Two novel gene-specific markers at the Pik locus facilitate the application of rice blast resistant alleles in breeding
2021-05-23TlANDagangCHENZiqiangLlNYanCHENZaijieLUOJiamiJlPingshengYANGLimingWANGZonghuaWANGFeng
TlAN Da-gang ,CHEN Zi-qiangLlN YanCHEN Zai-jieLUO Jia-miJl Ping-sheng,YANG Li-ming,WANG Zong-hua,WANG Feng
1 Fujian Key Laboratory of Genetic Engineering for Agriculture/Biotechnology Research Institute,Fujian Academy of Agricultural Sciences,Fuzhou 350003,P.R.China
2 State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops,College of Life Science,Fujian Agriculture and Forestry University,Fuzhou 350002,P.R.China
3 College of Biology and the Environment,Nanjing Forestry University,Nanjing 210037,P.R.China
4 Department of Plant Pathology,University of Georgia,Tifton,GA 31793,USA
Abstract Blast,a disease caused by Magnaporthe oryzae,is a major constraint for rice production worldwide. Introgression of durable blast resistance genes into high-yielding rice cultivars has been considered a priority to control the disease. The blast resistance Pik locus,located on chromosome 11,contains at least six important resistance genes,but these genes have not been widely employed in resistance breeding since existing markers hardly satisfy current breeding needs due to their limited scope of application. In this study,two PCR-based markers,Pikp-Del and Pi1-In,were developed to target the specific InDel (insertion/deletion) of the Pik-p and Pi-1 genes,respectively. The two markers precisely distinguished Pik-p,Pi-1,and the K-type alleles at the Pik locus,which is a necessary element for functional genes from rice varieties.Results also revealed that only several old varieties contain the two genes,of which nearly half carry the K-type alleles.Therefore,these identified varieties can serve as new gene sources for developing blast resistant rice. The two newly developed markers will be highly useful for the use of Pik-p,Pi-1 and other resistance genes at the Pik locus in markerassisted selection (MAS) breeding programs.
Keywords:rice,blast disease,molecular marker,Pik-p,Pi-1,K-type alleles
1.lntroduction
Rice (Oryza sativaL.) is one of the most important cereal crops in the world and provides a staple food source for over half of the world’s population (Khush 2005). One major obstacle encountered in rice production is the frequent and widespread occurrence and damage of rice blast,caused by the ascomycete fungusMagnaporthe oryzae(Jeunget al.2007). Cloning and utilizing resistance genes in rice breeding is an important and effective approach to enhance cultivar resistance in an economical and safe manner (Zeigleret al.1994). To date,more than 100 resistance (R) genes have been found,and at least 28 of these genes have been successfully cloned (Maet al.2015;Denget al.2017;Liet al.2019). Three of them are important clusters of blastR-genes that have been reported to confer durable and broadspectrum resistance (Chenet al.2016;Denget al.2017;Wang B Het al.2017). One such cluster is thePiklocus on chromosome 11,which has been commonly considered to be the major locus in control of rice blast disease since it contains at least six identified blast R genes,includingPi-1(Huaet al.2012),Pik-h(Zhaiet al.2014),Pi-k(Zhaiet al.2011),Pik-m(Ashikawaet al.2008),Pik-s(Wanget al.2009),andPik-p(Yuanet al.2011). Previous studies have shown that many R genes from thePiklocus confer broad-spectrum resistance to a diverse array ofMagnaporthe oryzaeisolates (Huaet al.2012). Due to this fact,thePiklocus has the potential to be used in a marker-assisted selection (MAS) breeding program for the improvement of blast resistance in rice.
Gene-specific markers developed from DNA polymorphisms among the alleles are particularly useful for MAS breeding and screening large amounts of germplasm for allelic diversity(Brar and Khush 1997;Xuet al.2005;Shomuraet al.2008;Kumariet al.2013;Nawazet al.2017). Such markers have been developed for genes includingPit,Pi-2,Pi-9,andPig-m(Hayashiet al.2010;Tianet al.2016;Wanget al.2019). In the case of thePiklocus,which displays high similarity amongPi-1,Pik-h,Pi-k,Pik-m,Pik-s,Pik-p,and wild rice clusters from the same region (Zhaiet al.2011;Huaet al.2012),it is difficult to differentiate these genes from each other by simple sequence repeat (SSR) markers. Though several derived cleaved amplified polymorphic sequences (dCAPS) have been developed that allow for screening ofPi-1,Pik-p,Pik-m,andPi-kgenes in cultivars (Zhaiet al.2011;Huaet al.2012;Chenet al.2016),it is inconvenient to apply dCAPS markers in large-scale breeding practices since they need to be digested with PCR products and restriction endonucleases,which is laborious and time-consuming.
The objectives of this study were:1) to analyze polymorphism in thePiklocus and developPi-1,Pik-p,and K-type allele marker systems;2) to use the newly developed marker systems to identify the presence of two blastR-genes in a large collection of 531 rice varieties;and 3) to verify the resistance of representative rice varieties by pathogenicity tests. Development of the gene-specific markers has great potential applicability in MAS breeding programs.
2.Materials and methods
2.1.Plant materials and pathogen isolates
The 531 rice varieties were maintained at Fujian Provincial Key Laboratory of Genetic Engineering for Agriculture,Fujian Academy of Agricultural Sciences,Fuzhou,China.Six monogenic lines,IRBLkh-K3-Pik-h (Pik-h),IRBLkm-Ts-Pik-m (Pik-m),IRBLkp-K60-Pik-p (Pik-p),IRBLk-Ka-Pi-k (Pi-k),IRBLks-F5-Pik-s (Pik-s) and IRBL1-CL-Pi-1(Pi-1),were developed by the International Rice Research Institute (IRRI) (Tsunematsuet al.2000). SevenM.oryzaeisolates,KJ201,CHE86061,CHNOS60-2-3,FJ86,PO6-6,CHL2110 and CHL768,were kindly provided by Dr.Pan Qinghua (Laboratory of Plant Resistance and Genetics,South China Agricultural University,China) and Dr.Guoliang Wang (Department of Plant Pathology,Ohio State University,USA).Magnaporthe oryzaeisolates were cultured on oatmeal agar plates in the dark for 7 days at 28°C and then grown under light and dark cycles of 12 h/12 h for 5 days.
2.2.Rice blast inoculation assay
Rice blast inoculation assays were conducted in a greenhouse with an automated fogging system to maintain a suitable temperature of around 28°C and relative humidity of >95%.About 2 weeks after sowing,20 rice seedlings of each of 25 rice varieties were spray-inoculated with spore suspensions of the seven isolates separately at a concentration of 5×105spores mL–1until run-off. After inoculation,the cement tank was completely covered with a tent,and the inoculated seedlings were maintained under dark conditions for 24 h.The tent was then removed from the tank,and the seedlings were grown in the greenhouse under high humidity (>95%)for an additional week for symptom evaluation. The 0–5 standard scale was used to score disease severity following Campbeet al.(2004). Plants with lesion scores 0,1 and 2 were considered to be resistant,while 3,4 and 5 were considered as moderately resistant,susceptible and highly susceptible,respectively.
2.3.DNA extraction
Leaf tissues of rice plants were collected for DNA extraction.About 50 mg of leaf samples collected in Eppendorf tubes were frozen in liquid nitrogen and were processed into a fine powder with a TissueLyser (Qiagen,Haan,Germany).Rice genomic DNA was extracted from ground leaf tissue by the cetyltrimethyl ammonium bromide (CTAB) method as described by Murray and Thompson (1980).
2.4.Development of allele-specific markers for Pi-1 and Pik-p
Based on the multiple sequence alignment ofPi-1(HQ606329),Pik-h(HQ662330),Pi-k(HM048900),Pik-m(GU811852),Pik-s(HQ662329),andPik-p(HM035360),Pikp-Del and Pi1-In markers were designed specifically targeting the InDel polymorphisms ofPik-pandPi-1genes amongPi-1,Pik-h,Pi-k,Pik-m,Pik-s,andPik-p. PCR was performed in a 25 μL reaction that contained 12.5 μL of 2×mix buffer (Mg2+Plus),1 μL of each primer (10 mmol L–1),and 0.2 μL Taqase (5 U mL–1). For each reaction,20–200 ng of genomic DNA was used. PCR was performed with the following thermal profile:initial denaturation at 94°C for 5 min,followed by 34 cycles of denaturation at 94°C for 30 s,55°C for 30 s,and extension at 72°C for 30 s,and a final extension at 72°C for 10 min. The PCR products were separated in 3% agarose gels and then visualized by ethidium bromide staining.
2.5.Genotyping diverse rice varieties with allelespecific markers
Genomic DNA of the 531 varieties was subjected to genotyping the presence ofPi-1,Pik-p,and K-type alleles using the Pi1-In and Pikp-Del markers. K-type allele origins from the BAC clone Ts18H12 ofcv.Tsuyuake,which is the necessary element for functional genes at the Pik locus,and the corresponding locus was referred to as N-type,being from the reference sequence ofcv.Nipponbare. In addition,two previously reported dCAPS markers,Pi1FNP and dCAPS-795 (Zhaiet al.2011;Huaet al.2012),which targeted the functional regions ofPi-1andPik-p,respectively,were used for verification. For genotyping using the Pi1FNP marker,PCR amplicons were digested withRsaI. For genotyping using the dCAPS-1879 marker,PCR amplicons were digested withNdeI. All the above products were separated electrophoretically in 3% agarose gels and then visualized by ethidium bromide staining.
3.Results
3.1.Functional markers for Pi-1/Pik-p/K-type genotypes
To determine the genotypes ofPi-1andPik-p,an extensive sequence alignment was conducted to analyze the InDel polymorphisms among the sequences ofPi-1,Pi-k,Pik-m,Pik-p,Pik-s,andPik-m. A conserved 19-bp deletion at the–1 015-bp location ofPikp-1,which is located at the promoter region ofPikp-1,and a 11-bp insertion positioned at +6 816-bp ofPi1-5-terminal were identified (Fig.1-A). To further verify the two identified InDel polymorphisms,two pairs of specific primers,Pikp-Del and Pi1-In,were designed to develop a molecular marker that targeted the regions containing the identified InDel (Table 1). PCR fragments corresponding to the two regions ofPi-1,Pik-p,Pi-k,Pik-m,Pik-h,Pik-s,and non-functional N-type alleles were amplified from rice monogenic lines IRBL1-CL-Pi1,IRBLkp-K60-Pik-p,IRBLk-Ka-Pik,IRBLkm-Ts-Pik-m,IRBLkh-K3-Pik-h,IRBLks-S-Pik-s,and Nipponbare with specific primers for the Pikp-Del and Pi1-In markers,respectively. Electrophoretic analysis showed that the band-types ofPik-pandPi-1were different from the others,and the N-type from Nipponbare had invisible PCR product (Fig.1-D). Therefore,the two developed InDel markers could clearly distinguish the genotypes corresponding to thePi-1andPik-ploci from other functional and non-functional alleles.
3.2.Assessment of the Pi-1,Pik-p,and K-type alleles in 531 rice varieties
A total of 531 rice varieties were assessed for the presence of thePik-p,Pi-1,and K-type alleles using the newly developed markers. The 531 rice varieties consisted of 98indicarestorer lines of hybrid rice,49 maintainer/sterile lines of hybrid rice,328indicaconventional cultivars/breeding materials,and 56japonicaconventional cultivars/breeding materials (Appendix A). These varieties included 478 collected from the major rice-producing areas in China,among which 84,34,18,12,33,16,35,25,and 18 were from Fujian,Guangdong,Guangxi,Hubei,Hunan,Jiangsu,Sichuan,Zhejiang,and Yunnan;123 were from IRRI,80 and 53 were from other countries or unkonown areas,respectively. When using the Pikp-Del marker for screening,five out of 531 and 264 out of 531 varieties were separately identified displaying 368-and 377-bp PCR fragments,respectively (Appendix A),suggesting that the five varieties might contain thePik-presistance genes and the 264 varieties had the functional K-type alleles.When using the Pi1-In marker for screening,eight and 256 varieties produced specific patterns identical to those of thePi-1and K-type alleles,respectively. Taken together,these results suggest that the 256 varieties may contain the other resistance genes,except for the eight that containPi-1and the five that havePik-p.
The rice varieties containingPik-p,Pi-1and K-type identified by the Pi1-In and Pikp-Del markers were also confirmed with K-and N-type specific primer sets,two dCAPS markers of dCAPS-Pi1 and dCAPS-795.Genotyping results of those markers were consistent with the results using the Pikp-Del and Pi1-In markers. The five varieties were identified as having aPik-pgenotype corresponding to thePik-pallele (Fig.2-A;Appendix A),and Pi1FNP analysis in the eight varieties gave a restriction enzyme digestion pattern identical to thePi-1allele (Fig.2-C;Appendix A).
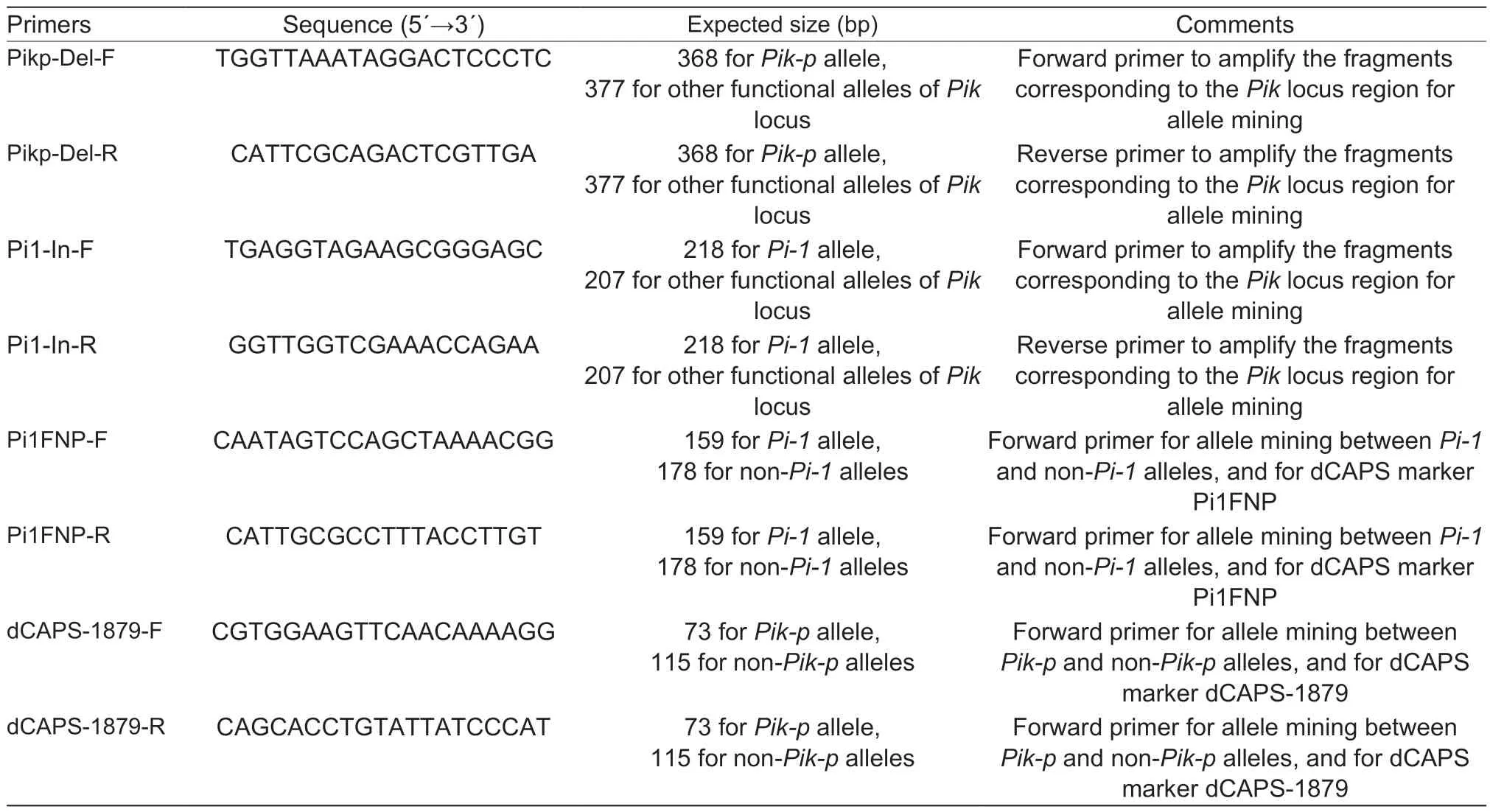
Table 1 List of primers in the present study
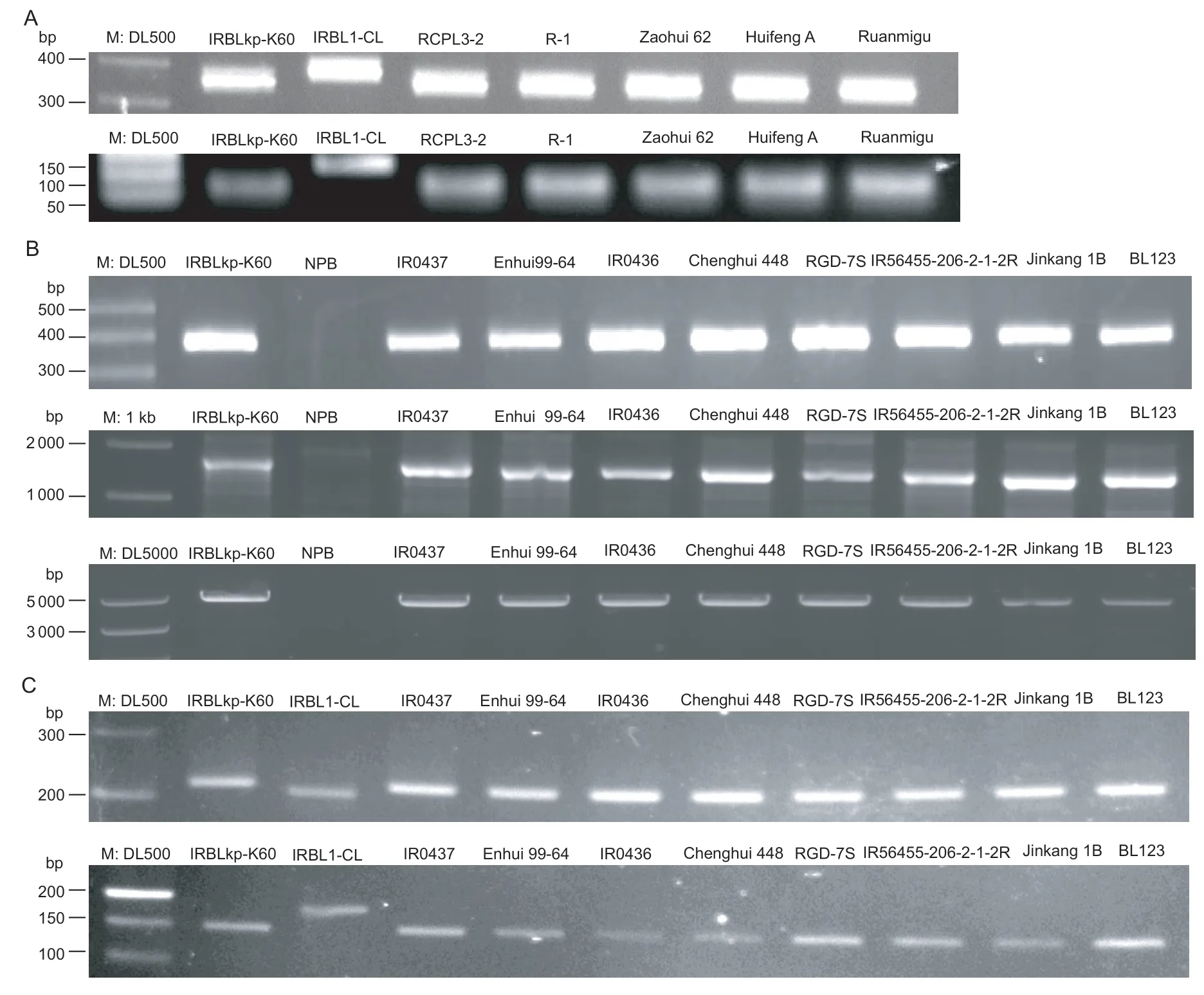
Fig.2 Electrophoresis profiles of the Pikp-Del and Pi1-In marker that differentiates the Pik-p and Pi-1 alleles. A,Pikp-Del and dCAPS-1879 markers that differentiate the Pik-p and non-Pik-p alleles. Upper panel shows the PCR products with Pikp-Del and lower panel shows the dCAPS-1879 PCR products digested with NdeI. B,Pikp-Del,Ku1-GT,and Ku2-GT markers that differentiate the K genotype and non-K genotype alleles. Upper panels show the PCR products with Pikp-Del,middle panels show PCR products with Kul-GT,and lower panels show PCR products with Ku2-GT. C,Pi1-In and Pi1FNP markers that differentiate the Pi-1 and non-Pi-1 alleles. Upper panels show the PCR products with Pi1-In and lower panels show the PCR products of Pi1FNP digested with RsaI.
3.3.Disease reactions of varieties possessing Pi-1 and Pik-p genotypes
To investigate whether thePik-p,Pi-1,and K-type varieties showed corresponding specific resistance,34 representative rice varieties withPi-1,Pik-p,K-type,and N-type alleles were selected and inoculated with seven blast pathogen isolates:KJ201,CHNOS60-2-3,CHE86061,FJ86,FJ501-3,CHL768(AvrPi-1),and CHL2110 (AvrPik-p). The last two isolates were for resistance identification of the corresponding R genes,and the other five isolates were used for resistance comparison of the 34 representative varieties.
As shown in Table 2 and Fig.3,the fourPik-pgenotype varieties showed high resistance to CHL2110,FJ86,FJ501-3,CHL768,and CHNOS60-2-3,but moderate resistance to CHE86061,which was similar to or even higher than the disease reactions in thePik-p-carrying line IRBLkp-K60(Table 2). The eightPi-1-type varieties also appeared equivalent to or had stronger resistance than thePi-1-carrying near-isogenic line IRBL1-CL. The nine K-type and six N-type varieties showed differences in the resistance spectrum compared to thePik-p,Pik-m,Pik-h,orPi-1donor lines,suggesting that these varieties likely possess other blast R genes.
4.Discussion
4.1.Pik locus contains multiple broad-spectrum R-genes to rice blast
ThePiklocus has been considered to confer broad spectrum blast resistance. It contains at least six resistance genes(Kiyosawa 1987;Ashikawaet al.2008;Wanget al.2009,2010;Zhaiet al.2011;Huaet al.2012).Pik-pcontributes to higher resistance responses in many regions in China including Guangdong,Jiangsu,and Sichuan provinces,indicating that this gene has greater applicability in these regions (Wanget al.2009;Wang W Jet al.2017).Pi-1was regarded as having great potential for achieving broad-spectrum and durable resistance against rice blast(Liet al.2005;Yanget al.2008;Huet al.2010;Zhanget al.2010;Jianget al.2012;Wang W Jet al.2017). The other resistance genes at the locus also have different resistance spectra to the blast fungal pathogen (Huaet al.2012).
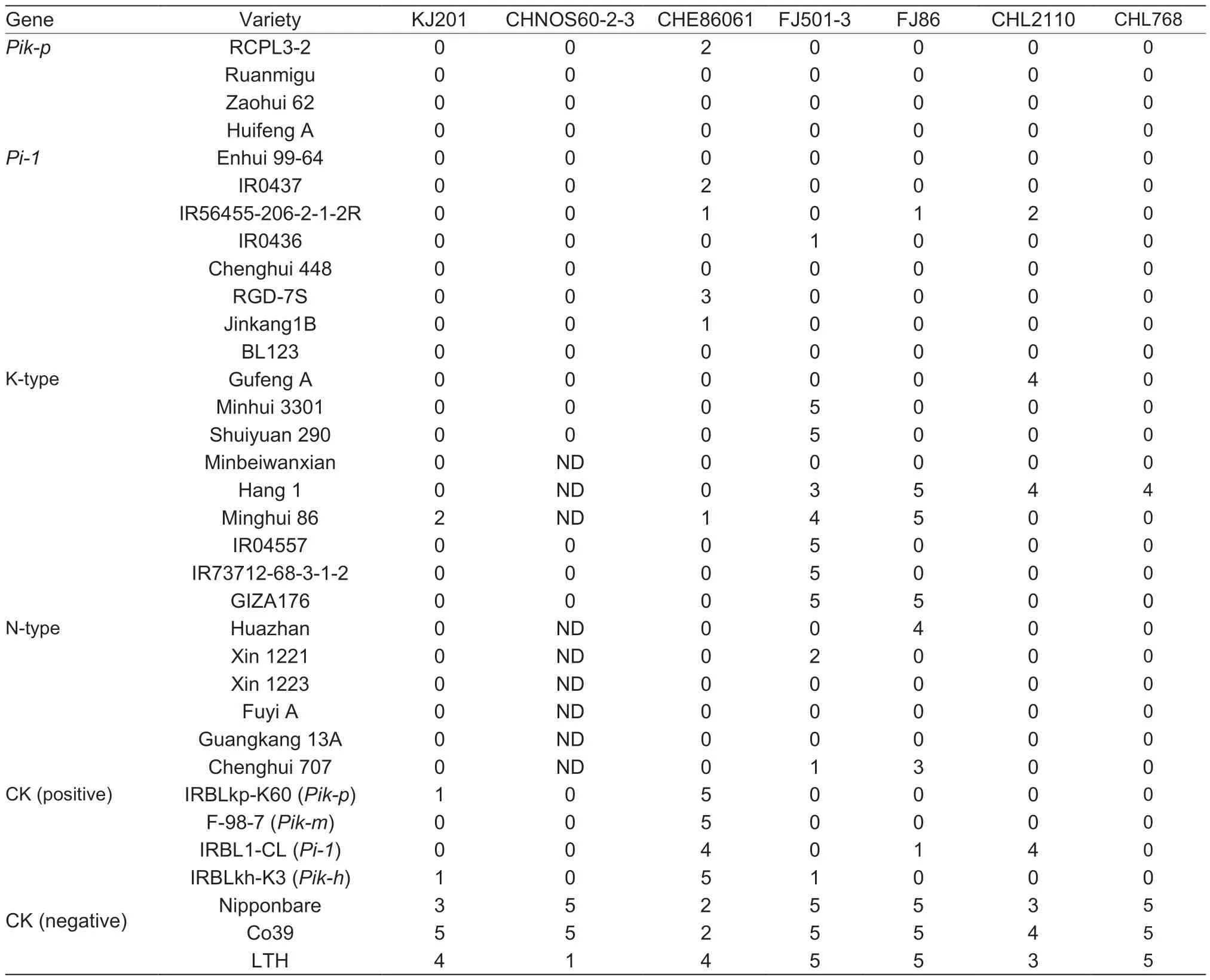
Table 2 Disease reactions of representative Pi-1,Pik-p,K-type-containing varieties using diverse Magnaporthe grisea strains
4.2.Molecular markers improve the availability of R genes
The utilization of molecular markers has made it feasible to test for the presence of different R genes,which was previously unattainable whenever several R genes conferred resistance to overlapping spectra of race-resistance in an inoculation test. With the availability of sequence data for functional R genes,gene-specific DNA markers have been developed for several blast R genes,such asPit(Hayashiet al.2010),Pi54(Ramkumaret al.2011),Pi-2/Piz-t/Pi-9/Pigm(Huaet al.2015;Tianet al.2016,2020),andPita(Heet al.2014) by using bioinformatics strategies. Not only can these gene-specific markers be used for screening for novel alleles in germplasm sets,but they can also be an effective selection tool for multiple allelic pyramid breeding.
4.3.Development and utilization of the Pikp-Del and Pi1-ln markers

Fig.3 Co-segregation analysis on Pikp-Del and Pi1-In with blast resistance using rice varieties containing Pik-p and Pi-1. A total of eight varieties containing Pi-1,four containing Pik-p and three isogenic lines as check varieties were used for inoculation identification by Magnaporthe oryzae isolates CHL768 (contain AvrPi-1) and CHL2110 (contain AvrPik-p),respectively.
Since thePiklocus containsPik-p,Pik-m,Pik-h,Pi-1,Pi-k,andPik-salleles,which confers broad-spectrum and durable resistance to rice blast disease (Huaet al.2012),it is of great significance to accelerate its application in breeding practices. The current study developed Pikp-Del and Pi1-In markers based on the gene-specific states of thePik-pandPi-1genes,respectively. The Pikp-Del marker lies in betweenPik-p1(K1) andPik-p2(K2),only 830-and 2 576-bp away from the K1 and K2 markers,respectively. The Pi1-In marker is located at +6 816 bp away from the start position of Pi1-5 (Fig.1). These two markers were further utilized to assess the genotypes of 531 rice varieties. All in all,the present study provides a unique opportunity to swiftly screen large scale germplasm resources for the target gene with high accuracy,which is very different from the former dCAPS marker method which required digesting PCR products with restriction endonuclease. Therefore,the newly developed markers have the ability to fix alleles in a population with higher efficiency,which will be particularly useful in MAS breeding methods.
Additionally,we found thatPi-1orPik-ponly exists in several old local or bred varieties,such as RCPL3-2,Ruanmigu and Zaohui 62,and few newly-bred varieties contain them. Thus,we can infer that these two genes have not been widely applied in rice breeding. In addition,the six representative K-type allelic varieties also showed different resistance responses from the four isogenic lines,which implies that there may be some newRalleles in these particular varieties.
Accurate identification of R genes in diverse elite germplasm panels through the use of suitable genetic markers and diverse blast races is the key to correctly utilizing the R gene in MAS for breeding programs(RoyChowdhuryet al.2012). Thus,these results will be beneficial to rice breeders who seek appropriate and even new R genes for the improvement of blast resistance from a wider range of germplasm,and contribute towards the development of novel molecular breeding practices.
5.Conclusion
In this study,we developed two gene-specific markers forPik-pandPi1,and identified the presence of the two blast R genes in 531 rice varieties based on the two new developed markers. The results revealed thatPi-1orPik-ponly exists in several older local or bred varieties,and nearly half of the identified varieties carry the K-type alleles,supporting the idea that the two blast R genes and the K-type alleles can be important gene sources for breeding of blast resistant in the future. Furthermore,the Pikp-Del and Pi1-In markers will be highly useful in MAS breeding for blast resistance.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (31640006),the China Postdocotral Science Foundation (2019M662219),the Youngth Program of Fujian Academy of Agricultural Sciences (YC2019004),and the Projection of Public Welfare of Fujian Province(2017R1019-10). We thank Dr.Pan Qinghua (Laboratory of Plant Resistance and Genetics,South China Agricultural University,China) and Dr.Guo-liang Wang (Department of Plant Pathology,Ohio State University,USA) forMagnaporthe oryzaeisolate assistance.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendixassociated with this paper is available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Low glycemic index:The next target for rice production in China?
- Effect of side deep placement of nitrogen on yield and nitrogen use efficiency of single season late japonica rice
- Advancements in plant regeneration and genetic transformation of grapevine (Vitis spp.)
- Indica rice restorer lines with large sink potential exhibit improved nutrient transportation to the panicle,which enhances both yield and nitrogen-use efficiency
- Effects of nitrogen management on the ratoon crop yield and head rice yield in South USA
- Response of grain-filling rate and grain quality of mid-season indica rice to nitrogen application
