Functional analysis of the nitrogen metabolism-related gene CsGS1 in cucumber
2021-05-23XlNMingQlNZhiweiYANGJingZHOUXiuyanWANGLei
XlN Ming,QlN Zhi-wei,YANG Jing,ZHOU Xiu-yan,WANG Lei
1 College of Horticulture and Landscape Architecture,Northeast Agricultural University,Harbin 150030,P.R.China
2 College of Resources and Environment,Northeast Agricultural University,Harbin 150030,P.R.China
Abstract Glutamine synthetase (GS) plays an important role in nitrogen (N) metabolism in cucumber. In this study,we cloned and sequenced the CsGS1 gene,and analyzed the expression patterns and subcellular localization of the GS1 protein in response to different N conditions in order to determine its role in low-nitrogen (LN) tolerance. CsGS1 was abundantly expressed in the leaves of the low N-requiring cultivar D0328,while the high N-requiring cultivar D0422 showed similar expression levels across different tissues including leaves,shoots and roots. Furthermore,the GS1 protein was primarily localized in the cytoplasm of plant cells. Both cultivars were then transformed with the CsGS1 coding sequence or antisense sequence via Agrobacterium tumefaciens in order to overexpress and silence GS1 expression,respectively. Overexpression of CsGS1 significantly improved LN tolerance and photosynthetic parameters,and increased chlorophyll b content,biomass,plant height,root length,N accumulation and GS activity under LN condition compared to the control. CsGS1 silencing on the other hand significantly reduced the above indices. Taken together,CsGS1 is crucial for maintaining N metabolism in cucumber plants during N deprivation,and is a promising target for generating novel transgenic breeds with increasing nitrogen utilization efficiency.
Keywords:cucumber,CsGS1,low nitrogen,functional verification
1.lntroduction
Nitrogen (N) is an essential plant macronutrient,and N fertilizers are routinely used to increase crop yield (Okaet al.2012). However,the excessive use of N fertilizers in the last decades has accumulated N content in soils to toxic levels(Tianet al.2020). Given the agricultural non-point source pollution and food safety issues caused by the widespread use of fertilizers and pesticides,there is an international consensus on controlling the application of agrochemicals(Yang and Lin 2019).
Due to their shallow roots,cucumber (Cucumis sativusL.) plants require significantly higher fertilizer content than other vegetables,especially that of N,to maintain normal growth. There is a considerable interest in developing novel cucumber breeds with increasing tolerance to N deprivation in order to reduce dependency on nitrogenous fertilizers and increase N use efficiency (NUE),while ensuring high yield and quality. This in turn requires a greater understanding of the genetic and molecular basis of the N metabolic pathway in plants (Foyer and Ferrario1994).
Glutamine synthetase (GS,EC 6.3.1.2) is a key enzyme in N metabolism that catalyzes condensation of ammonia and glutamate in an ATP-dependent pathway to produce glutamine in non-leguminous plants. Both glutamine and glutamate are the primary sources of organic N for the synthesis of proteins,nucleic acids and chlorophyll (Liet al.2016). Since mutations in theGSgene limit N incorporation in proteins,it is safe to surmise that GS is crucial for N assimilation (Avila-Ospinaet al.2015). Two GS isozymes have been identified in plants - the cytosolic GS (GS1) and chloroplastic GS (GS2). The gene and amino acid sequences of these isoenzymes are conserved across different species,and comprise a gene family (Bernardet al.2008). WhileGS2primarily metabolizes the ammonium resulting from nitrate reduction in leaves,GS1mediates glutamine synthesis,protein accumulation (Zhanget al.2017),N remobilization in senescing leaves,resistance to drought (Liet al.2016) and other abiotic stresses (Chuet al.2017),and N assimilation and recycling (Guoet al.2013). Not surprisingly,GS activity is closely related to various agronomic traits and crop yield.For instance,both GS1 and GS2 play an important role in grain protein accumulation (Zhanget al.2017),and their overexpression can improve N utilization (Tianet al.2015).GS1overexpression increased seed number in maize by 30%(Martinet al.2006) and improved the ammonia uptaken by soybean roots (Hirelet al.1997). Thus,GS plays a crucial role in N uptake and assimilation,and regulates plant growth and development.
The soil N content significantly affects the height,fresh weight,root length and chlorophyll content of cucumber plants (Xuet al.2006). The cucumber cultivars D0328 and D0422 are respectively most resistant and sensitive to N deprivation (Yuet al.2011) based on the chlorophyllbcontent. Although the total N content in these cultivars have been examined in terms of their ability to assimilate N (Jianget al.2012),little is known regarding the molecular mechanisms. Fenget al.(2012)first cloned the cucumber chloroplasticGS1gene (CsGS1) and compared it with that of other plant species,but its exact role in cucumber growth remains unknown. In this study,we analyzed the expression patterns ofCsGS1in the D0328 and D0422 cultivars in response to different N concentrations. Furthermore,we overexpressed and knocked downCsGS1to determine its role in N metabolism,as well as its correlation to the physiological and biochemical indices of N tolerance.
2.Materials and methods
2.1.Plant materials
The D0328 and D0422 cucumber cultivars with respectively high and low tolerance to N deprivation (Yuet al.2011) were used for the study. Plants of both cultivars were grown in normal (NN,14 mmol L–1) and low (LN,3 mmol L–1) N nutrient solutions at 70% humidity over a 16/8 h light/dark period. The composition of the nutrient solutions is listed in Appendix A. The solutions were prepared using deionized water and replaced twice weekly.
2.2.CsGS1 expression pattern analysis
Total RNA was extracted from roots,stems and leaves using TRIzol (Invitrogen,Carlsbad,CA,USA) and reverse transcribed using the QuantiNova Reverse Transcription Kit (Qiagen,Hilden,Germany). The cDNA samples were quantified using a SMA3000 UV spectrophotometer(Beijing,China) and subjected to qRT-PCR on the iQ5(Bio-Rad) thermocycler using SYBR®Green Master Mix(ToYoBo,Japan). The primers were designed online based on theCsGS1gene (https://www.genscript.com),and the sequences are listed in Appendix B.CsEF1α(XM_004138916) was used as the housekeeping gene.Three biological replicates (five plants with similar growth comprised one replicate) and four technical replicates were tested for each treatment. Relative expression ofCsGS1was calculated using themethod,and DPS 7.051 software (Liuet al.2018) was used for data analysis.
2.3.CsGS1 cloning and transformation
The full-length CDS of sense (S) or antisense (AS)CsGS1was cloned into the PCXSN-1250 plasmid (Chenet al.2009). The resulting PCXSN-CsGS1-S and PCXSNCsGS1-AS constructs were transformed intoAgrobacterium tumefaciensLBA4404 (BioVector NTCC Inc.,Beijing,China)using the freeze-thaw method (Jyothishwaranet al.2007).The D0328 and D0422 plants were respectively infected with the transformed bacteria as previously described (Zhanget al.2014). Differentiated buds appeared 28 days after transformation,and were pruned and transferred into rooting medium. The ensuing seedlings with the main and fibrous roots were transplanted into the NN solution.
2.4.PCR and qRT-PCR
DNA was isolated from the leaves of young transgenic plants,and the PCXSN-CsGS1sequence was amplified using specific primers (Appendix C). The PCXSN-CsGS1plasmid and wild-type cucumber DNA were the positive and negative controls,respectively. The PCR reaction mix consisted of 2 μL 10× PCR buffer,2 μL dNTPs,0.5 μL PCXSCN1250-F,0.5 μL PCXSCN1250-R,0.2 μLTaqand ddH2O to 20 μL. The cycling conditions were as follows:94°C for 3 min,followed by 30 cycles of 94°C for 1 min,55°C for 1 min,and 72°C for 1 min and extension at 72°C for 10 min. The PCR products were detected by 1.5% agarose gel electrophoresis. RNA was isolated from the leaves of wild-type and transgenic plants,andGS1expression levels were determined by qRT-PCR as previously described. Four technical replicates were tested for each sample.
2.5.Subcellular localization of the CsGS1 protein
The GS1 open reading frame (ORF) was amplified and cloned into the transient expression vector pGIIeGFP (GFP) between theXmaI andBamHI sites. The resulting 35S-CsGS1-eGFP plasmid was transformed intoArabidopsis thaliana(Columbia ecotype) protoplasts using PEG. After 16–18 h incubation under light,the subcellular localization of GS1 was observed by confocal laser scanning microscopy (Leica,Germany).
2.6.Physiological analysis of transgenic plants
The growth and development indices of T2 generation transgenics were evaluated. The photosynthetic parameters were measured using Li-6400 photosynthetic apparatus(Li-Cor Inc.,Lincoln,NE,USA). The chlorophyllbcontent was determined by spectrophotometry (Lichtenthaleret al.1996),and the total N using the Kjeldahl method (Kjeldahlet al.1883). The GS activity in leaves was determined as previously described (Fuenteset al.2001). Three biological replicates (four transgenic plants with similar growth) and four technical replicates were tested for each index.
3.Results
3.1.Upregulated CsGS1 under LN condition
TheCsGS1expression pattern was analyzed after growing cucumber seedlings for 24 d under normal and low N conditions. As shown in Fig.1,CsGS1transcript levels were 1.92 and 2.07-fold higher in the leaves of the D0328 cultivar compared to its roots and stems respectively under LN condition,but gradually levelled in all tissues (Fig.1-A).In addition,the relative expression ofCsGS1in leaves was significantly higher in the D0328 cultivar than in D0422 under LN condition,while the expression in other tissues of both cultivars showed similar levels. Unlike D0328,CsGS1expression did not undergo a time-dependent change in D0422 (Fig.1-B). Finally,no significant differences were observed between D0328 and D0422 under NN condition,indicating thatCsGS1is not responsive when N availability is sufficient.
3.2.Subcellular localization of CsGS1
To track the intracellular localization ofCsGS1 protein,we transformedArabidopsisprotoplasts with theCsGS1-GFP and hGFP vectors,and detected fluorescence signals.As shown in Fig.2,the cytoplasm of theCsGS1-GFPtransformed protoplast cells showed an intense green fluorescence,indicating thatCsGS1is a cytoplasmic protein.
3.3.Generation and identification of transgenic plants
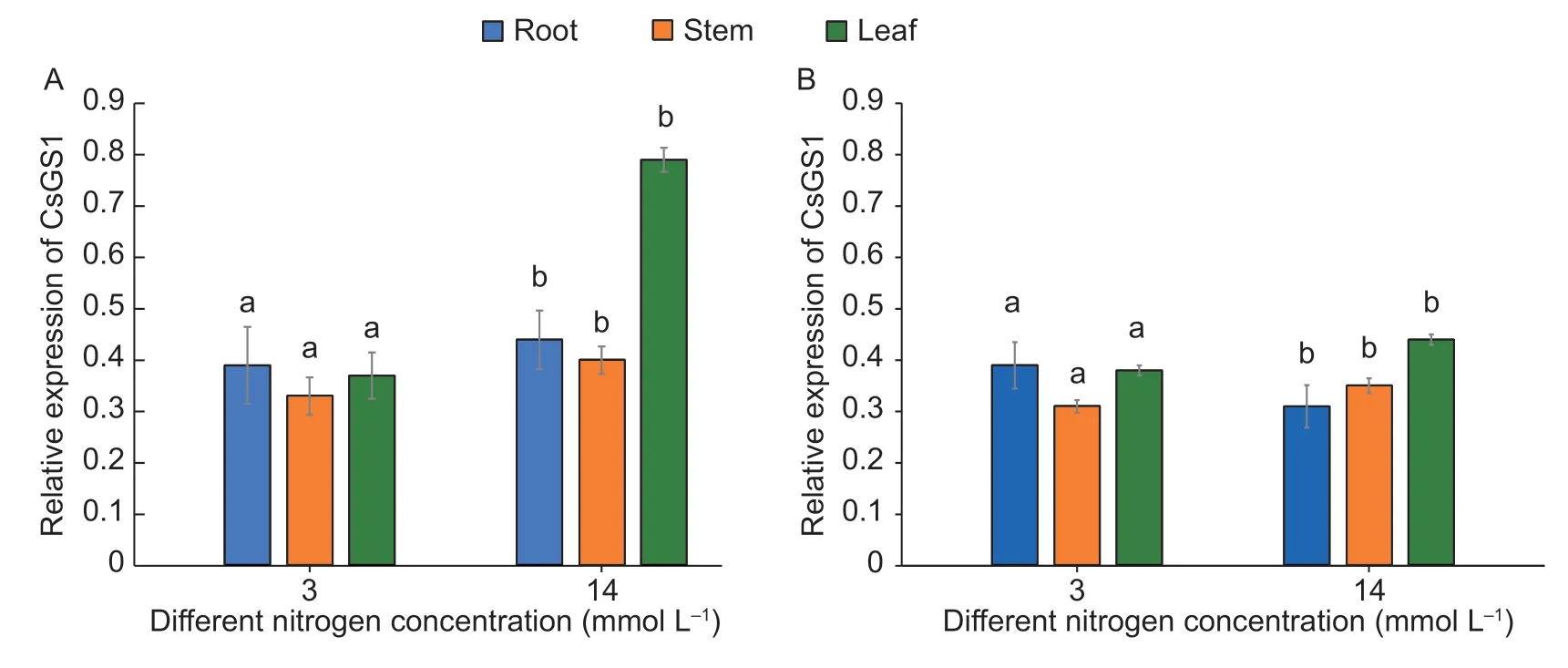
Fig.1 Expression of CsGS1 in different tissues of cucumber cultivars D0328 (A) and D0422 (B) under different N conditions. NN and LN,normal and low N nutrient solutions,respectively. Three biological replicates and four technical replicates were performed.Error bars indicate standard errors. Different lowercase letters indicate significant differences (P<0.05).
The young leaves of the transgenic D0328 and D0422 plants were tested forCsGS1on the basis of a clear band at 1 242 bp corresponding to the PCXSN-1250 sequence (171 bp)andCsGS1CDS (1 071 bp) (Fig.3). Furthermore,relative expression ofCsGS1were 3.1 and 3.7-fold higher in the D0328-S and D0422-S transgenics compared to the control plants (CK). In contrast,the D0328-AS and D0422-AS plants showed 9.5-and 11.6-fold lessCsGS1relative to control(Fig.4). A total of 425 cucumber plants were assessed,of which 63 wereCsGS1-overexpressing and 35 wereCsGS1-silenced,with an overall genetic transformation rate of 23.1%.
3.4.CsGS1 promoted photosynthesis under LN conditions
The effects ofCsGS1on photosynthesis were evaluated in terms of photosynthetic rate,stomatal conductance,intercellular CO2concentration and transpiration rate(Table 1). Under LN condition,the photosynthetic rates ofCsGS1-overexpressing D0328-S plants were 2.5 and 1.63-fold higher than those of theCsGS1-silenced (D0328-AS)and control (D0328-CK) plants,respectively. Likewise,D0422-S showed 2.55 and 1.72-fold higher photosynthesis compared to D0422-AS and D0422-CK,respectively. The overexpression ofCsGS1during N deficiency increased stomatal conductance and transpiration rate from 0.16 to 0.32 mol H2O m–2s–1and 1.13 to 3.50 mmol H2O m–2s–1,respectively,in the D0422 cultivar,while decreasing the intercellular CO2concentration from 315.60 to 223.05 mmol mol–1. As compared to the D0328-CK plants,D0328-S increased the stomatal conductance and transpiration rate to 0.21 mol H2O m–2s–1and 2.91 mmol H2O m–2s–1respectively from 0.11 mol H2O m–2s–1and 1.22 mmol H2O m–2s–1,while decreased the intercellular CO2concentration from 347.67 to 242.12 mmol mol–1. In contrast,the stomatal conductance and transpiration rate in theCsGS1-silenced D0328 plants were reduced by 36.36 and 13.11%,respectively,and the intercellular CO2concentration was increased by 2.91%under LN condition. The stomatal conductance of theCsGS1-silenced D0422 plants was reduced by 12.5%,while the transpiration rate and intercellular CO2concentration were increased by 7.03 and 7.08%,respectively,under LN condition. None of the above photosynthetic parameters were affected in either cultivar under NN condition. Taken together,CsGS1promotes photosynthesis in cucumber plants during N deprivation,and its absence has a an inhibitory effect.
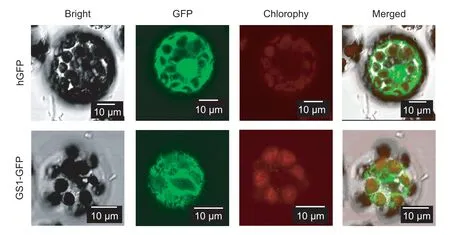
Fig.2 Subcellular localization of the CsGS1-GFP protein in Arabidopsis protoplasts. GFP and fusion protein CsGS1-hGFP were observed under white light,UV light,and chlorophyll-absorbing wavelengths. The merged image is the result of superimposition.Bars=10 μm.
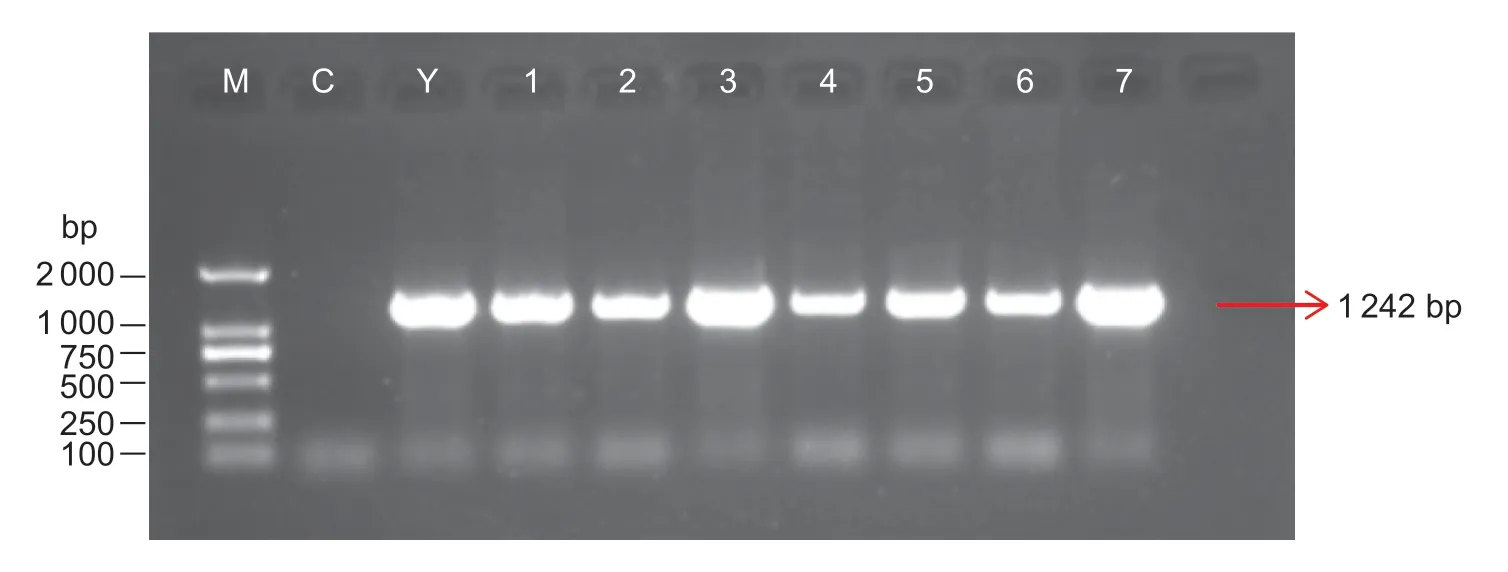
Fig.3 PCR identification of CsGS1 overexpression and anti-expression cucumber leaves. M,DNA Marker DL2000. Y,plasmid control. C,control plants. W,water control. 1–4,identification of overexpression plants by PCR. 5–7,identification of antiexpression plants by PCR.
3.5.CsGS1 increased chlorophyll b content under LN condition
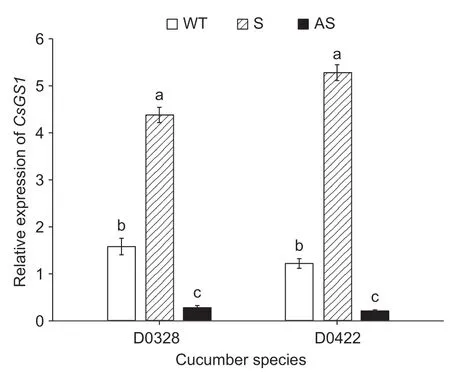
Fig.4 qRT-PCR analysis of CsGS1 in leaves of transgenic D0328 and D0422. Three technical replicates were performed.Error bars indicate standard errors. Different lowercase letters indicate significant differences (P<0.05).
Chlorophyllbis an indicator of low nitrogen stress in cucumber (Yuet al.2011). Under LN condition,overexpression ofCsGS1increased the chlorophyll b content in both the D0328 and D0422 plants compared to their respectiveCsGS1-silenced and control counterparts.As shown in Table 2,the chlorophyllbcontent in the D0328-S and D0422-S plants were 3.92-and 5.26-fold higher than that in theCsGS1-silenced plants and 2.15 and 4.44-fold higher than that in the control plants,respectively. However,CsGS1did not significantly affect the chlorophyllbcontent under NN condition (Table 2).
3.6.CsGS1 increased total N content under LN condition
Total N content in plants is a direct measure of its assimilation and metabolism (Zhenget al.2011). As shown in Table 3,N deprivation increased the total N content in the D0328-S plants by 1.88-and 1.35-fold compared to that in D0328-AS and D0328-CK,respectively,and increased total N in D0422-S by 2-and 1.32-fold relative to D0422-AS and D0422-CK,respectively. The total N content was unaffected under NN condition regardless ofCsGS1expression. Taken together,CsGS1can increase N utilization in cucumber plants exposed to LN stress,indicating a functional link betweenCsGS1and N metabolism.
3.7. Fresh weight,height and root length measurements
N absorption significantly affects morphological indices such as fresh plant weight,height and root length. As shown in Fig.5,the shoot and root fresh weight increased significantly by 32.82 and 16.39% in the D0328-S plants under LN conditions,and were 1.53-and 1.64-fold higher than that of D0328-CK,respectively. Likewise,the shoot and root fresh weight of D0422-S also increased by 24.46 and 23.68%,while that of D0422-AS decreased by 23.4 and45.93%,respectively,compared to the control plants under LN condition. Furthermore,the height and root length of the D0328-S plants were 1.21-and 1.14-fold higher,and that of the D0422-S plants were 1.22-and 1.50-fold higher than that of their respective controls. In contrast,CsGS1silencing significantly decreased the height and root length by 15.94 and 24.91% respectively in D0328,and by 26.42 and 23.46% respectively in D0422 under LN condition (Fig.6).None of the above morphological indices were affected in the presence of sufficient N regardless of cultivars orCsGS1expression status. Taken together,CsGS1overexpression in cucumber can significantly increase the plant biomass,and lengthen the shoots and roots under LN condition. The findings so far clearly indicate thatCsGS1is a crucial factor in the adaptability of cucumber to N deprivation stress.

Table 1 Photosynthetic parameters of transgenic plants under different N concentrations
3.8.GS activity determination
GSis the rate-limiting enzyme of N metabolism pathwaysin plants,and its activity is a determining factor of N absorption. We found that GSactivity increased uponCsGS1overexpression and was suppressed byCsGS1silencing under LN condition (Fig.7). Under LN condition,GS activity increased by 1.31-and 1.25-fold in the D0328-S and D0422-S plants,respectively,and decreased by 37.75 and 25% in D0328-AS and D0422-AS,respectively,compared with the controls. However,neitherCsGS1overexpression nor silencing had any significant effect on GS activity under NN condition. Therefore,N deprivation induces GS activity which can be respectively augmented and inhibited byCsGS1overexpression and silencing.
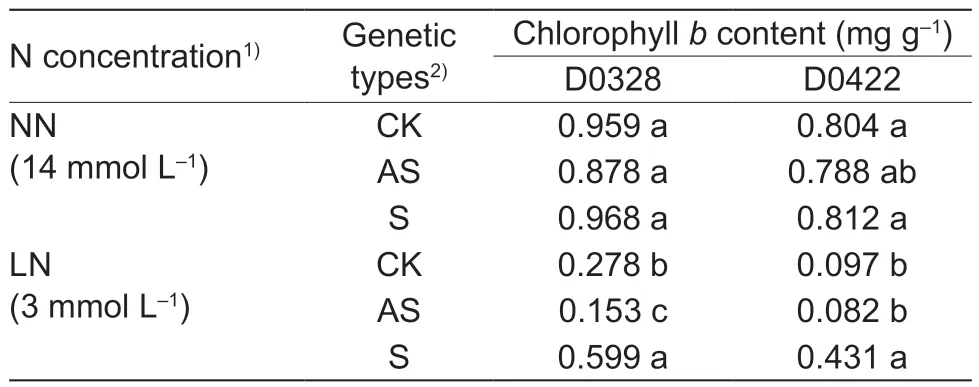
Table 2 The chlorophyll b content of transgenic plants under different N concentrations
4.Discussion
Excessive use of fertilizers and other agrochemicals often degrades soil quality and decreases agricultural productivity(Adegbeyeet al.2020),along with polluting the water bodiesand air (Shahet al.2019). In the EU,almost 80 million tons of N were released on average into the river basins per year from 1985 to 2005,mostly from agro-fertilizers(La Notteet al.2017). The fertilizer utilization per hectare in China is nearly twice as high as the safety limit (0.23 t ha–1) in developed countries (Yang and Lin 2019). In the OECD countries,the total losses from non-point source pollution caused by overfertilization exceeds billions of dollars annually. Despite numerous measures that have been taken to control the dosage of fertilizers,N pollution from agricultural production is still considerable. In addition to lowering the amount of N fertilizers,it is crucial to develop crop breeds with high NUE in order to reduce N demand.
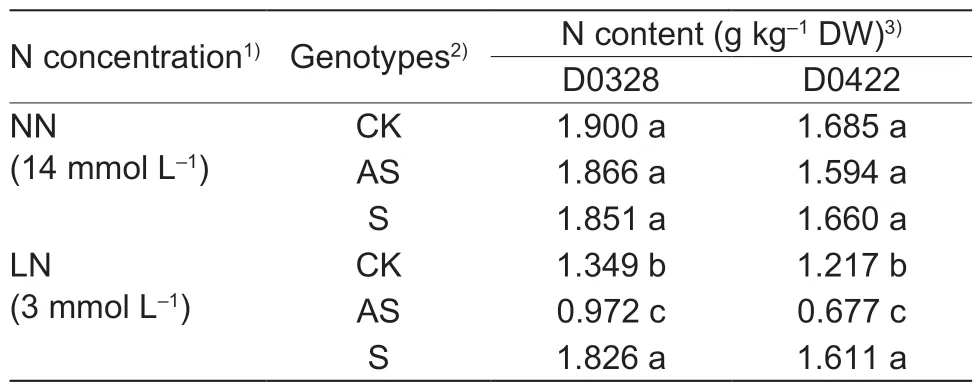
Table 3 The N content of transgenic plants under different N concentrations
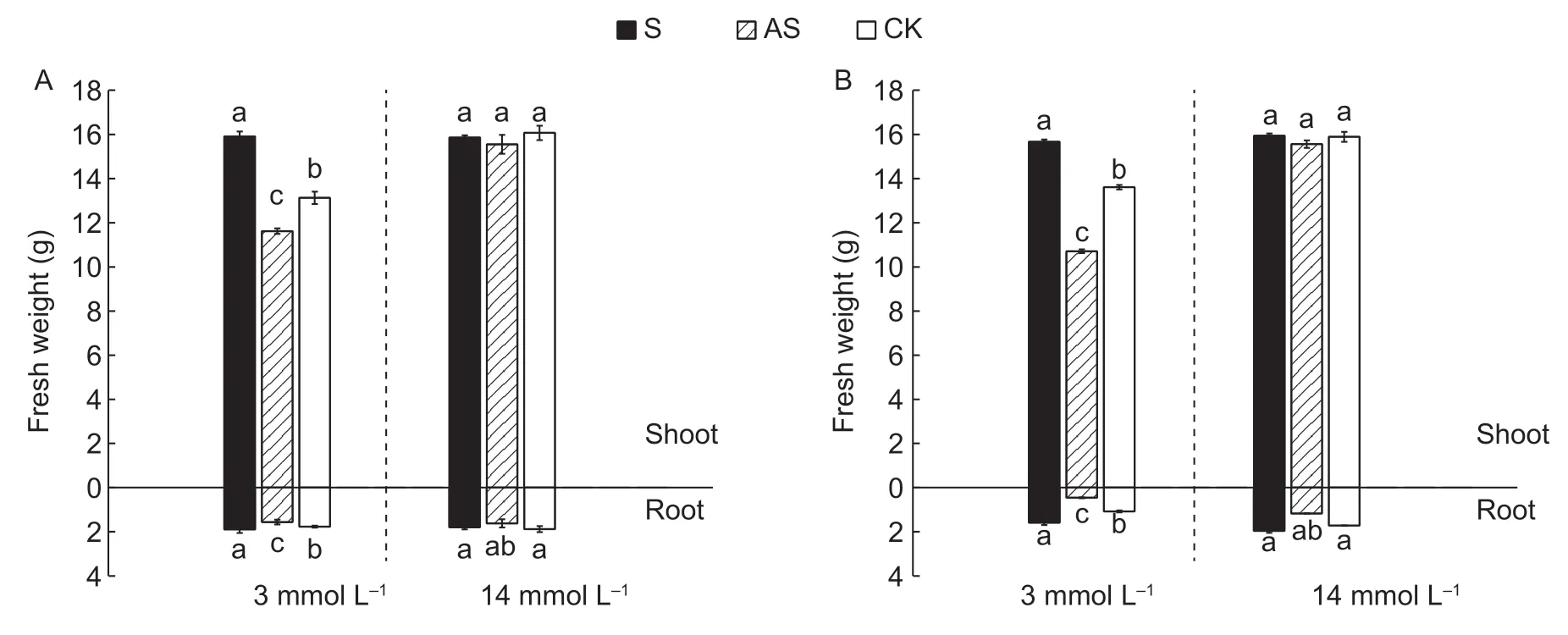
Fig.5 Fresh weight of transgenic plants under different N concentrations. A,fresh weight of D0328. B,fresh weight of D0422.Three biological replicates and four technical replicates were performed. Error bars indicate standard errors. Different lowercase letters indicate significant differences (P<0.05).

Fig.6 Root length and height of transgenic plants under different N conditions. A and C,plant root length and height of D0328.B and D,plant root length and height of D0422. Three biological replicates and four technical replicates were performed. Error bars indicate standard errors. Different lowercase letters indicate significant differences (P<0.05).
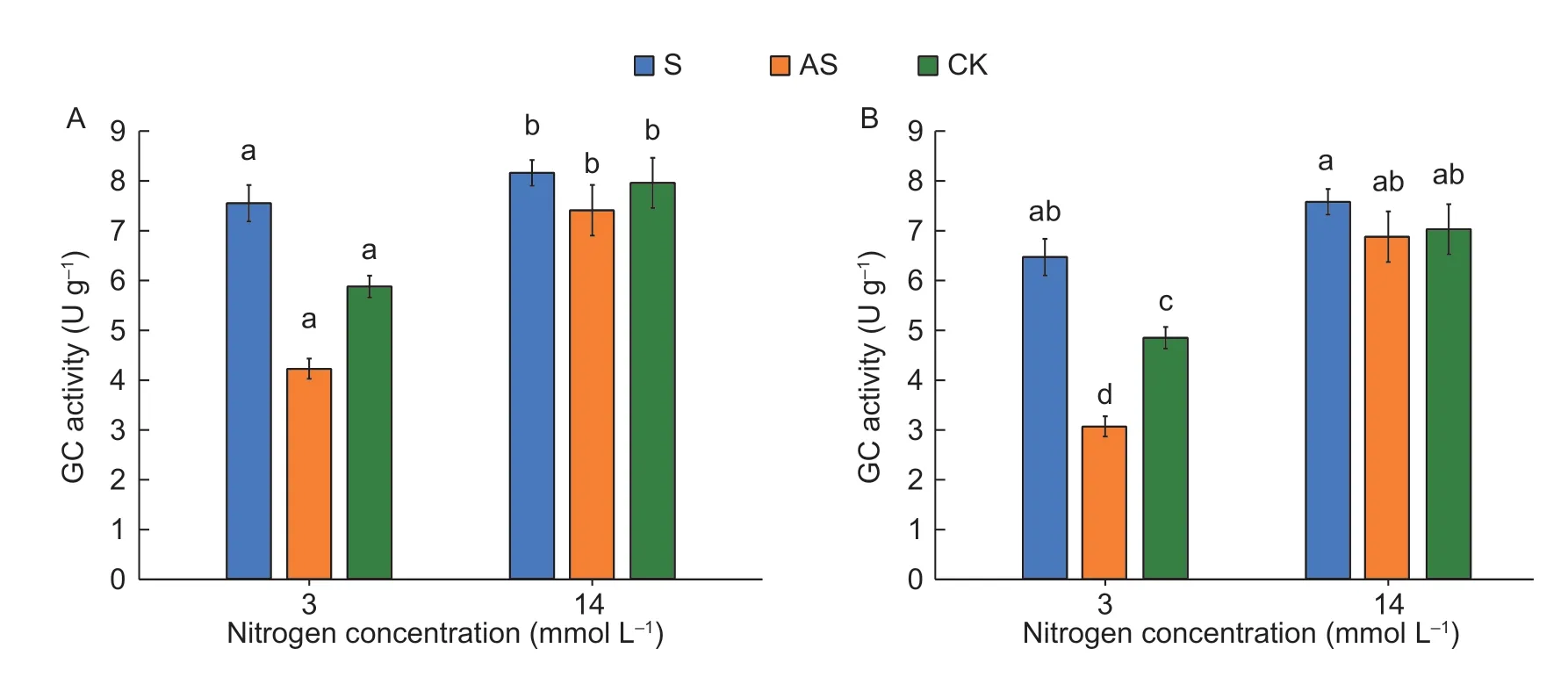
Fig.7 GS activity of transgenic plants under different N conditions. GS activities of D0328 (A) and D0422 (B). Three biological replicates and four technical replicates were performed. Error bars indicate standard errors. Different lowercase letters indicate significant differences (P<0.05).
GS plays a major role in plant N metabolism by converting ammonium to glutamine and glutamate that are essential for amino acid biosynthesis (Gadaletaet al.2014). Based on the anatomical distribution and physiological function,GS can be categorized into the GS1 and GS2 isoforms(Cren and Hirel 1999). In most higher plants,GS1 is primarily localized in the non-photosynthetic tissues wherein it synthesizes glutamic acid for N transfer. GS2 on the other hand assimilates and re-assimilates ammonia in the mesophyll cells (Huanget al.2010). Although severalGS2genes are recognized in soybean (Glycine max) and alfalfa(Zozaya-Garzaet al.1999),only a singleCsGS2gene has been identified in most higher plants. The cucumberCsGS1protein is localized in the cytoplasm. Furthermore,we found thatCsGS1was highly expressed in the leaves and less in the shoots and roots of both cultivars under N deprivation(Fig.1). In many plants,GSgenes are expressed in multiple tissues,such as seedlings (Caputoet al.2009),flag leaves(Zhanget al.2017) and shoots (Tianet al.2015),which can be attributed to their role in complex metabolic pathways and development (Gadaletaet al.2014).
Although the function ofGS1in N metabolism has been established in many plant species,little is known regarding the function in cucumber. The cucumberCsGS1gene,cloned and sequenced in 2012 (Fenget al.2012),shows 97% homology with that of melon (Cucumis meloL.). We found that the relative expression ofCsGS1was depressed by increasing N concentration,indicating that it is triggered by N deprivation and likely protects the plants against LN stress. To this end,we generated theCsGS1-overexpressing andCsGS1-silenced cucumber transgenics to determine the function ofCsGS1in LN adaptation.
N metabolism is closely related to photosynthesis(Chuet al.2017).GS1overexpression in tobacco plants maintained photosynthesis under N deprivation,compared to the 40–50% decrease in controls (Fuenteset al.2001).In addition,mutated plastidGScan impair photorespiration,eventually leading to plant death (Ferreiraet al.2019). We found that overexpression ofCsGS1significantly increased the photosynthetic parameters in cucumber under LN condition,in addition to increasing N accumulation,plant biomass,plant height and root length. Thus,CsGS1is crucial for maintaining N assimilation in cucumber plants that are grown in N deficient condition,andCsGS1overexpression can enhance their adaptability to LN stress.A previous study found that the chlorophyllbcontent of cucumber is a reliable biological indicator of LN stress because it had the strongest positive correlation with N metabolism (Yuet al.2011). Furthermore,the total N content in plant biomass is a gauge of NUE. In this study,both chlorophyllband total N content were significantly higher in the cucumber plants overexpressingCsGS1,indicating thatCsGS1enhances N assimilation and metabolism even under LN condition. Consistent with our findings,previous studies have shown thatGSmediates plant resistance to various abiotic stresses,including low N (Yanget al.2019),cold (Guet al.2018),drought (Liet al.2016) and high salt (Hoshidaet al.2000). However,the effect of overexpressingGS1varies across different plants species.For instance,Fuenteset al.(2001) showed that under LN condition,GS1overexpression increased GS activity by 6-fold compared to the wild type control. Studies on tobacco have also reported a similar augmentation in GS activity in theGS1-overexpressing plants during N deprivation,which corresponded to increased growth rate,yield and biomass.However,GS1overexpression was redundant when the plants were grown with sufficient N (Goodet al.2004). In this study as well,overexpression ofCsGS1significantly increased GS activity in cucumber under LN condition,and improved all morphological indices of N utilization. SilencingCsGS1not only reduced GS activity but also sensitized the LN-tolerant D0328 cultivar to N deprivation. Since both the high N-requiring D0422 and low N-requiring D0328 cultivars responded to changes in N concentration,we can surmise that the LN environment is more conducive toCsGS1function,and therefore improved the NUE of cucumber plants when the gene was overexpressed.
Compared toA.thaliana(Ferreiraet al.2019),wheat(Gadaletaet al.2014;Zhanget al.2017) and corn (Prinsi and Espen 2015),little is known regarding the genes related to N metabolism in cucumber. We have shown for the first time thatCsGS1overexpression improved photosynthesis,morphological indices,total N and chlorophyllbcontents of cucumber plants,and enhanced tolerance to LN stress by increasing GS activity. Nevertheless,since overexpression ofGS1and other isogenes do not result in a consistent phenotype in plants,there is a possibility that regulatory pathways upstream or downstream of GS affect its activity.Further research is needed to determine the large-scale and long-term feasibility of developingGS1-overexpressing transgenics in order to improve crop NUE.
5.Conclusion
CsGS1overexpression in cucumber improved NUE under LN condition,which in turn significantly improved agronomic traits and N accumulation. Therefore,GS1is a highly promising target for developing improved cucumber breeds.
Acknowledgements
We acknowledge the funding support from Heilongjiang Postdoctoral Scientific Research Developmental Fund,China (LBH-Q16021),“Academic Backbone”Project of Northeast Agricultural University,China (18XG06),the National Science Foundation of Heilongjiang Province,China (LH2019C033).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
杂志排行
Journal of Integrative Agriculture的其它文章
- Low glycemic index:The next target for rice production in China?
- Do cooperatives participation and technology adoption improve farmers’ welfare in China? A joint analysis accounting for selection bias
- Impacts of household income on beef at-home consumption:Evidence from urban China
- The water-saving potential of using micro-sprinkling irrigation for winter wheat production on the North China Plain
- Changes in bacterial community and abundance of functional genes in paddy soil with cry1Ab transgenic rice
- Synergistic effect of Si and K in improving the growth,ion distribution and partitioning of Lolium perenne L.under saline-alkali stress
