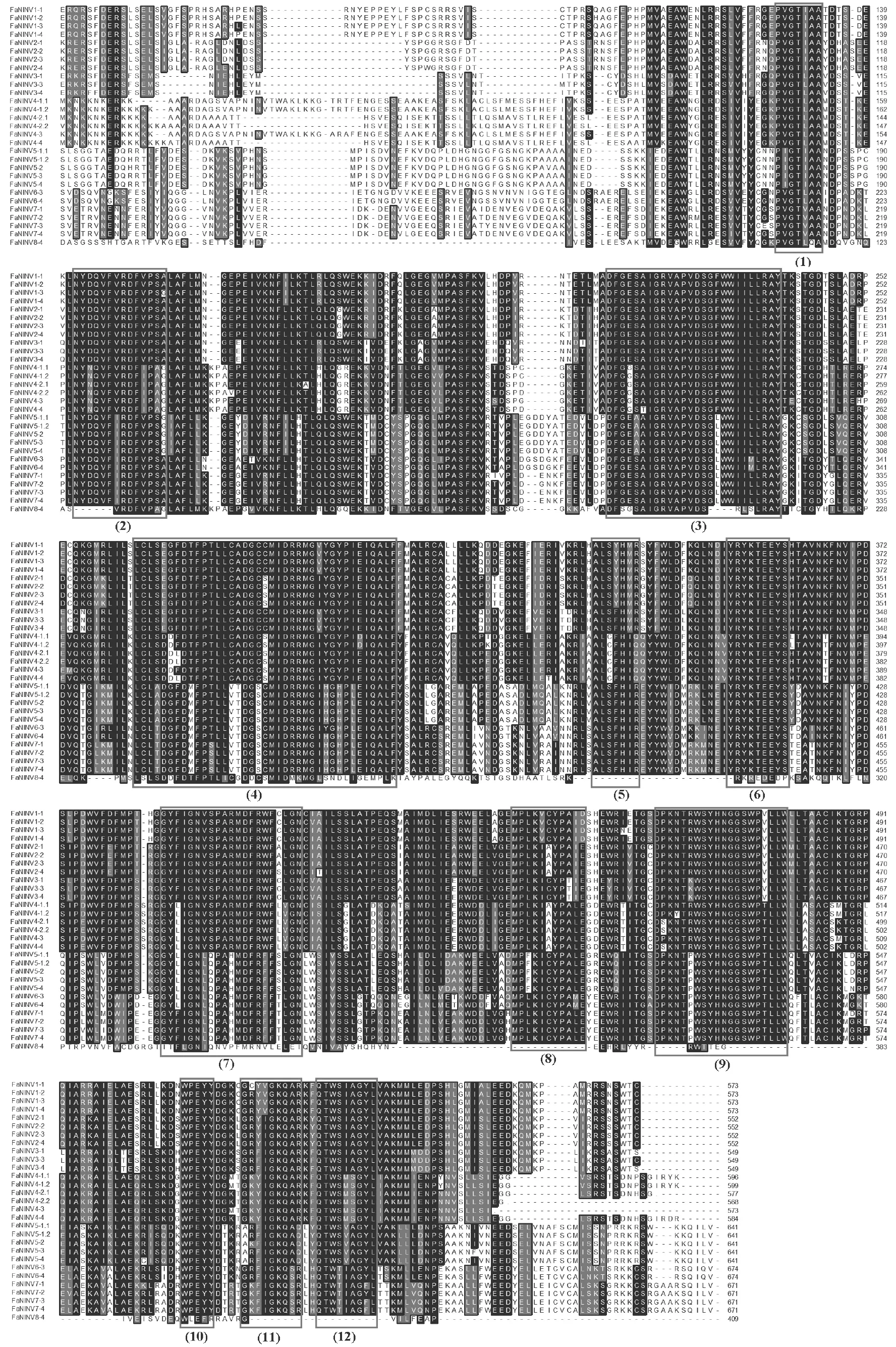
Fig.1 Multiple sequence alignments of FaNINVs.The black boxes indicate the 12 well-conserved regions.

Fig.2 Multiple sequence alignments of FvCWINVs and FvVINVs.The black boxes indicate the 13 well-conserved regions.
Next,the phylogenetic relationships and gene structures of strawberry INVs were explored.Strawberry INVs can be divided into two groups:AI branch and NINV.In addition,the AI branch can be divided into a CWINV clade and a VINV clade (Fig.3).The copies located on the fourF.×ananassasubgenomes possessed similar genetic structures and notably close evolutionary relationships.All of theFaINVscontained four to seven introns,except for the suspected pseudogenes which had fewer introns.Interestingly,except for the two incompleteFaCWIVs(FaCWINV2-1andFaCWINV2-2),the remainingFaCWIVswere observed to have one 9-bp exon,which is similar to the INVs in pepper(Shenet al.2018) and sugarcane (Wanget al.2017).
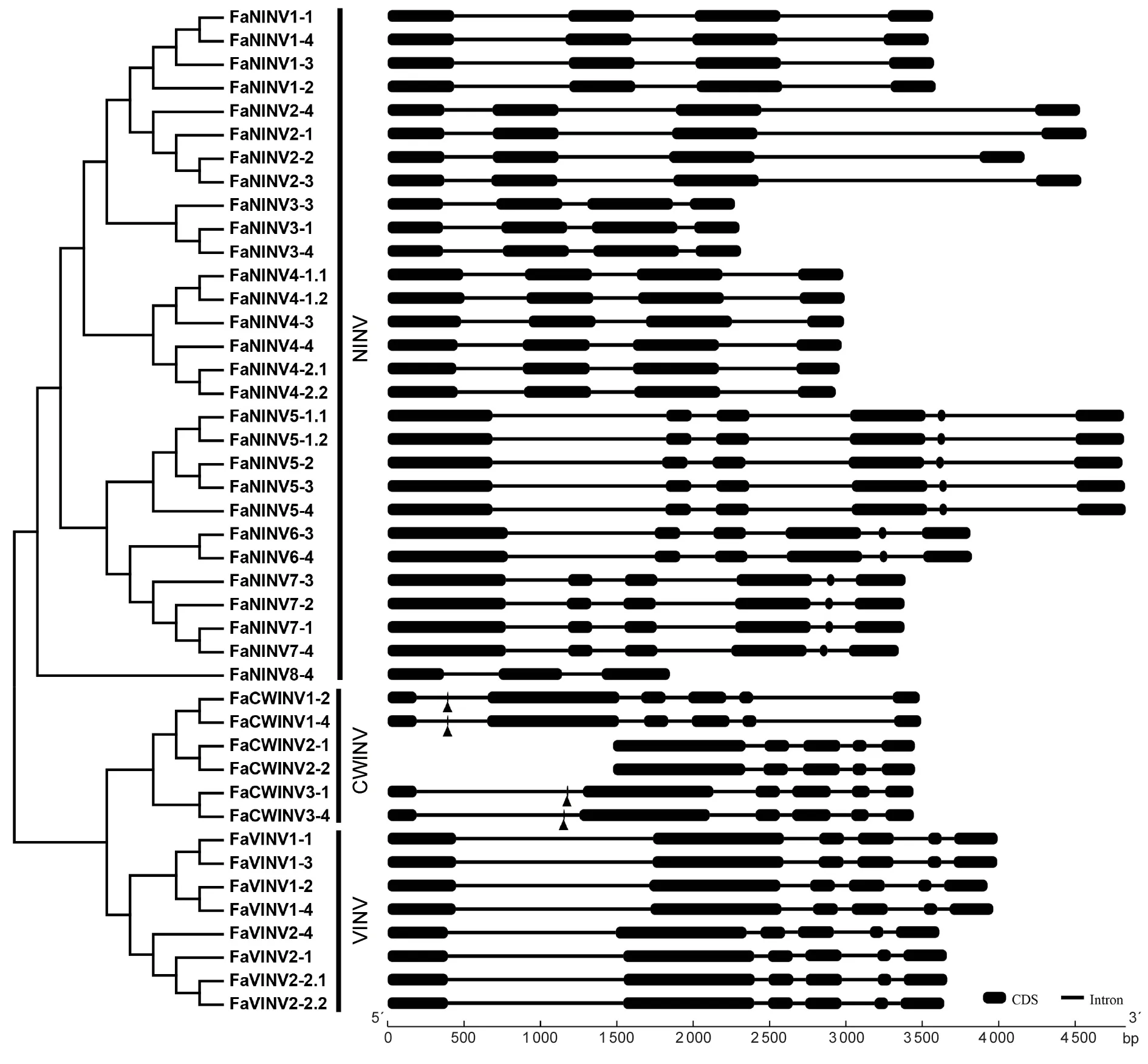
Fig.3 Phylogenetic analysis and exon–intron structures of Fragaria×ananassa INV genes.Fragaria×ananassa INV protein sequences were used to generate the phylogenetic neighbour-joining tree (1 000 bootstrap) using MEGA-X.The exon–intron structures of the F.×ananassa INV genes were determined using the GSDS2.0 tool.CDS,coding sequences.The black boxes and black lines represent exons and introns,respectively.The triangles indicate the 9-bp exon.
3.3.Chromosomal locations and gene duplication
Cultivated strawberry (F.×ananassa) has four parental subgenomes donated from the four diploid progenitor species.Fragaria vescawas deemed to be one of the four extant diploid progenitor species.Therefore,we compared the chromosomal locations ofF.×ananassaandF.vesca INVsand found that allFaINVsandFvINVswere distributed on six of the seven chromosomes,except chromosome 1(Fig.4).Compared withFvINVs,all orthologous genes were distributed at similar positions on the same chromosomes in theF.×ananassafour subgenomes.Our results showed that no orthologous genes forFvCWINV1–5were distributed in some of theF.×ananassafour subgenomes,especially forFvCWINV4andFvCWINV5;however,more than one orthologous gene for someFvNINVs(such asFvNINV4andFvNINV5) was found in one of the fourF.×ananassasubgenomes.TheFaNINVswere uniformly present on all six chromosomes;allFaCWINVswere present on chromosome 6;and theFaVINVswere distributed on chromosome 4 and chromosome 7,including a distinct tandem duplicate gene cluster with three tandem genes(FaVINV2-2.1,FaVINV2-2.2andFaVINV2-2.3).
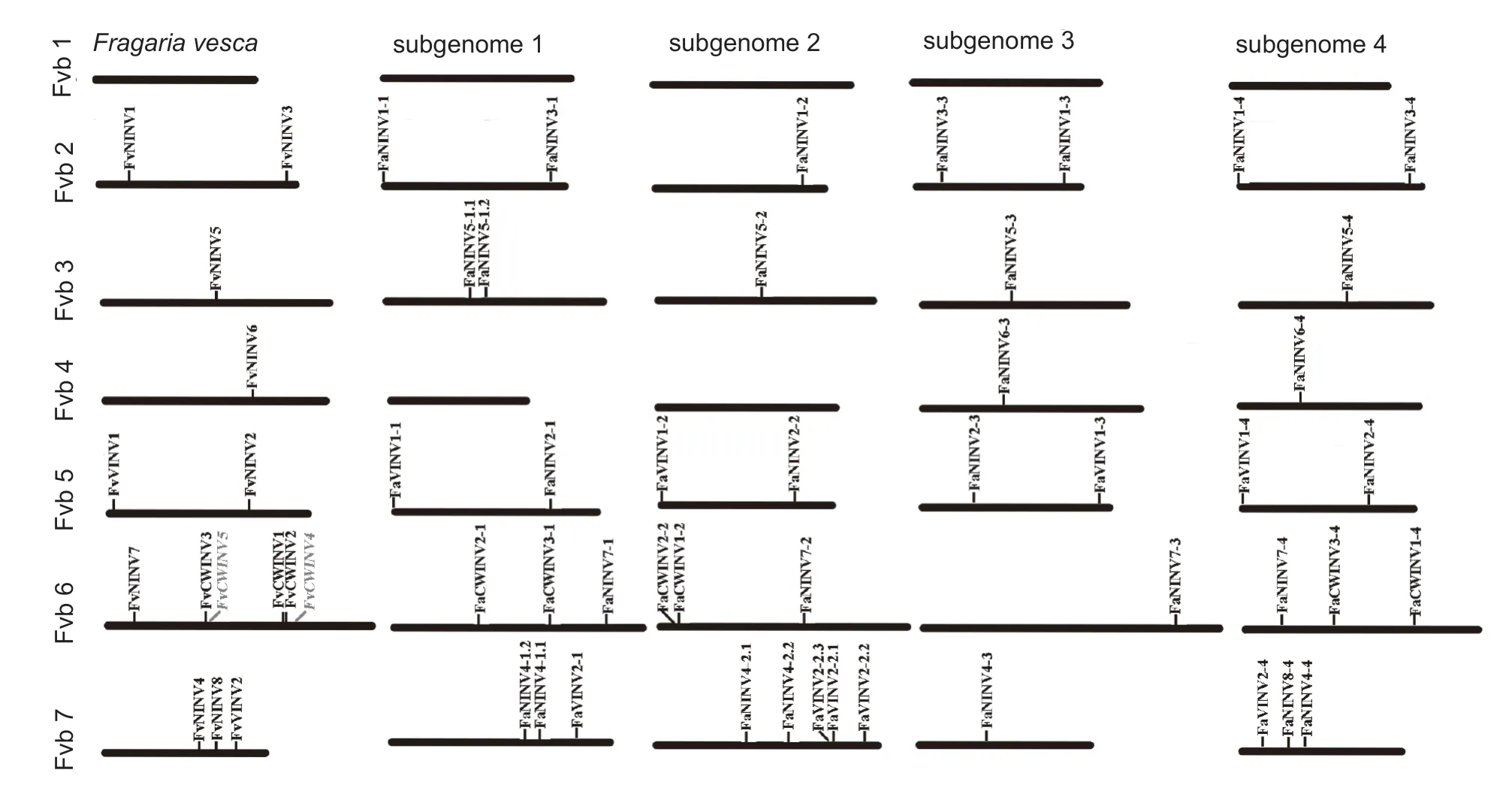
Fig.4 Chromosomal locations of invertase genes on the Fragaria vesca and F.×ananassa genomes.
We compared the sequence differences betweenF.×ananassa INVtranscripts and the correspondingF.vesca INVtranscripts.The molecular weights and isoelectric points ofF.×ananassaINVswere similar to those of the orthologous proteins inF.vesca.Additionally,the deduced protein lengths of someF.×ananassaINVs were the same as those of the orthologous proteins inF.vesca.The comparative analyses of whole protein sequence similarity showed that the overall similarities ofF.×ananassaandF.vescaINV orthologous protein pairs were notably high,e.g.,FaVINV1/FvVINV1 (98.27%).Next,we examined the phylogenetic relationships of strawberry (F.×ananassaandF.vesca) INV family members.As shown in Fig.5,the evolutionary relationships ofF.×ananassaandF.vescaINVs were notably close,and these members could also be divided into NINV,CWINV and VINV branches.Most of the FaINV/FvINV proteins showed a 1:4 orthologous relationship,such asFvVINV1fromF.vescaand the gene pair fromF.×ananassa,FaVINV1-1,FaVINV1-2,FaVINV1-3andFaVINV1-4.The phylogenetic relationships betweenF.×ananassaandF.vescaINV proteins also indicated thatF.×ananassawas an octoploid with four subgenomes.
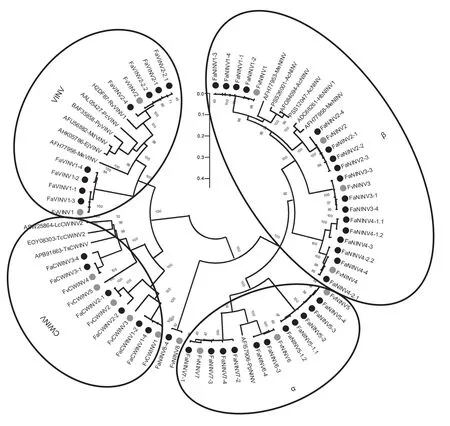
Fig.5 Phylogenetic tree of the INV proteins from Fragaria×ananassa,F. vesca and other plant species.This phylogenetic tree was generated by the neighbour-joining method (1 000 bootstrap) with MEGA-X.
3.4.Dominant expression in octoploid strawberry(F.×ananassa)
A BLASTn search of SMLR transcripts (119 897 records),which are generated by single-molecule long-read sequencing,confirmed the transcriptional activity of mostF.×ananassa INVs.Interestingly,half of theF.×ananassa INVtranscripts had a notably shorter coding sequence by skipped exon,exon variation and intron retention,which resulted in an early termination codon compared to model genes in public databases (Fig.6-A).For example,F01.PB4604 was predicted to translate a short protein(254 aa) by skipped exon,while F01.PB18447 and F01.PB17999 had an intron retention that resulted in an early termination codon (Fig.6-B).To elucidate the functions of these abundant transcripts,we compared the expression levels ofINVtranscripts containing whole coding sequences with those containing shorter coding sequences in strawberry fruit.Overall,INVtranscripts with a premature stop had lower or even no expression levels (RPKM),while the expression levels (RPKM) of transcript variants with whole coding sequences were considerably higher in strawberry fruit (Fig.6-C).Previous studies have shown that the expression profiles of many genes in allopolyploidy do not appear as simple additive combinations of the parental genes (Liet al.1981).Given the high degree of sequence identity of copy genes in the fourF.×ananassasubgenomes,it was challenging to distinguish transcripts for eachF.×ananassaINVgene.However,our data also showed thatINVgenes exhibit strong expression dominance in octoploid strawberry.Only a fewINVtranscripts from orthologous pairs were highly expressed,and the others exhibited low or no expression.For example,only F01.PB71124 and F01.PB101880 fromFaNINV2pairs (FaNINV2-1,FaNINV2-2,FaNINV2-3andFaNINV2-4) was highly expressed,and the remaining transcripts fromFaNINV2pairs were expressed at low or even non-existent levels (Fig.6-D).
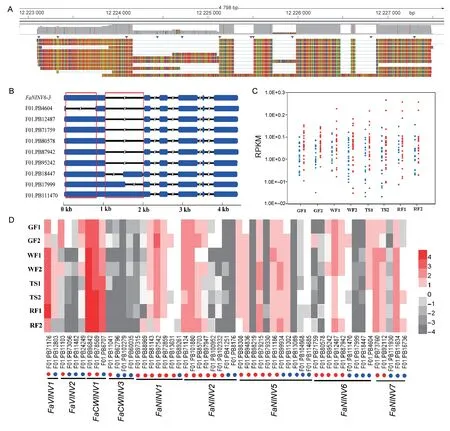
Fig.6 Dominant expression patterns in Fragaria×ananassa.A,alignment of FaNINV6-3 transcripts from the octoploid strawberry reference transcriptome (SMLR) to the F.×ananassa FaNINV6-3 genomic DNA sequence by the genome browser IGV.B,schematic showing that half of the F.×ananassa invertase transcripts (FaNINV6-3) had a very short coding sequence by skipped exon,exon variation and intron retention.In the gene structure diagrams,blue bars indicate exons,thin lines indicate introns,and red boxes indicate alternatively spliced regions.C,scatterplot showing the RPKM values of expression for invertase transcripts with either a whole coding sequence (red dots) or a shorter coding sequence (blue dots).D,heat map showing lnRPKM values of expression for invertase transcripts during strawberry fruit development.The blue dots represent transcript variants with a premature stop,and the red dots represent transcript variants with a whole coding sequence.
3.5.FaCWINV1 plays an important role in sugar accumulation during strawberry fruit development
Given the high sequence and functional similarity of copy genes in the fourF.×ananassasubgenomes,this approach was unable to design optimal qRT-PCR primers that were specific for each gene.Therefore,we designed only a pair of qRT-PCR primers for each of 12 sets ofINVgene pairs.The expression levels ofINVgenes were assessed using qRT-PCR along seven successive fruit developmental stages of cultivated strawberry (G1,G2,G3,G4,G5,G6,and G7).The qRT-PCR analysis revealed thatF.×ananassa INVgenes were differentially expressed in strawberry fruit(Fig.7-A).FaNINV3,FaNINV4,FaCWINV2andFaCWINV3were expressed at low or absent levels during the entire fruit development process.In contrast,FaNINV2andFaNINV5showed relatively higher expression during the entire process of fruit development.FaNINV1was more strongly expressed during periods of green fruit,and its expression decreased slightly during the fruit maturation period.Overall,these results indicate that the expression ofFaCWINV1rapidly increased during fruit development and was strong during the fruit maturation period (G5,G6 and G7).
CWINVs are proposed as core enzymes in the sucrose unloading pathway (Foyer 1987).To examine whetherFaCWINV1plays an important role in the sucrose unloading pathway,we measured the sugar content in strawberry fruit.Sucrose,glucose,and fructose were the three major soluble sugars in strawberry fruit (Fig.7-B).All three major soluble sugars accumulated extensively during strawberry fruit development.In the early stage of strawberry fruit development,glucose is the main soluble sugar;however,the content of sucrose increased rapidly and exceeded glucose and fructose in the late stage of strawberry fruit development (Fig.7-B).Correlational analyses indicated that a significant correlation existed between total sugar content and the expression level ofFaCWINV1during strawberry fruit development (Fig.7-C).In general,FaCWINV1may play an important role in sugar accumulation during strawberry fruit development.
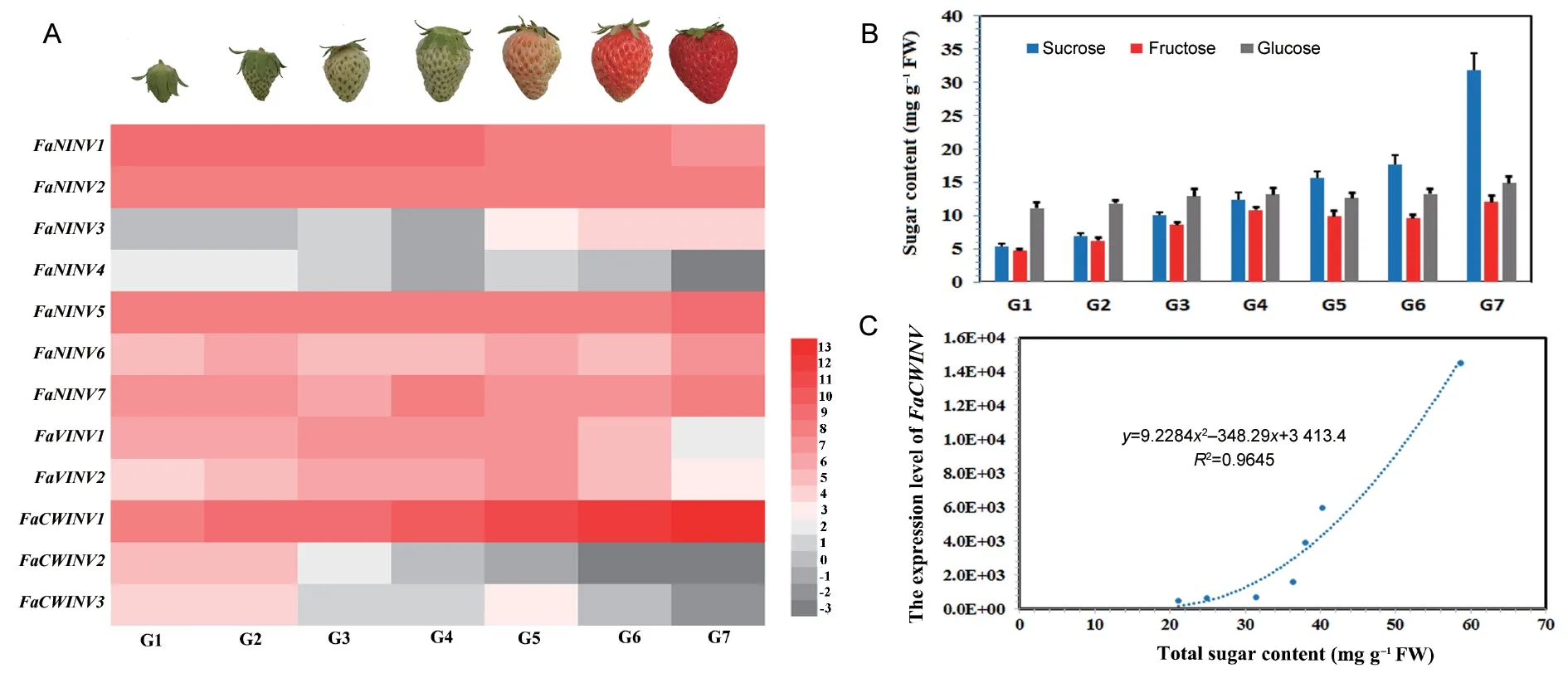
Fig.7 FaCWINV1 plays an important role in sugar accumulation during strawberry fruit development.A,real-time PCR analysis of FaINV expression during strawberry fruit development.Data were calculated by the 2–ΔCT method and log2(fold change).The strawberry ubiquitin gene was selected as the internal standard for normalization of gene expression.The expression level of the strawberry ubiquitin gene was assumed to be 1e+5.The colour scale representing the relative RNA level is shown to the right of the heat map.B,changes in sugar content during strawberry fruit development.C,correlational analyses between the total sugar content and expression level of FaCWINV1 during strawberry fruit development.
4.Discussion
4.1.Identification and characterization of the invertase gene family in strawberry
INVgenes play an important role in the regulation of sugar accumulation in plants and have been well-studied in many species (Goetzet al.2001;Goelet al.2006;Zanoret al.2009).In the sucrose storage organ,CWINVs play an important role in phloem unloading and sink strength,while VINVs are mainly responsible for coordinating cell expansion,sucrose accumulation,and the ratio of sucrose to hexose in vacuoles (Koch 2004).However,relevant research on the gene function ofINVsin strawberries is lacking to date.With the completion of whole-genome sequencing for strawberry (F.×ananassa) (Edgeret al.2019),we performed a genome-wide identification ofFaINVsin the strawberry genome.Forty-four putative proteins belonging to theINVgene family were identified in strawberry,thus providing information on theINVgene family in strawberry for the first time (Table 1).Based on the sequence homology and evolutionary relationships,the 44 proteins were divided into two groups:15 AIs and 29 NINVs;in addition,the AIs contained nine VINVs and six CWINVs.Among these genes,FaCWINV2-1andFaCWINV2-2were missing the first exon and portions of the second exon because of the incomplete genome assembly.FaVINV2-2.3andFaNINV8-4were predicted to be pseudogenes with imperfect or shorter open reading frames.Pseudogenes most likely regulate parental gene expression by generating non-coding RNA and antisense RNA that act as RNA sponges for miRNA (Anet al.2017).Therefore,we hypothesized thatFaVINV2-2.3andFaNINV8-4may have biologically relevant functions in the transcriptional regulation ofINVgenes.
Generally,FaAIs showed a slightly higher molecular weight range than FaNINVs,which is consistent with the reported INVs in other plants (Chenet al.2015).FaVINVs possessed a lower theoretical isoelectric point (4.66–6.41);FaNINVs had a slightly higher isoelectric point (5.57–8.53);and FaCWINVs gave the highest isoelectric point (7.60–9.15).All the FaAIs possessed 13 well-conserved regions,except for the suspected pseudogene,and these regions included NDPNG,RDP,and WECP(V)D motifs (Fig.2).Gene structure analyses showed one 9-bp exon that was translated into the tripeptide DPN located in the fructosidase motif (NDPNG) (Figs.2 and 3).This tripeptide is a typical structural feature of CWINVs in plants (Wanget al.2017;Shenet al.2018).However,all the FaNINVs contained 12 intact conserved regions,except for the suspected pseudogene (Fig.1),which is consistent with reported acid INVs in other plants (Chenet al.2015;Wanget al.2017;Shenet al.2018).
4.2.Gene duplication and dominant expression in allopolyploids
Polyploid events,also known as whole-genome duplications,have a strong impact on the amplification of members of gene families.The number of INV proteins in diploidFragaria vescais similar to those inPopulus,sorghum,rice,Arabidopsisand pepper (Jiet al.2005;Chenet al.2015;Shenet al.2018) (Table 2).However,the number of INV proteins in strawberry is notably greater (by almost three or four times) than those in other species.The strawberry (F.×ananassa) is an allo-octoploid (2n=8x=56)involving four diploid progenitor species (F.vesca,F.nipponica,F.iinumae,andF.viridis) (Edgeret al.2019).A comparison of the chromosomal locations of INV proteins inF.×ananassaandF.vescawas performed becauseF.vescais one of the progenitors ofF.×ananassa.All of theF.vescaINV homologous genes were distributed on the same position of the same chromosomes in theF.×ananassafour subgenomes,which showed that polyploid events were the main method ofINVgene amplification inFragaria×ananassa(Fig.4).Segmental and tandem duplications are considered two of the main causal factors for gene family expansion in plants (Cannonet al.2004).Three groups ofF.×ananassa INVgenes (NINV4,NINV5andVINV2) may be involved in segmental and tandem duplications (Fig.4).Because of the reduction of gene selection pressure caused by the amplification ofINVgene family members,someINVgenes (CWVIN4andCWINV5)may have been lost during strawberry evolution while otherINVgenes (such asNINV8) evolved into pseudogenes(Figs.4 and 5).
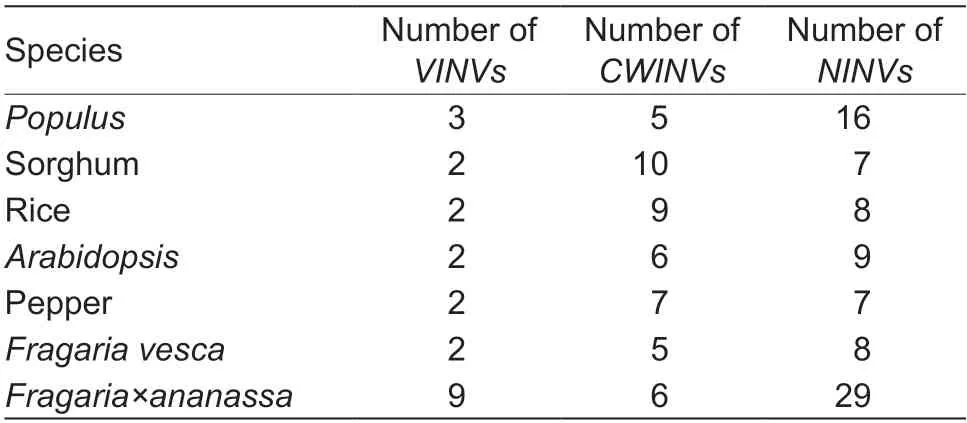
Table 2 Number of INV genes in the Populus,sorghum,rice,Arabidopsis,pepper, Fragaria vesca and F.×ananassa genomes
Recent studies have shown that gene expression patterns are dramatically altered by polyploidy (Hegartyet al.2006;Stuparet al.2007).The degree of influence varies considerably between species,although the duplicated genes in polyploids can present three main nonadditive expression patterns:dominant expression,in which the total gene expression level depends on one of the parents in a polyploidy;transgressive expression;and homeolog expression bias (Yooet al.2014).Our data showed that half ofF.×ananassaINVtranscripts have a premature termination codon which may cause a reduction,loss,or alteration in the function of the affected gene (Fig.6-A and B).Overall,F.×ananassaINVtranscripts with a premature stop had lower expression levels (FPKM) while the expression levels of transcript variants with whole coding sequence were much higher in strawberry fruit(Fig.6-C).Previous studies have shown that a dominant subgenome exists in octoploid strawberry (F.×ananassa),which has markedly higher gene content,gene expression abundance,and biased exchanges between homoeologous chromosomes than those of the other three subgenomes(Edgeret al.2019).Combined with the above analysis,we concluded thatINVgenes showed dominant expression patterns inF.×ananassa.
4.3.FaCWINV1 is involved in sugar accumulation in strawberry fruit
Sucrose,glucose,and fructose are the three major soluble sugars and rapidly accumulate during strawberry fruit development.In our study,glucose is the main soluble sugar in the early stage of strawberry fruit development;however,the content of sucrose increases rapidly and exceeds that of glucose and fructose in the later stages of strawberry fruit development,especially in the ripe stage.(Fig.7-B).Sugar accumulation in fruit is involved in a series of genes associated with sugar biosynthesis,metabolism,and transportation (Sturmet al.1999).CWINVsare involved in the hydrolysis of sucrose to fructose and glucose in sink organs to maintain sucrose concentration gradients,which are considered to be a driving force for sugar transport,partitioning,and storage from the source leaves to sink organs (Foyer 1987;Lemoineet al.2013).
To study the functions of the strawberryINVs,we analysed the temporal and spatial expression trends during strawberry fruit development using qRT-PCR along seven successive fruit developmental stages.Our qRT-PCR analysis showed that the expression level ofFaCWINV1rapidly increased during fruit development and was strongly expressed during the fruit maturation period (G5,G6 and G7) (Fig.7-A).The expression levels ofFaCWINV1were 20times higher than those of the otherF.×ananassaINVgenes in ripe strawberry fruit,indicating thatFaCWINV1may be involved in strawberry fruit development.Overall,the results of this study indicate that there is a significant correlation between the total sugar content andFaCWINV1expression level during strawberry fruit development (Fig.7-C).
5.Conclusion
We provided a comprehensive analysis of theINVgene family in octoploid strawberry (F.×ananassa),including the gene structure,chromosomal locations,conserved domains,and gene evolution and expression profiles during strawberry fruit development.Our study revealed that polyploid events resulted in theINVgene being abundantly amplified (almost three-or four-fold) in theF.×ananassagenome,and these amplifiedINVgenes showed dominant expression in strawberry fruit.The expression ofFaCWINV1was markedly upregulated during fruit development and strongly expressed in ripe fruit.Moreover,a significant correlation was observed between the total sugar content andFaCWINV1expression level.Given thatCWINVfunctions in sucrose unloading and sink strength establishment,we conclude thatFaCWINV1is the key factor in sugar accumulation during strawberry fruit development.
Acknowledgements
This work was funded by the Major Project for Breeding New Varieties of Jiangsu Province,China (PZCZ201721),the National Horticulture Germplasm Resources Center,China(NHGRC2020-NH16),and the National Crop Germplasm Resources Protection of the Ministry of Agriculture and Rural Affairs of China (19200361).
Declaration of competing interest
The authors declare that they have no conflict of interest.
杂志排行
Journal of Integrative Agriculture的其它文章
- Assessing the impact of non-governmental organization’s extension programs on sustainable cocoa production and household income in Ghana
- Food safety inspection and the adoption of traceability in aquatic wholesale markets:A game-theoretic model and empirical evidence
- Bacterial diversity and community composition changes in paddy soils that have different parent materials and fertility levels
- lncreased ammonification,nitrogenase,soil respiration and microbial biomass N in the rhizosphere of rice plants inoculated with rhizobacteria
- Regional distribution of wheat yield and chemical fertilizer requirements in China
- Changes in organic C stability within soil aggregates under different fertilization patterns in a greenhouse vegetable field
