Bacterial diversity and community composition changes in paddy soils that have different parent materials and fertility levels
2021-08-12MAXinlingLlUJiaCHENXiaofenLlWeitaoJlANGChunyuWUMengLlUMingLlZhongpei
MA Xin-ling ,LlU Jia ,CHEN Xiao-fen ,Ll Wei-tao ,JlANG Chun-yu ,WU Meng,LlU Ming,Ll Zhong-pei
1 State Key Laboratory of Soil and Sustainable Agriculture,Institute of Soil Science,Chinese Academy of Sciences,Nanjing 210008,P.R.China
2 University of Chinese Academy of Sciences,Beijing 100049,P.R.China
3 Soil and Fertilizer &Resources and Environment Institute,Jiangxi Academy of Agricultural Sciences,Nanchang 330200,P.R.China
4 CAS Key Laboratory of Tropical Forest Ecology,Xishuangbanna Tropical Botanical Garden,Chinese Academy of Sciences,Mengla 666303,P.R.China
Abstract Parent materials and the fertility levels of paddy soils are highly variable in subtropical China.Bacterial diversity and community composition play pivotal roles in soil ecosystem processes and functions.However,the effects of parent material and fertility on bacterial diversity and community composition in paddy soils are unclear.The key soil factors driving the changes in bacterial diversity,community composition,and the specific bacterial species in soils that are derived from different parent materials and have differing fertility levels are unknown.Soil samples were collected from paddy fields in two areas with different parent materials (quaternary red clay or tertiary sandstone) and two levels of fertility (high or low).The variations in bacterial diversity indices and communities were evaluated by 454 pyrosequencing which targeted the V4–V5 region of the 16S rRNA gene.The effects of parent material and fertility on bacterial diversity and community composition were clarified by a two-way ANOVA and a two-way PERMANOVA.A principal coordinate analysis (PCoA),a redundancy analysis (RDA),and multivariate regression trees (MRT) were used to assess changes in the studied variables and identify the factors affecting bacterial community composition.Co-occurrence network analysis was performed to find correlations between bacterial genera and specific soil properties,and a statistical analysis of metagenomic profiles(STAMP) was used to determine bacterial genus abundance differences between the soil samples.The contributions made by parent material and soil fertility to changes in the bacterial diversity indices were comparable,but soil fertility accounted for a larger part of the shift in bacterial community composition than the parent material.Soil properties,especially soil texture,were strongly associated with bacterial diversity.The RDA showed that soil organic carbon (SOC) was the primary factor influencing bacterial community composition.A key threshold for SOC (25.5 g kg–1) separated low fertility soils from high fertility soils.The network analysis implied that bacterial interactions tended towards cooperation and that copiotrophic bacteria became dominant when the soil environment improved.The STAMP revealed that copiotrophic bacteria,such as Massilia and Rhodanobacter,were more abundant in the high fertility soils,while oligotrophic bacteria,such as Anaerolinea,were dominant in low fertility soils.The results showed that soil texture played a role in bacterial diversity,but nutrients,especially SOC,shaped bacterial community composition in paddy soils with different parent materials and fertility levels.
Keywords:microorganisms,soil biodiversity,soil fertility,soil texture,soil nutrients
1.lntroduction
Microbial diversity and community composition play important roles in soil ecosystem processes and functions because they drive nutrient cycling,maintain soil structure,and transform organic carbon (Ibekweet al.2002;Brussaardet al.2007).They are also important for agricultural sustainability (Bhat 2013).
The importance of the impacts of parent material on bacterial diversity and community composition have been compared to soil depth (Shenget al.2015),land use (Denget al.2015),age of the plant stand (Guoet al.2015),nutrient application,and straw return (Sunet al.2016).Parent material has a considerable effect on microbial diversity indices (Denget al.2015).It is also considered to be the primary factor influencing microbial community composition (Guoet al.2015;Sunet al.2016).For example,parent material has a stronger impact on the soil microbial community during the earlier stages of ecosystem development (Wagaiet al.2011).It is also thought that soils developed from the same parent materials usually accommodate similar microbial communities (Guoet al.2015).
Soil properties have important effects on bacterial diversity and community composition.For example,soil pH has been generally shown to have crucial effects on bacterial community structure in many studies (Fierer and Jackson 2006;Lauberet al.2009;Griffithset al.2011).Several studies have found that soil carbon (C),nitrogen(N),phosphorus (P),base ions,salinity and elemental ratios affect bacterial diversity and community composition(Faoroet al.2010;Ramirezet al.2010;Kuramaeet al.2012;Zhanget al.2019).Soil texture is known to influence microbial biomass (Müller and Höper 2004;Chodak and Niklinska 2010;Caoet al.2016),enzymatic activity,and respiration (Meleroet al.2007;Chodak and Niklinska 2010;Józefowskaet al.2017).It also affects microbial diversity(Chauet al.2011;Naveedet al.2016) and community composition (Johnsonet al.2003).
Paddy fields cover 30% of the arable land in China and 90% of them are distributed in tropical or subtropical China(Jiaet al.2010).Parent materials in subtropical China vary considerably (Xu and Cai 2007).The different parent materials and their weathering stage determine soil texture,clay mineral contents,and nutrient availability to biota (Ulrich and Becker 2006),all of which affect soil microorganisms(Denget al.2015).Fertilization regimes,crop exudates,and residue return also make fertility highly variable across the paddy soils in this region.However,the contributions made by different parent materials and fertility to bacterial diversity and community composition in paddy soils are unclear.In this study,it was hypothesized that both the parent materials and soil fertility affect soil properties and ultimately determine the bacterial diversity and community composition of paddy soils.Therefore,soil samples were taken from paddy fields with different parent materials and fertility in a red soil region of subtropical China.Bacterial diversity and community composition were measured with 454 pyrosequencing.The objectives of this study were to:(1) clarify the critical soil factors driving the changes in bacterial diversity and community composition,and (2)identify the dominant bacterial species in paddy soils that were specific to certain parent materials and fertility levels.
2.Materials and methods
2.1.Site description and soil collection
The sampling sites were located in Yingtan City (28°12´–28°13´N,116°55´E),Jiangxi Province,subtropical China.The climate in this region is typical subtropical monsoon,and has an annual precipitation of 1 795 mm,an annual evaporation of 1 318 mm,and a mean annual temperature of 17.6°C.Quaternary red clay and tertiary sandstone were identified as the typical parent materials in this region.All the paddy fields belonged to a state farm and double rice was continuously cultivated before the sampling period.The agronomic practices,such as flood irrigation and fertilizer application,have been almost identical across the sampling plots in recent years.The fertilizer application rates were approximately 300 kg N ha–1yr–1,66 kg P ha–1yr–1,and 248 kg K ha–1yr–1for each plot.After consulting local experts,low-and high-fertility soils derived from quaternary red clay were sampled from newly cultivated (about 5 years)and old (>100 years) paddy fields.Low-and high-fertility soils derived from tertiary sandstone were also sampled from newly cultivated (about 15 years) and old (>80 years)paddy fields.As a result,there were four soil groups,namely quaternary red clay parent materials and high fertility (RCH),quaternary red clay parent materials and low fertility (RCL),tertiary red sandstone parent materials and high fertility(RSH),and tertiary red sandstone parent materials and low fertility (RSL).For each soil group,three paddy fields (~0.2 ha each) that were about 100 m apart were selected as replicated sampling plots.Five surface soil subsamples (0–15 cm) were collected from each sampling plot immediately after late rice cultivation at the end of October,2014.They were collected using a soil auger (3 cm in diameter) and a zigzag sampling pattern.The subsamples were thoroughly mixed and the composite soil samples were used for the chemical-physical property and microbial analyses.The average values of soil chemical–physical properties and rice yields for each soil group are shown in Appendix A.
2.2.Chemical property measurements
The soil pH was measured by a potentiometer,and the SOC was determined by the potassium dichromate oxidation method (Mebius 1960).The soil samples were digested with 0.8 mol L−1K2Cr2O4and concentrated H2SO4(v/v,1:1)at 150°C for 30 min.Then the SOC was titrated against 0.5 mol L−1ferrous iron solution.The total N (TN) was determined using the Kjeldahl method (Kjeldahl 1883).The soil sample was heated and boiled with concentrated H2SO4.Then,the TN was absorbed by 2% boric acid solution and titrated against 0.1 mol L−1sulfuric acid.The available N(AN) was hydrolyzed by 1 mol L−1sodium hydroxide and measured using micro-diffusion methods (Conway 1948).After the soil had been melted with anhydrous sodium carbonate or extracted with 0.5 mol L−1sodium bicarbonate,the total P (TP) and available P (AP) were determined by the molybdenum-blue colorimetric method (Dickman and Bray 1940).Soil total K (TK) and available K (AK) were determined by the modified flame emission spectrometric method after the soil had been digested in concentrated HF/HClO4(v/v,2:1) or extracted using 1 mol L−1ammonium acetate,respectively (Pansu and Gautheyrou 2006).Another set of soil samples were dispersed in 0.5 mol L−1sodium hydroxide and soil texture was determined using the pipette method (Pansu and Gautheyrou 2006).
2.3.DNA extraction,PCR amplification and 454 pyrosequencing
Soil microbial genomic DNA was extracted from 0.5 g fresh soil using a FastDNA™ SPIN Kit (MP Biomedicals,Santa Ana,CA,USA),and the quantity and quality of the extracted DNA were measured by an Eppendorf BioSpectrometer Basic (Eppendorf Corporate,Hamburg,Germany).The DNA concentration was >100 ng μL−1and the OD260/OD280was >1.6 for each DNA sample,which meant that they could be used in the following analyses.The primers (F515 and R907) and target regions of the bacterial 16S rRNA genes were as described in Shenet al.(2013).The PCR cycling conditions were 94°C for 5 min,then 33 cycles of 94°C for 30 s,55°Cfor 30 s,and 72°C for 45 s.There was a final extensionat 72°C for 10 min.The PCR products were purified using an E.Z.N.A.Cycle-Pure Kit (Omega,Norcross,GA,USA) and run on a Roche FLX 454 pyrosequencing platform (Roche Diagnostics Corporation,Branford,CT,USA).About 4 175 high-quality 16S rRNA reads for each sample were obtained after sequencing.The raw data produced by the sequencing were analyzed according to Caporasoet al.(2010).About 1 630 high-quality sequences were screened out for each sample after short sequences(<200 bp),ambiguous bases,barcodes,homopolymers and low quality sequences (quality score Q<25) were eliminated.Sequences with ≥97% similarity were assigned to the same operational taxonomic unit (OTU) by UCLUST (Edgar 2010).The most abundant sequence in each OTU was selected as the representative sequence.Then,the representative OTU sequences were classified by RDP classifier with a confidence threshold of 80% and the taxonomic information was annotated (Wanget al.2007).The sequence data were submitted to the Sequence Read Archive (SRA) of the NCBI with the accession number SRP267058.
2.4.Data analysis
Bacterial diversity and community were analyzed by R version 3.2.3 (R Core Team 2014).The phyloseq package was used to calculate the Bacterial Chao1 and Shannon indices.The vegan package in R was used to perform a principal coordinate analysis (PCoA) and a redundancy analysis (RDA).Removal of the autocorrelation variables and significance testing of the soil factors followed Liuet al.(2018) when the RDA was carried out.The ‘mvpart’ package was used to generate multivariate regression trees (MRT)(De’Ath 2002).Any significant effects of parent material and fertility on bacterial diversity and community composition were determined with two-way ANOVA and two-way PERMANOVA using Past 3 (Hammeret al.2001).Significant differences among soil groups were determined atP<0.05.Correlations between soil properties and bacterial diversity indices were also determined with the two-tailed Pearson correlation statistic using Past 3.A co-occurrence network analysis of bacterial genera and specific soil properties was carried out using the plugin CoNet in Cytoscape 3.5.1(Shannonet al.2003).Strong correlations between two genera or between genera and soil properties were defined as having Spearman’s correlation coefficients >0.75 atP<0.01.The topology parameters of the network were determined by Cytoscape 3.5.1 using NetworkAnalyzer (Shannonet al.2003),and bacterial genus abundance differences among soil groups were determined by a statistical analysis of metagenomic profiles (STAMP) (Parkset al.2014).
3.Results
3.1.Bacterial diversity and community composition in soils with different parent materials and fertility levels
Both parent material and fertility significantly affected bacterial richness and the diversity indices.The order for the Chao1 and Shannon index changes was as follows:RSL~RCH>RSH>RCL.The parent material contributed 23.0 and 24.4% to the total variation in the Chao1 and Shannon indices,respectively (Table 1),whereas fertility explained 22.8 and 24.0% of the total variation in the Chao1 and Shannon indices,respectively (Table 1).
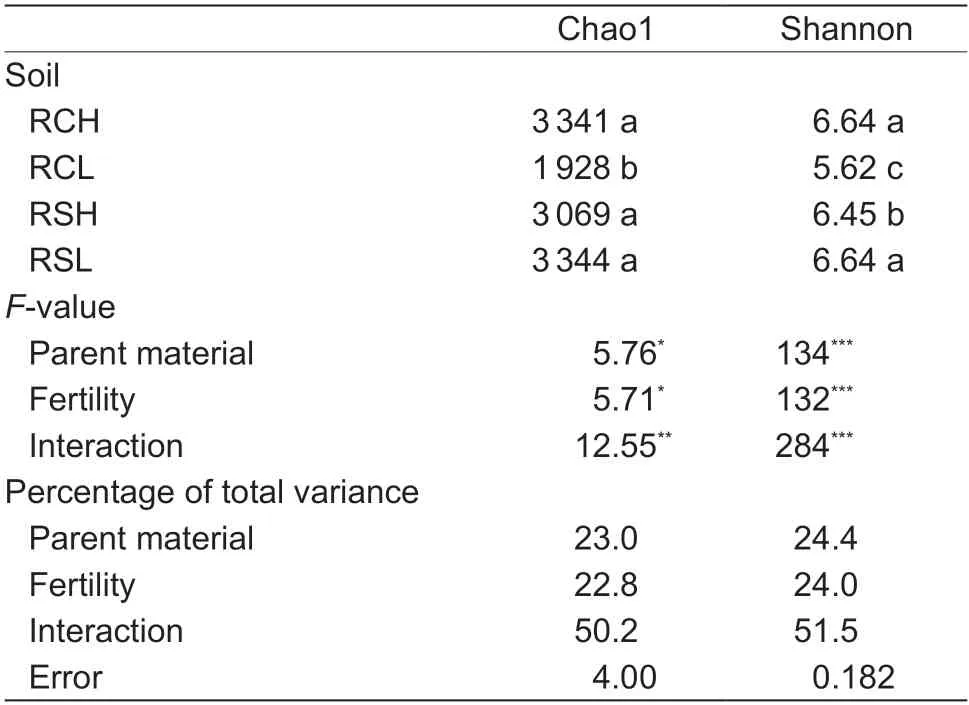
Table 1 Bacterial diversity indices for paddy soils with different parent materials and fertility levels1)
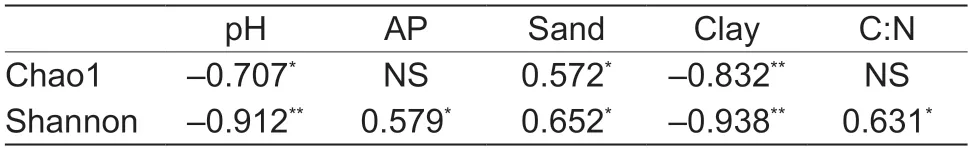
Table 2 Correlations between bacterial diversity,and soil chemical and physical properties1)
For a given parent material,the relative abundances of Proteobacteria,Acidobacteria,Actinobacteria,Chlorobi,Bacteroidetes,Planctomycetes,and Gematimonadetes in high-fertility soils were significantly higher than those in low-fertility soils (Fig.1).On the contrary,Chloroflexi and Nitrospirae were remarkably lower in high-fertility soils than in low-fertility soils (Fig.1).At a given soil fertility level,the relative abundances of Chloroflexi and Acidobacteria in red clay soils were significantly higher than those in red sandstone soils (Fig.1).In contrast,Proteobacteria,Chlorobi,Bacteroidetes,Gematimonadetes,and Spirochaetae were significantly lower in red clay soils than in red sandstone soils (Fig.1).
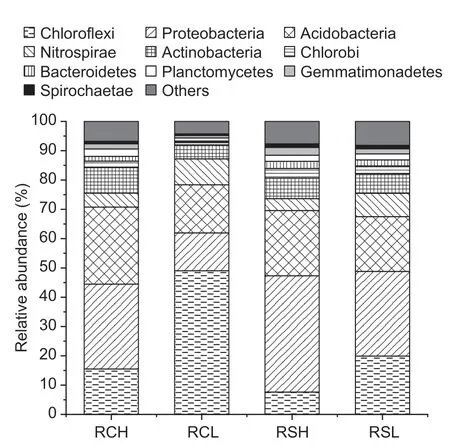
Fig.1 Relative abundances of the major bacterial phyla in paddy soils with different parent materials and fertility levels.RCH,soils with quaternary red clay parent materials and high fertility;RCL,soils with quaternary red clay parent materials and low fertility;RSH,soils with tertiary sandstone parent materials and high fertility;RSL,soils with tertiary sandstone parent materials and low fertility.
Axes 1 and 2 of the PCoA assigned 35.6 and 16.1% of the variation to bacterial community composition,respectively(Fig.2).The bacterial communities in soils with different parent materials and fertility levels could be separated(Fig.2).The two-way PERMANOVA also indicated a significant difference in community composition between the different parent materials (P<0.01) and fertility levels(P<0.001) (Appendix B).Soil fertility explained about 45.4%of the total variation in bacterial community composition,whereas parent material explained 23.9% (Appendix B).
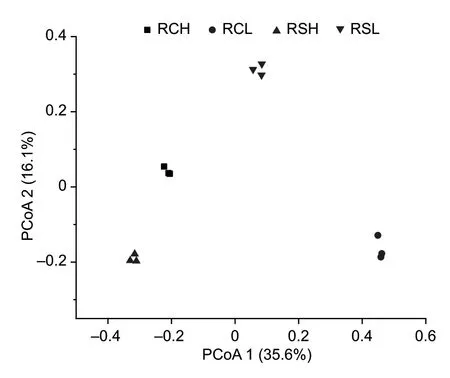
Fig.2 Principal coordinate analysis (PCoA) of the variation in bacterial community composition,based on the Bray-Curtis distance,for paddy soils with different parent materials and fertility levels.RCH,soils with quaternary red clay parent materials and high fertility;RCL,soils with quaternary red clay parent materials and low fertility;RSH,soils with tertiary sandstone parent materials and high fertility;RSL,soils with tertiary sandstone parent materials and low fertility.
3.2.lmpact of soil properties on bacterial diversity and community composition
The bacterial Chao1 and Shannon indices were negatively related to soil pH and clay content,but positively associated with sand content (Table 2).The Shannon diversity index was particularly positively associated with AP and the carbon:nitrogen (C:N) ratio (Table 2).
The redundancy analysis results showed that axes 1 and 2 accounted for 42.2 and 16.8% of the variation in bacterial community composition between the parent materials and fertility,respectively (Fig.3).Bacterial community composition was significantly influenced by SOC,AP,and pH (Fig.3).In particular,SOC explained 28.5% of the changes in bacterial community composition and separated the different soils along axis 1.The bacterial community compositions in the different soils were divided by the SOC thresholds derived from the multivariate regression tree analysis (Fig.4).These were SOC<25.5 g kg–1and ≥25.5 g kg–1for the low-fertility soils (RCL and RSL)and high-fertility soils (RCH and RSH),respectively (Fig.4).
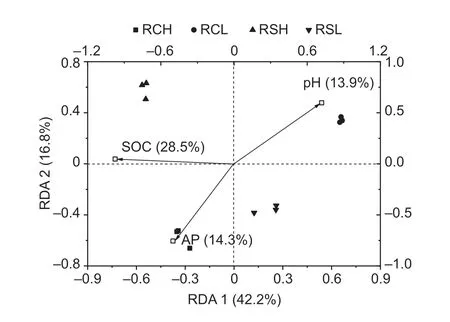
Fig.3 Redundancy analysis (RDA) of the soil property effects on the composition of bacterial communities in paddy soils with different parent materials and fertility levels.RCH,soils with quaternary red clay parent materials and high fertility;RCL,soils with quaternary red clay parent materials and low fertility;RSH,soils with tertiary sandstone parent materials and high fertility;RSL,soils with tertiary sandstone parent materials and low fertility;SOC,soil organic carbon;AP,available phosphorus.
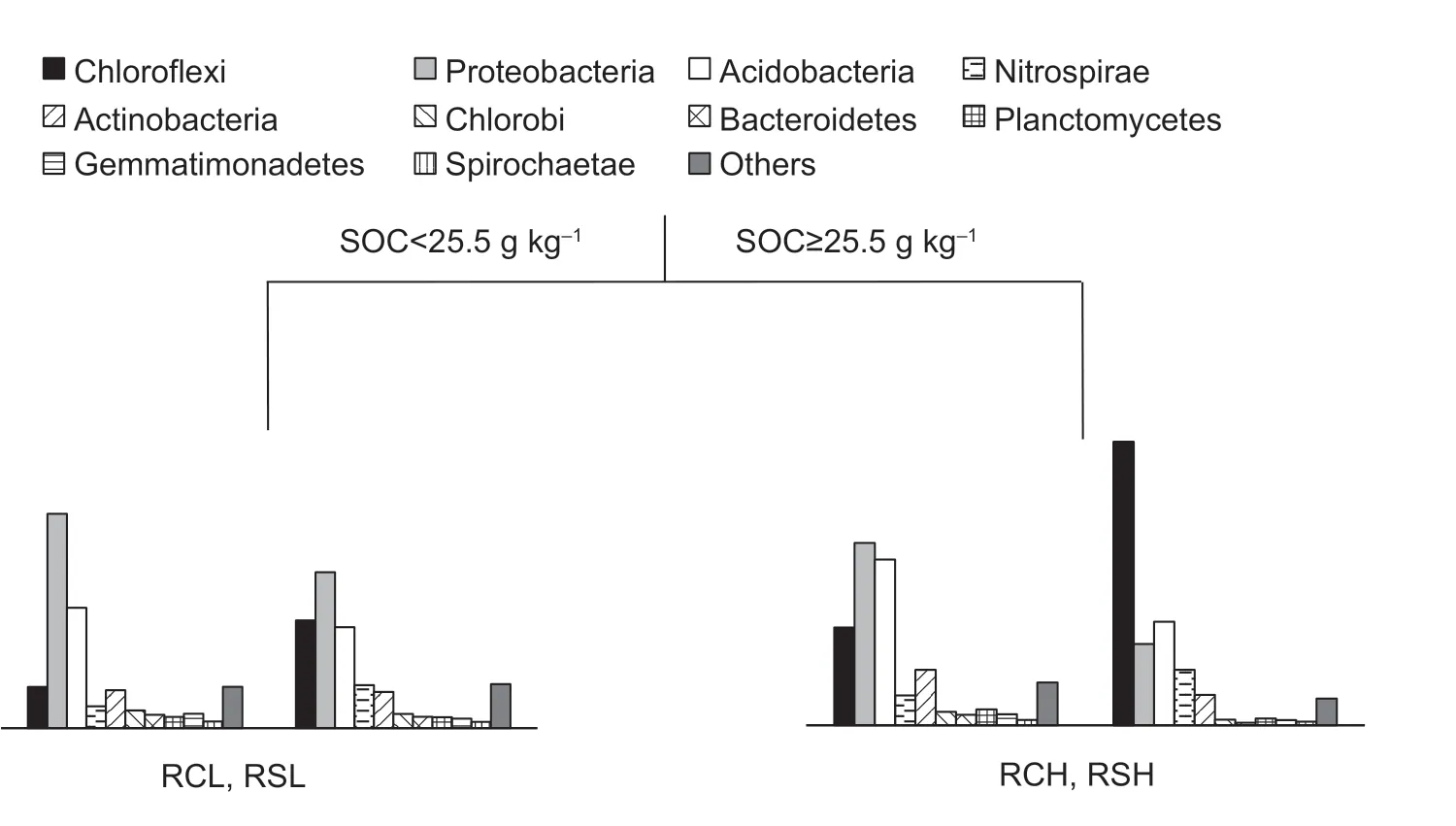
Fig.4 Multivariate regression tree for bacterial community compositions in paddy soils with different parent materials and fertility levels when partitioned by soil organic carbon.Bar plots indicate the relative abundances of the bacterial phyla.SOC,soil organic carbon;RCH,soils with quaternary red clay parent materials and high fertility;RCL,soils with quaternary red clay parent materials and low fertility;RSH,soils with tertiary sandstone parent materials and high fertility;RSL,soils with tertiary sandstone parent materials and low fertility.
The co-occurrence network analysis showed there were 21 nodes and 114 edges in the network (Fig.5).The average clustering coefficient (avgCC),betweenness centrality,and closeness centrality of the network values were 0.774,4.52,and 0.703,respectively (Appendix C).The generaMassilia,Rhodanobacter,Tumebacillus,Rhodomicrobium,Gemmata,Haliangium,Marmoricola,Nevskia,Tahibacter,MucilaginibacterandCandidatus Planktophilawere positively related to SOC (Fig.5).The proportions of positive and negative edges for the above genera were about 33.0 and 26.6%,respectively (Appendix C).Genera ofAnaerolinea,Leptolinea,Kedonobacter,Actinospica,Desulfovibrio,Geobacter,SinomonasandAcidothermuswere negatively related to SOC (Fig.5).Their proportions of positive and negative edges were about 16.0 and 21.3%,respectively (Appendix C).
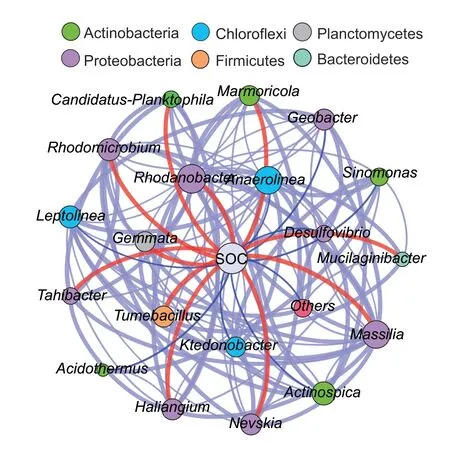
Fig.5 Co-occurrence network analysis of the bacterial communities at the genus level.The node colors are based on bacterial taxonomic information at the phylum level.Edges that are directly related to soil properties are shown in order to visualize the potential effects of soil properties on bacterial co-occurrence patterns.The red lines represent positive interrelationships,the blue lines represent negative interrelationships,and the grey lines represent interrelationships between genera.The size of each node represents the connecting degree,and the width of each line represents the strength of the correlation.
3.3.Differences in bacterial genus abundance between high-and low-fertility soils
The multivariate regression tree results divided the bacterial communities in the different soil samples into two groups.The bacterial communities in the RCH and RSH soils belonged to group 1,and those in the RCL and RSL soils were classified as group 2.The STAMP analysis indicated that three bacteria genera were significantly different in abundance between groups 1 and 2.MassiliaandRhodanobacterwere dominant in group 1,whereasAnaerolineawere much more abundant in group 2 (Fig.6).
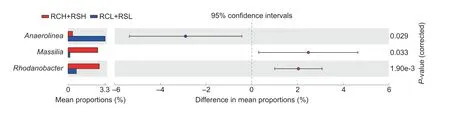
Fig.6 Differentially abundant bacterial genera in paddy soils with different fertility levels.Significant differences between groups were calculated by Welch’s test and the corrected P-values were calculated by the Bonferroni multiple test correction.The error bars indicate the standard deviations of triplicated samples and the circles represent 95% confidential intervals.RCH,soils with quaternary red clay parent materials and high fertility;RCL,soils with quaternary red clay parent materials and low fertility;RSH,soils with tertiary sandstone parent materials and high fertility;RSL,soils with tertiary sandstone parent materials and low fertility.
4.Discussion
In the current study,the contributions made by parent material to bacterial richness and diversity index variations were comparable to soil fertility.Parent material and soil fertility also significantly affected the relative abundances of major bacterial phyla.However,parent material explained less of the variation in bacterial community composition than fertility.The influence of parent material on bacterial diversity and community composition is usually restricted by nutrient availability (Shenget al.2015).For example,in paddy field surface soils,fertilizer application and crop exudation increase soil C,N,P,and K availability and soil fertility,both of which lead to improved microbial growth and shape community structure (Shenget al.2015).Different rice cultivation times contributed to the discrepancies in soil fertility recorded in this study,which was in agreement with Chen X Fet al.(2018).The significant interaction effect in this study of parent material and fertility on bacterial diversity and community composition also indicated that the impact of parent material might be moderated by soil fertility.
Generally,bacterial richness and diversity increase in acidic to neutral soils (Fierer and Jackson 2006).In the current study,there were apparent negative relationships between soil pH and the bacterial diversity indices.However,the path analysis suggested that pH positively and directly affected bacterial richness and diversity.The negative effect was probably caused by the indirect influences of other soil factors (Appendices D and E).The results also showed that bacterial diversity was positively associated with soil nutrients,such as AP and the C:N ratio.Similarly,a previous study also found that bacterial diversity was closely related to soil nutrients,such as soil ammonium N (Sunet al.2015).Previous reports have also shown that soil texture is more stable than other soil properties and has important effects on soil microorganisms (Denget al.2015;Caoet al.2016).For example,bacterial richness indices increase when the sand (Chauet al.2011) or coarse soil (Carsonet al.2010)contents are higher.Bacterial richness and diversity were positively correlated to soil sand content,but negatively correlated with clay content in this study.A possible reason is that the coarser soil had more isolated water films than the fine-textured soil.Therefore,the coarser soil had more isolated micro-habitats and less competitive pressure than the finer soil,which means that it could harbor more species(Chauet al.2011).
The soil chemical and physical properties also influence the bacterial community in soils with different parent materials(Chodak and Niklinska 2010;Chauet al.2011;Guoet al.2013;Shenget al.2015;Caoet al.2016).For example,as the terminal restriction fragment length polymorphism analysis showed,bacterial composition was significantly correlated with soil pH and available Mn in the top soil(Shenget al.2015).Sunet al.(2015) suggested that,in addition to soil pH,SOC and AP also significantly contribute to bacterial community variation.This study also found that SOC,AP,and pH significantly affected bacterial community composition,which confirmed the results reported by Sunet al.(2015) and Shenget al.(2015).In particular,SOC was the primary factor affecting the variation in bacterial community composition.A threshold of 25.5 g kg–1SOC led to significant variations in the bacterial communities between the high-and low-fertility soils.Sunet al.(2015) found that 10.1 g kg–1of soil total C significantly contributed to changes in the bacterial communities found in lime concretion black soils.The different thresholds for SOC between previous studies and this study were probably due to the different soil types,fertilities,and fertilizer application rates.
The network analysis indicated that the bacterial genera positively correlated with SOC had more positive edges.In contrast,the proportion of negative edges was higher among the bacterial genera negatively related to SOC.The positive edges suggested that there was mutual cooperation rather than competition within the bacterial assembly (Liet al.2020),whereas negative edges implied that inter-competitive exclusion occurred within the bacterial community (Fenget al.2017).The results from this study indicated that bacterial interactions tended towards cooperation when the soil environment (SOC and fertility level) improved,which confirmed the results of previous studies (Liet al.2019;Xieet al.2019).The network analysis also revealed that most of the genera positively related to SOC were copiotrophic bacteria.Furthermore,the STAMP analysis showed theMassiliaandRhodanobacterwere rich in soils with high fertility levels (SOC≥25.5 g kg–1),whereasAnaerolineadominated in the low-fertility soils (SOC<25.5 g kg–1).Some species ofMassiliacan use a wide range of sugars,organic acids,and amino acids as their sole carbon for growth (Zulet al.2008),andRhodanobacterare often isolated from the rhizosphere of many types of plants (Madhaiyanet al.2014;Wonet al.2015).MassiliaandRhodanobacterbelong to the phylum Proteobacteria,which generally contains copiotrophic bacteria.This means that they thrive where available nutrient levels are elevated,such as in high-fertility soils.Anaerolineabelong to phylum Chloroflexi,which are oligotrophic bacteria.A large number ofAnaerolineaare found in paddy soils and can decompose diverse C substrates under anaerobic conditions (Zhanget al.2019).Anaerolineahave also been shown to have a cellulolytic capacity (Xiaet al.2016).Therefore,we can conclude thatAnaerolineamight have proliferated in the low-fertility soils in our study because they could utilize recalcitrant C resources.
In our study,the rice yields were about 16 500 and 10 500–12 000 kg ha−1yr−1in the high-and low-fertility paddy fields,respectively.The differences in rice yield might indicate variations in the amounts of easily degradable carbon,e.g.,root exudates.The different crop yields had an indirect influence on the bacterial community by favoring different copiotrophic or oligotrophic microbial life strategies in the soil (Cederlundet al.2014).In contrast,the soil microbial community drives the decomposition of organic matter and nutrient cycling,which affect soil fertility and agrosystem sustainability (García-Oreneset al.2016;Chen Het al.2018;Zhouet al.2020).For example,it has been shown that microbial communities have a considerable influence on soil sustainability when maize (Ramírezet al.2017) or soybean (Merileset al.2009) are grown.One possible reason is that the microbes may regulate the mineralization of and competition for nutrients that sustain plant productivity(Heijdenet al.2008).Therefore,future studies should focus on the relationship between the changes in bacterial trophic type and rice production sustainability in red paddy soils with specific parent materials and fertility levels.
5.Conclusion
The effects of parent materials and soil fertility on bacterial diversity were approximately comparable in paddy soils in our study.But soil fertility contributed greater to the variation of bacterial community composition.Soil texture was the primary factor determining bacterial richness and diversity in soils with different parent materials and fertility levels.However,nutrients,especially SOC,determined bacterial community variations.Differences in SOC content led to changes of bacterial interactions and the growth and decline of specific copiotrophic and oligotrophic bacteria.Future work should focus on the relationship between shifts in bacterial trophic type and rice production sustainability in red paddy soils.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD0301104-01),the National Natural Science Foundation of China(41201242 and 41907041),the Major Research and Development Program for Science and Technology of Jiangxi Province,China (20182ABC28006),and the“135”Plan and the Field Frontier Project,China (ISSASIP1642).We thank International Science Editing (http://www.internationalscienceediting.com) for language editing.In addition,we are grateful to the anonymous reviewers and editors for their helpful comments.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Assessing the impact of non-governmental organization’s extension programs on sustainable cocoa production and household income in Ghana
- Food safety inspection and the adoption of traceability in aquatic wholesale markets:A game-theoretic model and empirical evidence
- lncreased ammonification,nitrogenase,soil respiration and microbial biomass N in the rhizosphere of rice plants inoculated with rhizobacteria
- Regional distribution of wheat yield and chemical fertilizer requirements in China
- Changes in organic C stability within soil aggregates under different fertilization patterns in a greenhouse vegetable field
- Follicle-stimulating hormone is expressed in ovarian follicles of chickens and promotes ovarian granulosa cell proliferation
