Long non-coding RNA NKILA mediates the progression of hepatocellular carcinoma through regulating miR-889-3p
2021-05-25WangHongmeiChenSiruiDuHongCaoJia
Wang Hongmei,Chen Sirui,Du Hong,Cao Jia
Abstract Objective:To explore the potential role of NKILA in the process of hepatocellular carcinoma (HCC) development.Methods:The relative expression of NKILA and miR-889-3p in HCC tissues and cells was quantified by quantitative real-time reverse transcription PCR (qRT-PCR).Cell growth and tumor angiogenesis were assessed by colony formation and angiogenesis assay,respectively.Western blotting was used to detect epithelial-mesenchymal transition (EMT)-related proteins.The association between NKILA and miR-889-3p was conducted by luciferase reporter analysis.A model of mouse xenograft tumor was established to detect the expression of related proteins by immunohistochemistry(IHC)analysis.Results:miR-889-3p was a direct target of NKILA,and NKILA negatively regulated miR-889-3p.qRT-PCR assay showed the NKILA expression was conspicuously down-regulated and miR-889-3p expression was up-regulated in HCC tissues and cells.NKILA overexpression suppressed HCC cell proliferation,EMT and tumor angiogenesis through directly sponging miR-889-3p.More importantly, in vivo model displayed NKILA significantly reduced the volume and weight of xenograft tumor,accompanied by E-cadherin increase and VEGF decrease.Furthermore,IHC revealed NKILA alone reduced the number of vascular endothelial cell markers (CD34 and CD105).Conclusion:NKILA overexpression curbed the development of HCC via regulating miR-889-3p,which may be a novel therapeutic target in patients with HCC.
Keywords long non-coding RNA;malignant neoplasm;epithelial-mesenchymal transformation;tumor angiogenesis
Introduction
Hepatocellular carcinoma (HCC),a primary or metastatic liver cancer,is the second leading cause of cancer-related death [1],and its incidence is increasing year by year.In 2018,there are approximately 841,080 new cases and 781,631 deaths of liver cancer worldwide [2].HCC is associated with multiple risk factors including hepatitis virus infection,obesity,alcohol consumption,poor diet,etc [3-4].At present,liver resection and liver transplantation are still the main treatment methods [5-6].Due to lack of effective diagnostic tools and treatment means,the prognosis of liver cancer is very unsatisfactory,and the fiveyear average survival rate is less than 10%[7].Therefore,it is very important to find an effective way to reduce the mortality of liver cancer.
Human genome studies demonstrate that long noncoding RNA (lncRNAs) has more than 200 nucleotides [8-9],which play an crucial role in a variety of biological processes,such as cell cycle,metabolism,survival and metastasis[10-11].It is worth noting that IncRNA is usually expressed in a unique way [12],which makes these molecules become targets for treatment and latent bio-markers for diagnosis and prognosis of cancer patients [13].NKILA is down-regulated in various cancers such as colorectal cancer [14],tongue squamous cell carcinoma [15]or malignant melanoma [16],and promotes the distant metastasis of highly malignant tumors [17].Nevertheless,the role of NKILA in HCC is still ambiguous.
In this study,we tested the expression levels of NKILA in human hepatocellular carcinoma tissues and cells,and investigated the functional role of NKILA in HCC.We found that NKILA inhibited the epithelial-mesenchymal transition (EMT) and tumor angiogenesis of hepatocellular carcinoma by targeting miR-889-3p.
Materials and methods
Tissue samples
A total of 46 HCC tissues and matched paracancerous normal tissues were obtained from Affiliated Hospital of Chengdu University of Traditional Chinese Medicine.The collected fresh tissue was immediately frozen in liquid nitrogen until used.All tissue specimens were examined by two pathologists.This study was approved by the Ethics Committee of Chengdu University of Traditional Chinese Medicine and obtained written informed consent from patients.
Cell culture
Human umbilical vein endothelial cells (HUVECs),human immortalized nontransformed normal liver cell line L-02 and the liver cancer cell line HepG2 were obtained from American type culture collection(ATCC,Manassas,VA,USA) and were cultured in Dulbecco's Modified Eagle Medium (DMEM,Gibco,CA,USA) supplemented with 10% fetal bovine serum (Gibco,CA,USA).All cells were maintained in a constant temperature incubator at 37°C of 5%CO2.
Plasmids construction and cell transfection
HepG2 cells with density of 1×104/well were seeded evenly in 6-well plate.When the cell growth density was 75%-85%,the cells were transferred according to the manufacturer’s instructions.After mixed with LipofectamineTM2000(Invitrogen,CA,USA),the transfection reagent was added to 1.5 mL serum-free medium,and the cells were incubated at 37°C.After 4-6 h of transfection,the culture medium was changed and unceasingly cultured for another 48 h,and then the follow-up experiments were carried out.
Luciferase reporter assay
Herein,wild-type NKILA (NKILA wt) and mutanttype NKILA (NKILA mut) luciferase vectors were constructed.Immediately,the vectors were co-transfected with the miR-889-3p mimics or miR-NC in the HepG2 cells using the LipofectamineTM2000 transfection reagent (Invitrogen,CA,USA).Lastly,luciferase activities were determined by a dual-luciferase reporter system using a luminometer (GloMax 20/20 Luminometer,Promega,WI,USA).
Clone formation assay
In short,the base containing 0.6% agarose was prepared in 6-well plate.Next,about 1.5×104single cells were mixed with 1 mL medium containing 0.3% soft Agar (Sigma,Missouri,USA).Then,the mixture was added to the 6-well plate.2 weeks later,cell colonies were fixed and stained with phosphate buffer saline(PBS) containing 4% formaldehyde and 0.005% crystal violet for 15 min.Finally,the images were captured and colony numbers were counted,clone formation rate=(clones/inoculated cells)×100%.
Tube formation assay
As described previously [18],about 1×105HUVECs were seeded on a Matrigel-coated plate and incubated with HepG2 for 18 h in the presence or absence of NKILA or miR-889-3p to evaluate the effect of NKILA or miR-889-3p on the formation of vascular endothelial growth factor (VEGF)-induced tubes.Images were obtained by a optical microscope (Leica,Weizlar,Germany),and the total number of branches was countered to quantify the tube formation.
Quantitative real-time polymerase chain reaction(qRT-PCR)
Total RNA from liver tissues and cells were extracted with TRIzol reagent (Invitrogen,CA,USA),and the cDNA was synthetized was synthesized using Prime-Script RT Kit (TaKaRa,Otsu,Japan) for RNA,or Prime-Script miRNA cDNA Synthesis Kit (TaKaRa,Otsu,Japan)for miRNA,according to the manufacturer’s instructions.qRT-PCR was performed using SYBR Green I nucleic acid gel stain (Sigma,Missouri,USA) The primers sequence as follows:NKILA,F:5'-GGGGTACCAGACCCGGCACCCGCGCAA-3',R:5'-CGGGATCCCCAGTTAAATTGAGATATACTTACAC-3';miR-889-3p,F:5'-GCGCGTTAATATCGGACAAC-3',R:5'-AGTGCAGGGTCCGAGGTATT-3';GAPDH,F:5'-AGGTCGGTGTGAACGGATTTG-3',R:5'-GGGGTCGTTGATGGCAACA-3';U6,F:5'-ACCCTGAGAAATACCCTCACAT-3';R:5'-GACGACTGAGCCCCTGATG-3'.GAPDH and U6 were employed as internal control.
Western blotting
HepG2 cells were cracked with RIPA(radio immunoprecipitation assay) lytic buffer(Beyotime,Shanghai,China),incubated on ice for 10 min and centrifuged at 12,000 r/min for 15 min.The protein concentration was determined by a bicinchoninic acid (BCA) kit(Sigma,Missouri,USA).Next,the protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE),and transferred to polyvinylidene fluoride (PVDF,Millipore,MA,USA) membrane.After that,the membranes were sealed with 5% skim milk at room temperature for 1 h,then incubated at 4°C with the corresponding primary antibody overnight.The primary antibodies used in this study were as below:Ki67 (1:500,ab16667,Abcam,Cambridge,UK),E-cadherin (1:1,000,ab40772,Abcam),N-cadherin(1:5,000,ab76011,Abcam),Fibronectin (1:300,ab2413,Abcam) and VEGF (1:500,ab39638,Abcam).The next day,the membranes were fully cleaned and incubated with goat anti-rabbit antibody coupled with horseradish peroxide (HRP) for 1 h at room temperature.Finally,the imprinted membranes were visualized with electrochemiluminescence (ECL) reagent (Thermo Fisher,MA,USA).GAPDH acted as the internal control,all antibodies are purchased from Abcam(Cambridge,UK).
Animal model
Male BALB/c nude mice (4-6 weeks old,n=10) were purchased from Guangdong Medical Lab Animal Center (Foshan,China),all animal operations were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals(National Academies Press,2011) and approved by the Animal Care and Use Committee of Hubei Medical College.HepG2 cells were stably transfected with lentiviral vector expressing NKILA or empty vector(Genechem,Shanghai,China),then transfected cells were injected subcutaneously into mice to obtain xenograft mice.30 days later,mice were sacrificed,tumors tissues and/or serum were immediately collected and stored at-80°C for the following assays.
Immunohistochemical(IHC)
Standard IHC protocol was followed to analyse E-cadherin (1:500,ab2313032,Abcam),VEGF (1:1,000,ab150375,Abcam),CD34 (1:100,ab81289,Abcam)and CD105 (1:100,ab221675,Abcam).Tumor tissues were fixed in 10% buffer formalin.After paraffin embedding,the samples were sliced into a thickness of 4 μm.In addition,the activity of endogenous peroxide was blocked by hydrogen peroxide block for 15 min.Next,the antigen was repaired using 10 mM heated citrate buffer for 10 min and primary antibodies were incubated at 4°C overnight.The following day,the corresponding second antibodies were incubated at room temperature for 1 h.Finally,the specimens were stained with diaminobenzidine (DAB) kit (Bost Biotechnology Co.,Ltd.,Wuhan,China).Images were captured under A confocal microscope (Leica,Wetzlar,Germany).
Statistical analyses
Data are presented as mean±standard deviation (SD),and at least three independent experiments were performed.GraphPad (La Jolla,CA,USA) Prism and SPSS (IBM,Chicago,USA) software were used for data analysis.Differences between two groups were determined by Student'st-test,and differences among more than two groups was used for one-way ANOVA analysis.P<0.05 indicated statistical significance.
Results
NKILA was down-regulated and miR-889-3p was up-regulated in HCC tissues and cell
The expressions of NKILA and miR-889-3p in 46 cases of HCC and paracancerous tissues were detected.The results showed that the mRNA level of NKILA in HCC tissues was significant lower than that in paracancerous liver tissues,while the expression of miR-889-3p was striking higher (P<0.01,Fig.1A-B).In addition,the expressions of NKILA and miR-889-3p in immortalized normal liver cell line L-02 and HCC cell line HepG2 were also detected.Similarly,the results were consistent with those in tissues (P<0.05,Fig.1C-D).Furthermore,the low expression of NKILA was markedly associated with tumor size,liver cirrhosis,lymph metastasis or BCLC stage,while no differences with age,gender or AFP(Table 1).

Figure 1 Related expression of NKILA and miR-889-3p in HCC.(A-B)The expression levels of NKILA and miR-889-3p in HCC and paracancerous tissues were examined by qRT-PCR;**P<0.01 vs.paracancerous tissues.(C-D)The expression levels of NKILA and miR-889-3p in HCC and normal cells were examined by qRT-PCR;*P<0.05 vs.HepG2 cells.Data are presented as mean±SD.
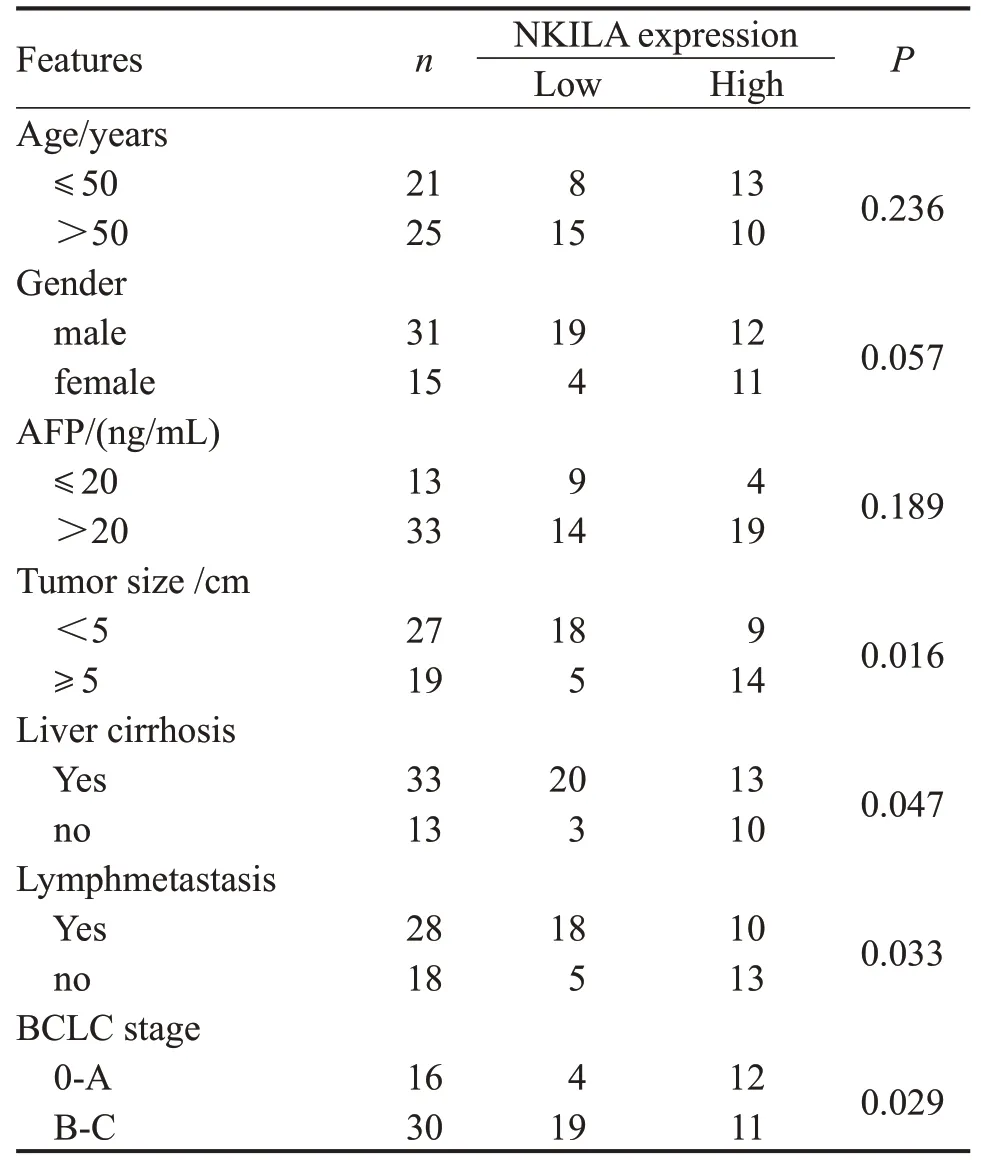
Table.1 Relationship between NKILA expression and clinicopathologic characteristics
NKILA negatively regulated miR-889-3p
Bioinformatics analysis (http://www.targetscan.org/)recommended that NKILA might be a target for miR-889-3p (Fig.2A).Hence,we transfected NKILA into HepG2 cell for 24 hours,and the data presented that the expression of NKILA was up-regulated(P<0.05,Fig.2B).Besides,the expression of miR-889-3p was significant down-regulation (P<0.05,Fig.2C).To further confirm this,and HepG2 cells were transfected with the miR-889-3p/NC mimic.We observed that the expression of miR-889-3p increased (P<0.05,Fig.2D),and the luciferase activity of wt 3’-UTRNKILA decreased by luciferase report assays,but the luciferase activity of mut 3’-UTR-NKILA was almost unchanged(P<0.05,Fig.2E).
NKILA abolished the inhibition of miR-889-3p on the proliferation of HCC cells
NKILA over-expression plasmids and miR-889-3p mimic were transfected into HepG2 cell,and the effects of NKILA on the proliferation of hepatocellular carcinoma cell was observed.As shown in Fig.3A-B,compared with the control,clone formation assay found that over-expression of NKILA markedly suppressed the proliferation abilities of HepG2 cell,and the protein level of Ki67 was significantly decreased(P<0.05).On the contrary,miR-889-3p mimic stimulated HepG2 cell viability and increased Ki67 level(P<0.05,Fig.3C).After co-transfected with NKILA and miR-889-3p mimic,NKILA abolished the inhibition of miR-889-3p mimic in cell proliferation (Fig.3A-C).
NKILA suppressed the effect of miR-889-3p on the EMT of HCC cells
We next examined the effect of NKILA on EMT in HepG2 cell,it was confirmed that miR-889-3p mimic changed cells from epithelial phenotype to mesenchymal phenotype(Fig.4A),and Fig.4B-C showed miR-889-3p mimic reduced the expression of epithelial marker (E-cadherin),while elevated the expression of other mesenchymal proteins (N-cadherin and Fibronectin),compared with the control(P<0.05).Post cotreated with NKILA and miR-889-3p or NKILA alone,NKILA reversed miR-889-3p-mediated EMT phenotype,concomitant with up-regulation of E-cadherin and the down-regulation of N-cadherin and Fibronectin in HepG2 cell(P<0.05,Fig.4A-C).
NKILA suppressed the effect of miR-889-3p on tumor angiogenesis
Parallelly,we evaluated biological function of NKILA in angiogenesis by a HUVECs model.MiR-889-3pmimic promoted tube formation,while over-expressed NKILA strongly inhibited the ability of HepG2 cell to induce tube formation of HUVECs,compared with the control (P<0.05,Fig.5A-B).In addition,the expression of VEGF was decreased after transfection of NKILA,miR-889-3pmimic eliminated the inhibitory effect of NKILA(P<0.05,Fig.5C).
Over-expression of NKILA restrained the growth of xenograft tumors in vivo
In vivoexperiments further showed that the over-expression of NKILA significantly reduced the volume and weight of HCC xenograft tumor (P<0.05,Fig.6A).In addition,IHC assay founded that the expression of E-cadherin was increased,while the expression of VEGF was weakened,compared with the control (P<0.05,Fig.6B).Also,Fig.6C revealed a decrease in the number of vascular endothelial cell markers CD34 and CD105 post-NKILA alone (P<0.05).
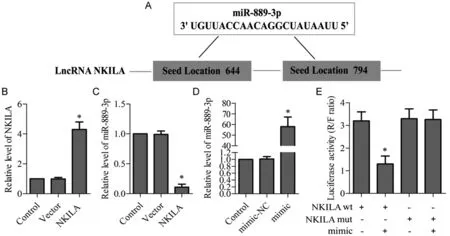
Figure 2 MiR-889-3p was negatively regulated by NKILA.(A)The target site between NKILA and miR-889-3p.(B-C)NKILA was transfected into HepG2 cell,the expression levels of NKILA and miR-889-3p was examined by qRT-PCR;*P<0.05 vs.Control.(D)MiR-889-3p NC or mimic was transfected into HepG2 cell,the expression level of miR-889-3p was examined by qRT-PCR;*P<0.05 vs. Control .(E) MiR-889-3p as a direct target of NKILA was performed by dual-luciferase reporter assay;*P<0.05 vs. NC mimic.Data are presented as mean±SD.

Figure 3 Effect of NKILA on proliferation capacity of HCC cell.(A-B)Cell proliferation was assessed by cloning formation assay,associated images were magnified to × 400.(C) The protein level of Ki67 was examined by western blotting.*P<0.05 vs. Control,#P<0.05 vs.mimic.Data are presented as mean±SD.

Figure 4 Effect of NKILA on the EMT of HCC cell.(A)Cells phenotype was observed,associated images were magnified to×200.(B)The expression of E-cadherin,N-cadherin and Fibronectin was examined by western blotting;*P<0.05 vs.Control,#P<0.05 vs.mimic.Data are presented as mean±SD.

Figure 5 Effect of NKILA on angiogenesis of HCC cell.(A-B)Tube formation was analysed,associated images were magnified to×400;*P<0.05 vs.Control,#P<0.05 vs.mimic.(C)The protein level of VEGF was examined by western blotting;*P<0.05 vs.Control,#P<0.05 vs.mimic.Data are presented as mean±SD.

Figure 6 Effect of NKILA in xenograft tumor.(A) HepG2 cells stably transfected with NKILA were injected subcutaneously into mice,tumor weight was recorded.(B) The expression of E-cadherin and VEGF were detected using IHC.(C) The expression of CD34 and CD105 was detected using IHC.Images were magnified to×100.*P<0.05 vs.Control.Data are presented as mean±SD.
Discussion
In recent years,the morbidity and mortality of HCC are still very high,and the disorder of IncRNAs has been confirmed to be closely related to the tumorigenesis,metastasis and prognosis of HCC [19].NKILA,as an NF-κB interacting LncRNA,was first found by Liu et al.[20]in breast cancer with low expression.In addition,the inhibitory effect of NKILA on tumor migration and invasion was further found in tongue squamous cell carcinoma [15],malignant melanoma [16]and non-small cell lung cancer [21].In this study,data showed that NKILA was down-regulated in HCC tissues and cells,overexpression of NKILA inhibited the cell proliferation,migration,and angiogenesis of HCC cells.In vivomodels showed that elevated NKILA inhibited tumor formation in tumor-bearing mice.Notably,we identified a novel tumor suppressor mechanism of NKILA through sponging miR-889-3p.lncRNAs has attracted more and more attention because of its important regulatory role in pathological processes such as cell proliferation,cycle,apoptosis,migration and invasion [22-24].Tumor invasion and metastasis is one of the typical features of cancer,and the intrahepatic and extrahepatic metastases are the main causes of high recurrence and poor prognosis in HCC [25].EMT is a biological process closely related to tumor progression and metastasis,which is considered the first step and major phenotype in cancer metastasis and invasion [26].Cells morphology receiving EMT initiation will change from an epithelial phenotype to a fibroblast-like mesenchymal phenotype as evidenced by the down-regulation of E-cadherin (epithelial marker) and up-regulation of vimentin,fibronectin or N-cadherin (mesenchymal markers),thereby promoting metastasis and invasion of malignant tumor cells [27-28].Previous reports have confirmed that NKILA interfered with the migration and invasion of tongue squamous cell carcinoma by EMT inhibition[29].In this study,we observed a cobblestone-like epithelial morphology and cell aggregation in miR-889-3p overexpressed HCC cells,which decreased E-cadherin expression while increased Ncadherin and Fibronectin expression.On the contrary,the fibroblast-like mesenchymal morphology and obvious cellular scattering were shown in the HCC cell with high NKILA expression.These findings suggested that low expression of NKILA in HCC was associated with EMT-mediated cancer invasion or metastasis.
In our study,we were the first to provide evidence that NKILAin vitroandin vivoattenuated tumor angiogenesis,which was necessary for aggressive tumor growth and metastasis [30].Currently,antiangiogenic therapy is used as an effective anticancer mean in cancer patients to achieve the desired effect [31].This study elucidated the role of NKILA in regulating angiogenesis of HUVECs models,as NKILA overexpression strongly inhibited HUVECs angiogenesis and VEGF expression,which was a key inducer of tumor angiogenesis [32].Our findings suggested that NKILA is a very effective target for antiangiogenic HCC treatment.Moreover,tumor angiogenesis is involved in primary tumor growth,local tissue infiltration,and distant metastasis [33].In vivoxenograft tumor model also showed that elevated NKILA inhibited tumor growth and reduced the number of vascular endothelial cell markers (CD34 and CD105).This is consistent with previous reports that NKILA is contributed to tumor development in multiple tumor types[14-17].
In short,our study demonstrated that NKILA was down-regulated and miR-889-3p up-regulated in HCC,and miR-889-3p was a direct target of NKILA.These data highlighted the tumor inhibition of NKILA in HCC by targeting miR-889-3p,accompanied by suppressing cell proliferation,EMT phenotype and tumor angiogenesis,it provided new horizon for clinical examination or treatment of HCC.
AcknowledgmentsThis work was supported by a grant from the Key projects of science and Technology Department of Sichuan Province(No.2019SZ0069)
