Changes in bacterial community and abundance of functional genes in paddy soil with cry1Ab transgenic rice
2021-05-23
Institute of Biotechnology,Fujian Academy of Agricultural Sciences/Fujian Provincial Key Laboratory of Genetic Engineering for Agriculture,Fuzhou 350003,P.R.China
Abstract A field experiment involving cry1Ab transgenic rice (GM) and its parental non-cry1Ab rice (M) has been on-going since 2014.The diversity of the bacterial communities and the abundance of the microbial functional genes which drive the conversion of nitrogen in paddy soil were analyzed during the growth period of rice in the fifth year of the experiment,using 16S rRNAbased Illumina MiSeq and real-time PCR on the amoA,nirS and nirK genes. The results showed no differences in the alpha diversity indexes of the bacterial communities,including Chao1,Shannon and Simpson,between the fields cultivated with line GM and cultivar M at any of the growth stages of rice. However,the bacterial communities in the paddy soil with line GM were separated from those of paddy soil with cultivar M at each of the growth stages of rice,based on the unweighted UniFrac NMDS or PCoA. In addition,the analyses of ADONIS and ANOSIM,based on the unweighted UniFrac distance,indicated that the above separations between line GM and cultivar M were statistically significant (P<0.05) during the growth season of rice. The increases in the relative abundances of Acidobacteria or Bacteroidetes,in the paddy soils with line GM or cultivar M,respectively,led to the differences in the bacterial communities between them. At the same time,functional gene prediction based on Illumina MiSeq data suggested that the abundance of many functional genes increased in the paddy soil with line GM at the maturity stage of rice,such as genes related to the metabolism of starch,amino acids and nitrogen. Otherwise,the copies of bacterial amoA gene,archaeal amoA gene and denitrifying bacterial nirK gene significantly increased (P<0.05 or 0.01) in the paddy soil with line GM. In summary,the release of cry1Ab transgenic rice had effects on either the composition of bacterial communities or the abundance of microbial functional genes in the paddy soil.
Keywords:cry1Ab transgenic rice,bacterial community,microbial functional gene,Illumina MiSeq Platform,real-time PCR
1.lntroduction
In 2018,26 countries planted 191.7 million hectares of biotech crops,which added 1.9 million hectares to the previous record of plantings in 2017,according to data published by International Service for the Acquisition of Agri-biotech Applications (James 2018). Planting transgenic crops has brought many economic benefits but has also raised some concern over the potential impact on the environment,such as on the soil microbial community(Luet al.2017). Microbial community diversity is directly related to the soil material energy cycle,and the stability and sustainability of the soil ecosystem. Studies have shown that plant species is an important factor affecting the diversity in the microbial community as a result of differences in the composition and amount of root exudates (Airaet al.2010;Inceogluet al.2010). The cultivation of transgenic crop varieties may alter the soil microbial community through root exudates and plant residues,over both the short and long term (Songet al.2014).
Previous studies have shown that the release of many transgenic plants may have varying influences on the soil microbial communities. For example,some measures,such as the diversity and abundance of indigenous soil bacteria and fungi,changed in soil with Bt transgenic cotton(Doneganet al.1995),and Bt transgenic maize residues had effects on the soil bacterial community (Castaldiniet al.2005). In other cases,Bt transgenic rice straw returning to field had no effect on the culturable microorganisms in soil (Wuet al.2004). There was no significant effect of Bt transgenic rice straw decomposition on the soil bacterial and fungal communities in the field (Luet al.2010),and many other studies show a similar lack of effects.
The inconsistent results among studies on the effects of transgenic plants on the soil microbial community are partially due to the extremely high complexity of the soil microbial community and the limitations of the detection technologies for studying that community. The development of high-throughput sequencing technologies has resulted in large-scale soil microbial gene sequencing,which has provided a wealth of information for research on soil microbial species,structure and diversity (Zheng and Jia 2013;Louet al.2014). High-throughput sequencing of microbes has been widely used to study microbial communities. A study found that the abundance of soil microbes in rotation cropping was far higher than that of continuous cropping by 16S rRNA-based Illumina MiSeq sequencing (Zhanget al.2014). The diversity of fungal in strawberry rhizosphere soil increased significantly in the twelfth year of continuous cropping in the greenhouse by ITS-based Illumina MiSeq sequencing (Li and Liu 2019).
We had previously analyzed the effect of plantingcry1Ac/cptitransgenic rice on the diversity and abundance of the bacterial or fungal communities in the paddy soil,using denaturing gradient gel electrophoresis (DGGE) and realtime PCR based on the 16S rRNA or SSU rDNA genes. The results showed that the release ofcry1Ac/cptitransgenic rice has no significant effect on the composition or abundance of bacterial or fungal communities in the paddy soil during the rice growth stages,at least in a short-term study of four years (Songet al.2014).
In this study,the impact of the release of transgenic rice with a different Bt gene (cry1Ab) on the diversity and composition of the bacterial communities in paddy soil was assessed by a 16S rRNA-based Illumina MiSeq Platform.In addition,we compared the abundances of microbial functional genes,which drive the conversion of nitrogen,in the paddy fields with cultivation ofcry1Abtransgenic rice versus its control rice cultivar during the rice growth stages,using the real-time PCR analysis ofamoA,nirSandnirKgenes.
2.Materials and methods
2.1.Field experiment and sampling
A field experiment for the safety test has been approved at Shoushan (26°11´N,119°16´E),located in Fujian Province,by the the Ministry of Agriculture and Rural Affairs,China,since 2014. The experimental site had two rice varieties,the line ofcry1Abtransgenic rice (OryzasativaL.) mfb-MH3301-1 (GM) and the corresponding parental noncry1Abrice MH3301 (M). Thecry1Abtransgenic rice line was initially generated by introducing thecry1Abgene fromBacillusthuringiensisinto the rice. A double T-DNA vector pCDMARUBb-Hyg of the syntheticcry1Abgene was constructed and transformed to rice restorer line MH3301 by theAgrobacteriummethod,and then the non-marked (with the hygromycin gene removed)cry1Abgene homozygous rice strain mfb-MH3301-1 was obtained through the detection and selection of multi-generation self-crossing.
The field soil properties included:organic C,15.77 g kg-1;pH 5.82,total N;1.28 g kg–1;total P,0.27 g kg–1;total K,18.35 g kg–1;available N,153.60 mg kg–1;available P,15.67 mg kg–1and available K,77.98 mg kg–1. Each treatment was replicated four times in 25 m2plots randomly distributed in the field. Pesticides were not applied to the rice during the experiment. The field experiment has been implemented repeatedly from 2014 until now,with the management of water and fertilizer according to the conventional methods in the area.
The soil adhering to the roots,at about 0–20 cm depth,was collected as the root zone soil in the fifth year of the experiment at the seeding,tiller,heading and maturity stages of rice. In each field replicate,the soil of five sites was excavated randomly and pooled to yield one sample per field plot. Soil samples were either stored at–70°C until molecular analysis or air-dried for chemical analysis.
2.2.DNA extraction and high-throughput sequencing
DNA was extracted using the FastDNA®SPIN Kit for Soil (QBIOgene,USA) and sent to MAGIGENE Co.,Ltd.,China for analysis by high-throughput sequencing(16S rRNA-based Illumina MiSeq). Gene-specific primers for the V3–V4 region of 16S rRNA were 338F(5´-ACTCCTACGGGAGGCAGCAG-3´) and 806R(5´-GGACTACHVGGGTWTCTAAT-3´) (Walterset al.2011).PCR reactions,containing 25 μL 2×PremixTaq(TaKaRa Biotechnology,Dalian Co.Ltd.,China),1 μL each primer(10 pmol mL–1) and 3 μL DNA (20 ng μL–1) template in a volume of 50 μL,were amplified by thermocycling:5 min at 94°C for initialization;30 cycles of 30 s denaturation at 94°C,30 s annealing at 52°C,and 30 s extension at 72°C;followed by 10 min final elongation at 72°C. PCR products were mixed in equivalent ratios,purified and sequenced on an Illumina MiSeq Platform (Guangdong Magigene Biotechnology Co.,Ltd.,Guangzhou,China).
Sequences with ≥97% similarity were assigned to the same OTU by Usearch Software (ver.10). Alpha diversity including PD_whole_tree,Dominance,Observed_species,Chao1,Shannon and Simpson was calculated with QIIME(ver.1.9.1). Permutation Test for Homogeneity of Multivariate Dispersions (PERMDISP) was performed using R Software based on Bray-Curtis distance. Nonmetric Multidimensional Scaling (NMDS),Principal coordinate analysis (PCoA) and the non-parametric analyses of ADONIS and ANOSIM were performed using R Software based on the weighted and unweighted UniFrac distance matrix. LDA Effect Size(LEfSe) analysis was used to find the biomarker of each group. Functional gene prediction was performed using PICRUSt based on the KEGG pathway.
2.3.Quantification of microbial functional gene abundance
The abundance of microbial functional genes was determined by real-time PCR on a PRISM7500 Real-Time PCR System (ABI,USA). Primers and thermal profiles for the real-time PCR are listed in Table 1. The standard curve for real-time PCR in this study was produced according to the methods in previously published papers (Heet al.2007;Song and Lin 2014). The ten-fold serial dilutions of the standard curves ranged from 9.09×108to 9.09×103copies μL–1for bacterialamoAgene,1.66×1010to 1.66×103copies μL–1for archaealamoAgene,3.67×109to 3.67×103copies μL–1fornirSgene and 8.07×109to 8.07×103copies μL–1fornirKgene. Amplification efficiencies of bacterial or archaealamoA,nirSandnirKgenes were 1.01,0.99,0.98 and 1.01,respectively.
2.4.Soil chemical properties and plant harvest
Soil chemical properties of each soil sample,including organic C,pH (1:2.5 H2O,soil:solution),alkali-hydrolyzable N,Olsen-P and NH4OAc-K,were analyzed by standard procedures (Bao 2007),and used as environmental factors for redundancy correspondence analysis (RDA) of the Illumina MiSeq data.
Plants from each plot were harvested and ten plants,excluding those in the boundary lines,were chosen for determinations of the agronomic characters,yield traits and plant nutrition levels,such as total N,P and K.
2.5.Statistics
Statistical significances of the differences between the two rice lines and among different growth stages in this study were assessed by analysis of variance (ANOVA) and least significant difference (LSD) using SPSS,ver.17.0.
3.Results
3.1.Diversity of bacterial communities in the paddy soils
In this study,a total of 67 271 OTUs was identified,with an average of 2 102±267 OTUs per soil replicate,based on Illumina MiSeq data. Alpha diversity indexes including PD_whole_tree,Dominance,Observed_species,Chao 1,Shannon and Simpson are shown in Table 2. The results indicated no significant differences in the indexes of Chao 1,Shannon or Simpson between the fields planted with cultivarM and line GM (Table 2). PD_whole_tree and Observed_species in the paddy soil with line GM were markedly higher(P<0.05) than those in the paddy soil with cultivar M at the tiller stage of rice;on the contrary,the former was markedly lower (P<0.05) than the latter at the heading stage of rice(Table 2). At the same time,the indexes of Dominance significantly decreased (P<0.05) in the paddy soil with line GM at the tiller stage of rice (Table 2). These results indicated that most of the indexes for bacterial community richness and diversity in the paddy soil had no obvious differences betweencry1Abtransgenic rice line GM and its wild type cultivar M in the rice growing season.

Table 1 Primers and real-time PCR conditions used for this study
The dimensionality reduction analysis of NMDS and PCoA,based on weighted UniFrac distance,showed that the four paddy soil replicates of cultivar M were separated from two replicates of line GM by NMDS1 (Fig.1-A) or PC1,which represented 80.03% of the total variation(Fig.1-B),and from the other two replicates of line GM by NMDS2 (Fig.1-A) or PC2,which only represented 5.77%of the total variation (Fig.1-B) at the tiller stage of rice,respectively. For the analysis based on the unweighted UniFrac distance,the paddy soil replicates of line GM were clearly separated from those of cultivar M by either NMDS2(Fig.1-C) or PC2 (Fig.1-D) at each growth stage of rice.For cultivar M,whether based on weighted or unweighted UniFrac distances,the four replicates of paddy soil at the tiller stage of rice were clearly separated from the four replicates of paddy soil at the seeding or heading stages of rice by NMDS1 (Fig.1-A and C) or by PC1 (Fig.1-B and D),respectively.
The analyses of ANOSIM and ADONIS differences,based on unweighted UniFrac distance,showed that the separations in paddy soil bacterial communities of line GM from those of cultivar M were statistically significant (P<0.05) at each of the growth stages of rice (Table 3). Based on weighted UniFrac distance,the analysis of ANOSIM differences also suggested the above separations were significant (P<0.05) at each of the growth stages of rice,while the analysis of ADONIS differences suggested they were significant (P<0.05) only at the tiller and maturity stages of rice (Table 3).
Additionally,the analyses of ADONIS and ANOSIM differences showed that the separation in paddy soil bacterial communities among the different growth stages of rice was statistically significant (P<0.01) in the field planted with cultivar M (Table 4),while the ANOSIM result suggested the separation was also statistically significant (P<0.05) in the field planted with line GM (Table 4).
The beta diversity of PERMDISP analysis illustrated that the differences in paddy soil bacterial communities between line GM and cultivar M were statistically significant at either the seeding (P<0.05) or maturity (P<0.01) stages of rice (Table 5),and the differences in paddy soil bacterial communities among different growth stages of rice were statistically significant (P<0.01) in the field planted with cultivar M (Table 6).
The most abundant phylum in the paddy soil of the two rice lines was Chloroflexi,followed by Proteobacteria and Acidobacteria,and then by Bacteroidetes,Patescibacteria,Nitrospirae,Firmicutes,Verrucomicrobia,Planctomycetes and Actinobacteria,and then the others (Fig.2). These major bacterial phyla accounted for over 93% of total abundance in the bacterial communities in the paddy soil of both cultivar M and line GM. Among these major phyla,the relative abundances of Proteobacteria,Bacteroidetes and Nitrospirae were significantly lower in the paddy soil with line GM compared with cultivar M at the rice growth stages of seeding (P<0.05),tiller (P<0.05) and heading (P<0.01),respectively (Fig.2-A,B and C),but the relative abundance of Acidobacteria was significantly higher in the paddy soil with line GM than that with cultivar M at the maturity stage of rice (P<0.01) (Fig.2-D).
In addition,bacterial biomarkers in the paddy soil with cultivar M or line GM were analyzed by LEfSe. The LDA value distribution histogram (Fig.3) shows the significant enrichment of species and their differences in each group of either cultivar M or line GM. The default LDA value was set to 2,and the LDA scores represented the influences of the different species. The results showedthat Acidobacteria (including Acidobacteriia,Acidobacteria,Acidobacteriales,Solibacteraceae_[Subgroup_3_],Solibacterales,Candidatus_Solibacter,Bryobacter,Koribacteraceae,Candidatus_Koribacter,Subgroup_2)and Chloroflexi (including Bacteria,Ktedonobacteria,Ktedonobacterales,Ktedonobacteraceae,HSB_OF53-F07,Anaerolineae) had significant enrichment of species in the paddy soil with line GM,while Bacteroidetes (including Bacteroidales and Bacteroidetes_vadinHA17),Chloroflexi(including RBG_13_54_9 andRBG_16_58_14) and Sericytochromatia (which belongs to Cyanobacteria) had significant enrichment of species in the paddy soil with cultivar M (Fig.3). For line GM or cultivar M,the enrichment of Acidobacteria or Bacteroidetes should have impacts onthe differences in the soil bacterial communities between line GM and cultivar M,respectively.
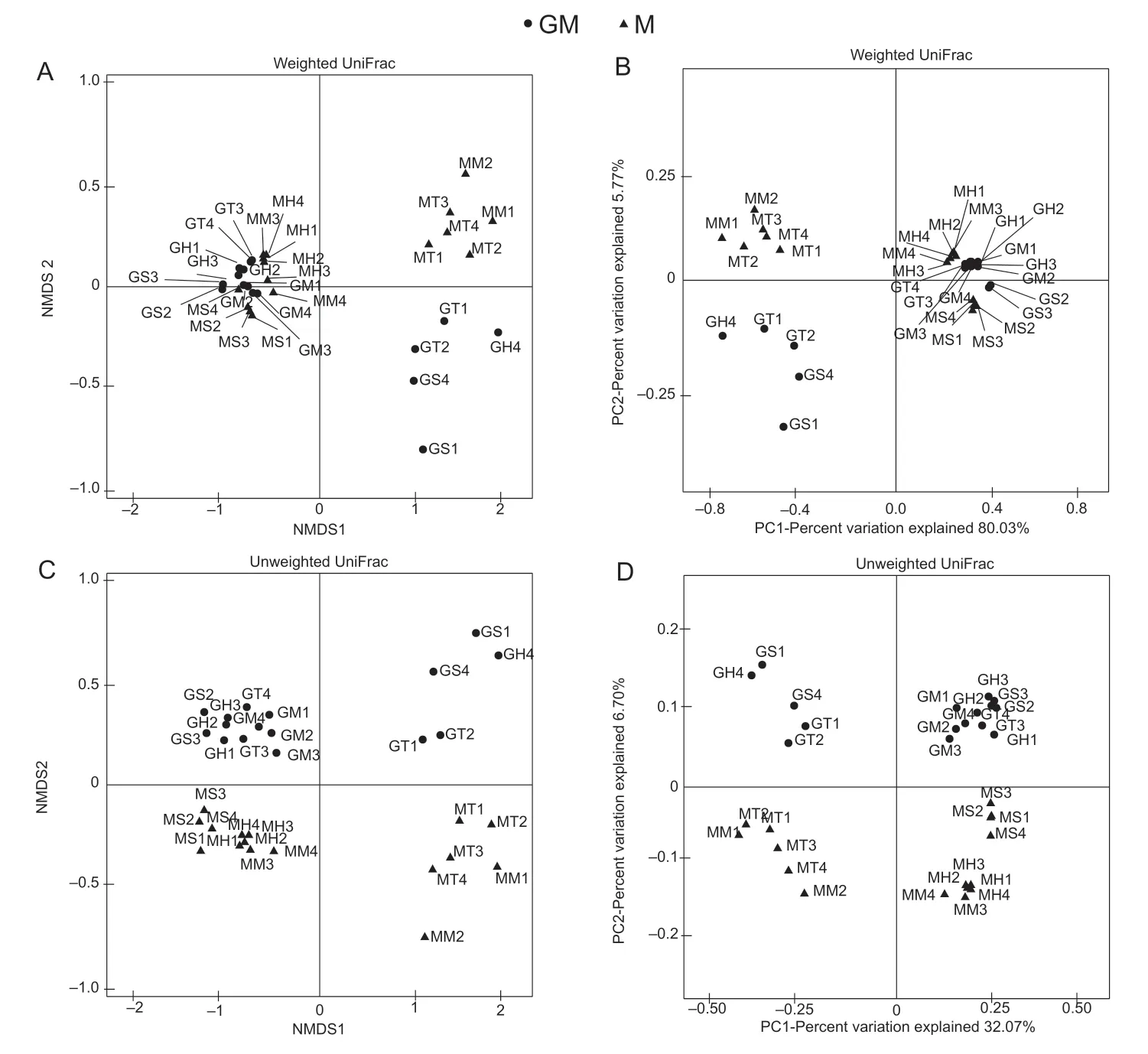
Fig.1 Non-metric multidimensional scaling (NMDS) and principal coordinate analysis (PCoA) of bacterial communities in the paddy soil with different rice lines. A and B are NMDS and PCoA plots based on weighted UniFrac distance. C and D are NMDS and PCoA plots based on unweighted UniFrac distance. MS1–MS4 and GS1–GS4,MT1–MT4 and GT1–GT4,MH1–MH4 and GH1–GH4,and MM1–MM4 and GM1–GM4 indicate the four replicates of cultivar M and line GM at the seeding,tiller,heading and maturity stages of rice,respectively. M,non-cry1Ab rice MH3301;GM,cry1Ab transgenic rice mfb-MH3301-1.
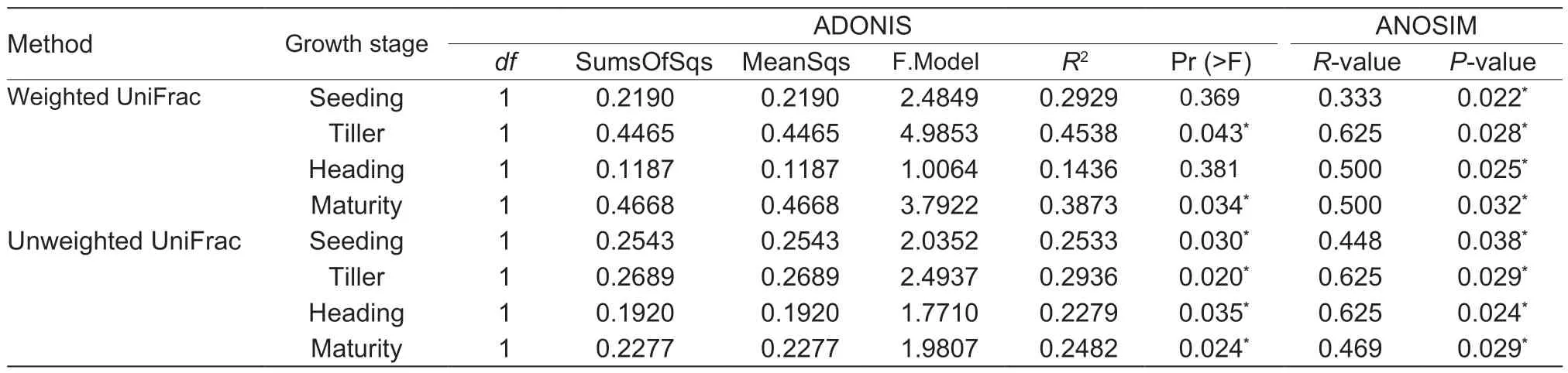
Table 3 ADONIS and ANOSIM differences in soil bacterial communities between non-cry1Ab rice MH3301and cry1Ab transgenic rice mfb-MH3301-1
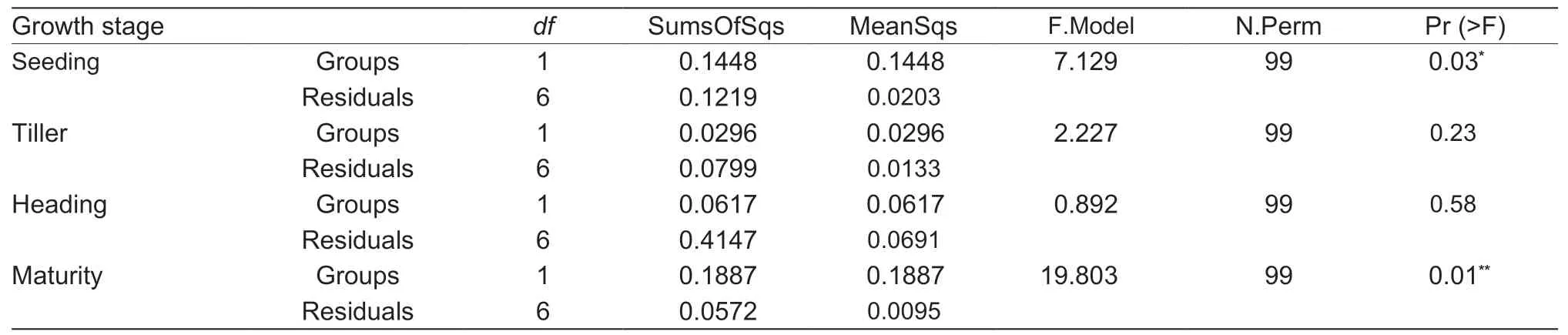
Table 5 PERMDISP analysis of the soil bacterial communities in paddy with non-cry1Ab rice MH3301and cry1Ab transgenic rice mfb-MH3301-11)
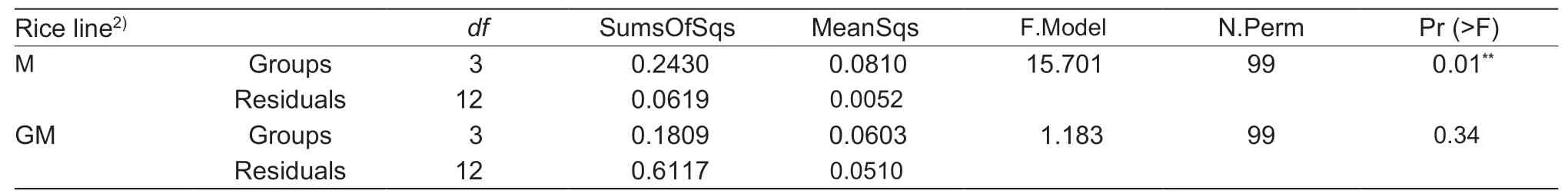
Table 6 PERMDISP analysis of the soil bacterial communities in paddy among the growth stages of seeding,tiller,heading and maturity1)
At the same time,functional gene prediction was performed using PICRUSt based on the KEGG pathway.Differences were found in the abundances of many functional genes between the paddy soil with line GM and cultivar M at the maturity stage of rice (Fig.4). A total of 80%of the top 50 most abundant functional genes significantly(P<0.05 orP<0.01) increased in the paddy soil with line GM,mainly including functional genes related to the metabolism of amino acids,starch and sucrose,nitrogen and other metabolites (Fig.4).
3.2.Abundance of microbial functional genes in the paddy soils
In the paddy soil,the bacterial and archaealamoAgene copy numbers ranged from 7.40×105to 2.19×106g–1dry soil and 2.18×107to 5.76×108g–1dry soil,respectively. In all treatments,the copy numbers of the archaealamoAgene were higher (P<0.01) than those of the bacterialamoAgene,and the ratios of archaea to bacteria ranged from 22.1 to 352.8. TheamoAgene copy numbers of bacteria and archaea in the paddy soil with line GM were higher (P<0.05 and 0.01) than those in the paddy soil with cultivar M at each of the growth stages of rice (Fig.5-A and B).
The abundance of denitrifying bacterialnirSgene ranged from 4.34×108to 6.57×108g–1dry soil,while the abundance of denitrifying bacterialnirKgene ranged from 5.81×106to 2.10×107g–1dry soil. ThenirSgene was richer (P<0.05)than thenirKgene in the paddy soil with either cultivar M or line GM. At the same time,there was no obvious difference in the abundance ofnirSgene between the paddy soils with cultivar M and line GM (Fig.6-A),but the abundance ofnirKgene in the paddy soil with line GM was higher (P<0.01)compared with that of cultivar M at each of the stages of rice growth (Fig.6-B).
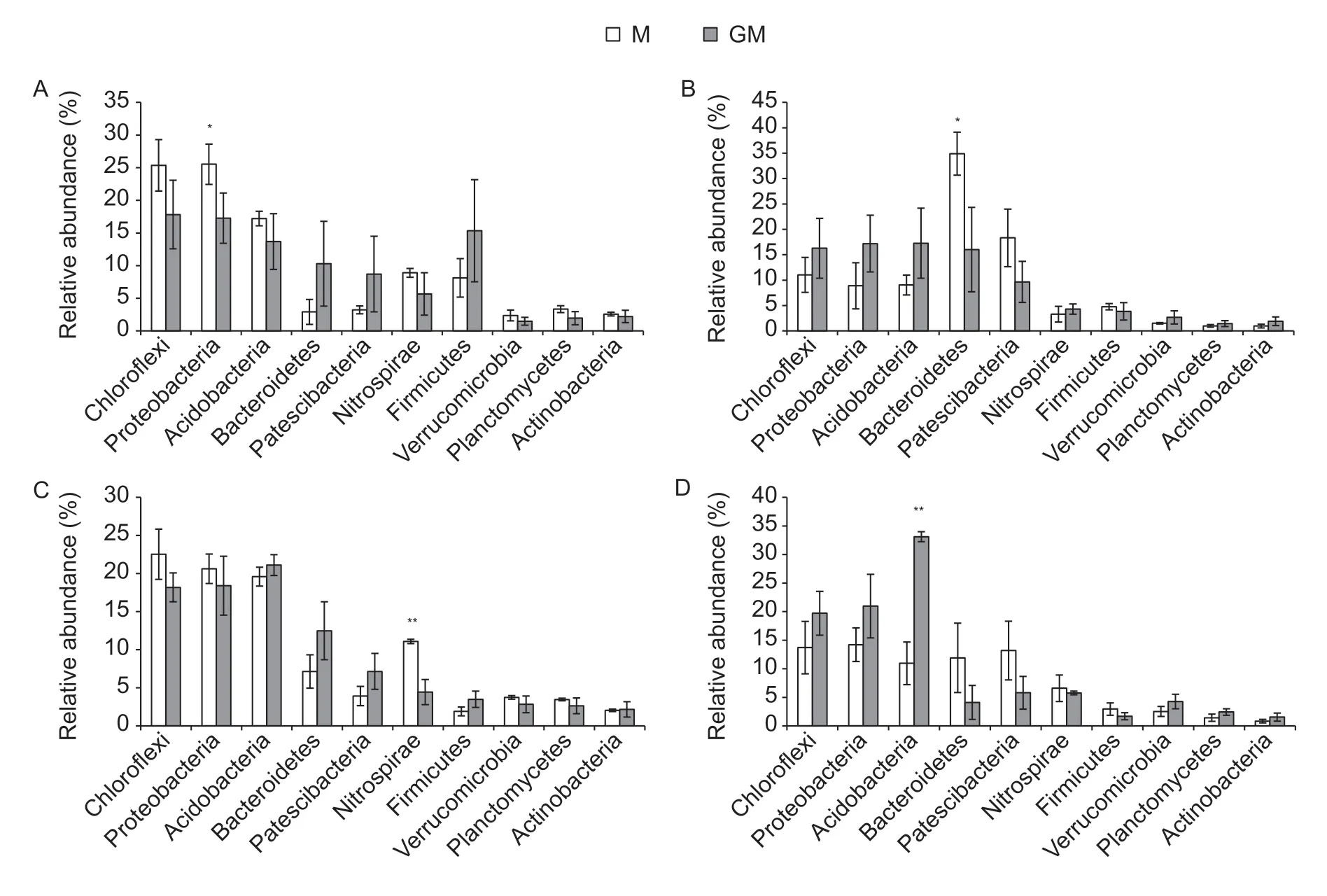
Fig.2 The relative abundances of bacteria at the phylum level in the paddy soil with different rice lines during the growth period of rice. A–D indicate seeding,tiller,heading and maturity stages of rice,respectively. M,non-cry1Ab rice MH3301;GM,cry1Ab transgenic rice mfb-MH3301-1. *,P<0.05;**,P<0.01. Bar indicates SD (n=4).
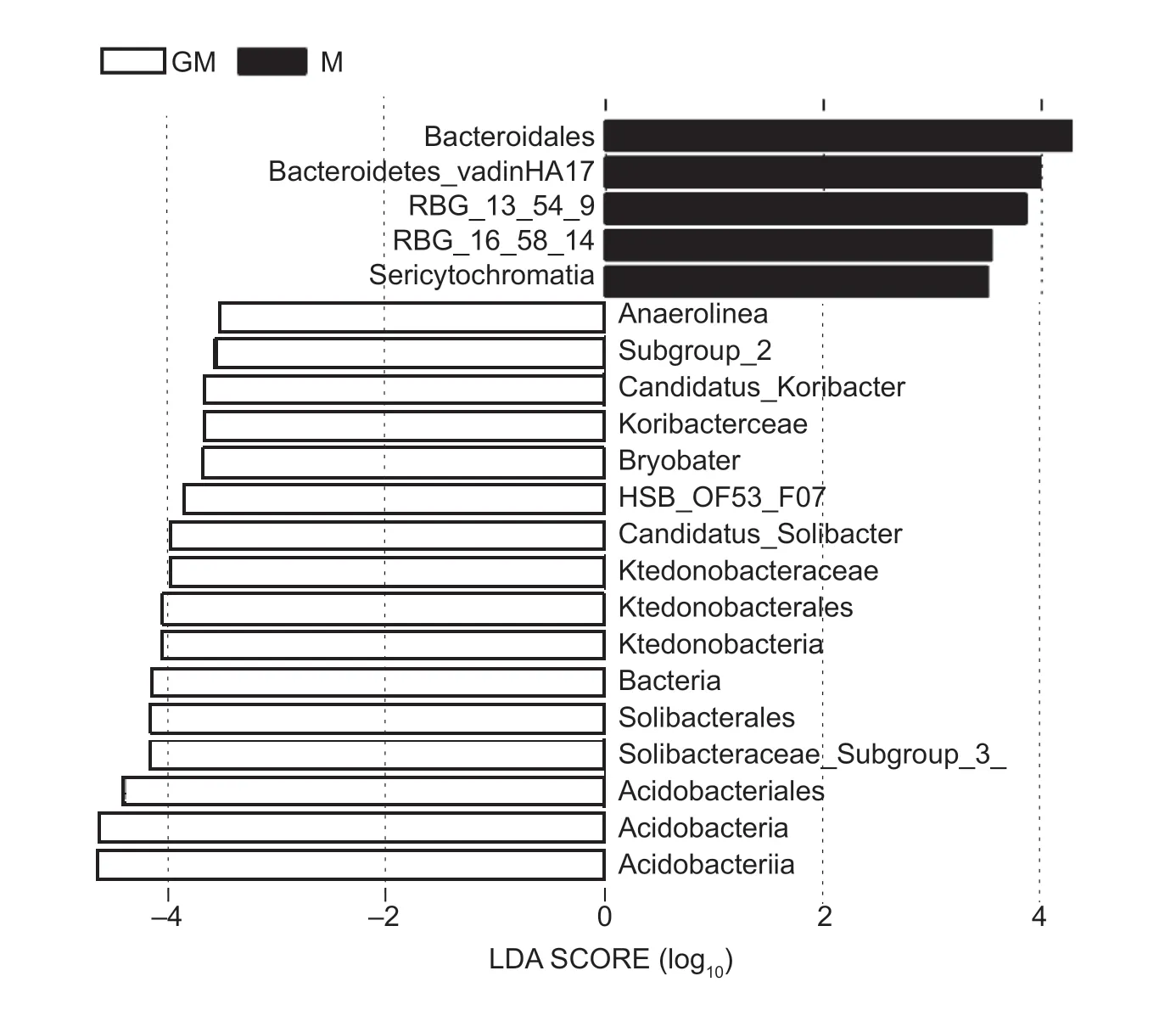
Fig.3 The latent dirichlet allocation (LDA) value distribution histogram of significant differences among species in the paddy soil with different rice lines. GM,cry1Ab transgenic rice mfb-MH3301-1;M,non-cry1Ab rice MH3301. Differences in species abundance are significant when the value of LDA is greater than 2 (default is set to 2) between cultivar M and line GM.
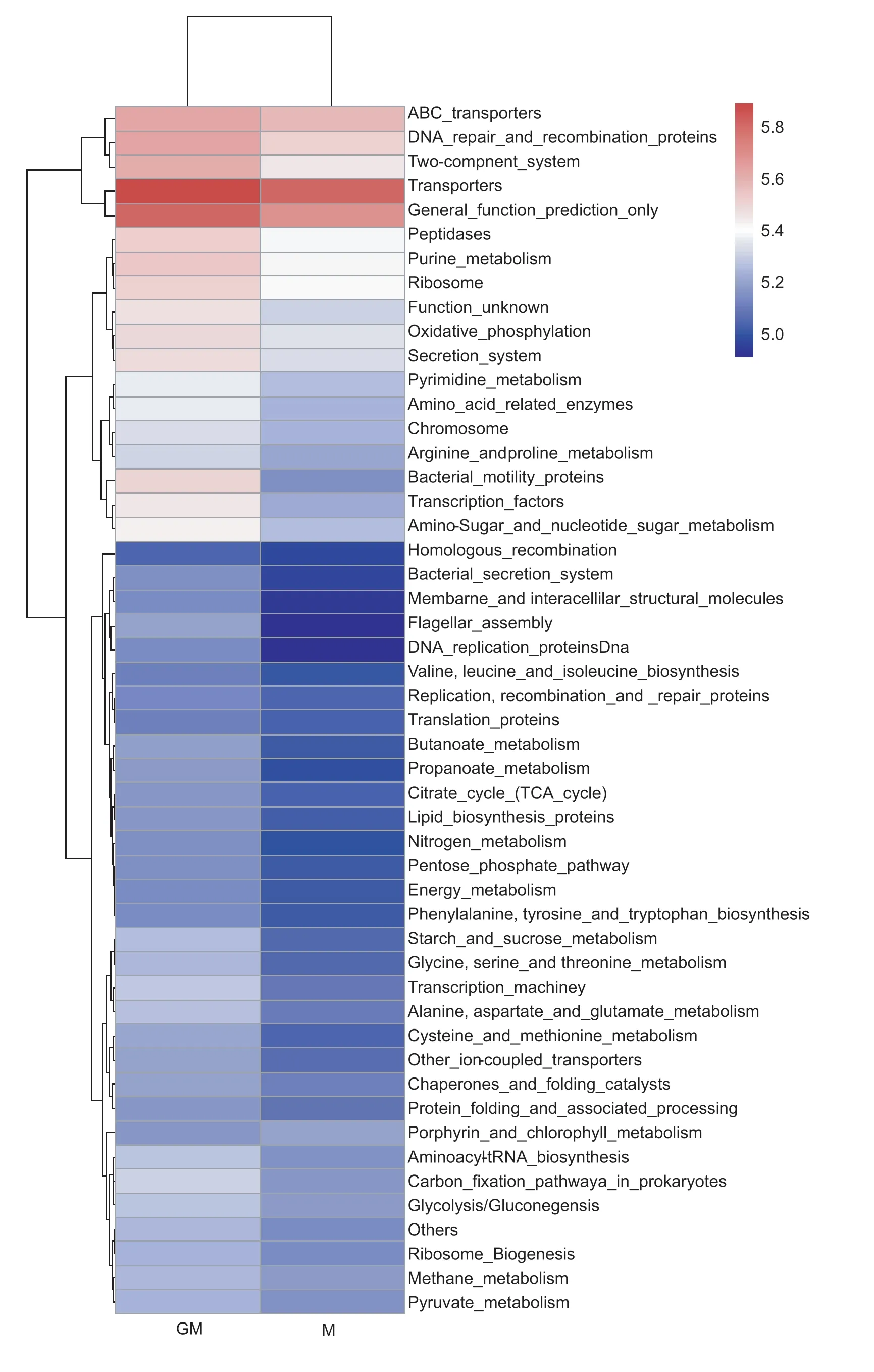
Fig.4 Heat map plot of the top 50 KEGG abundances in the paddy soil with different rice lines at the maturity stage of rice.GM,cry1Ab transgenic rice mfb-MH3301-1;M,non-cry1Ab rice MH3301.
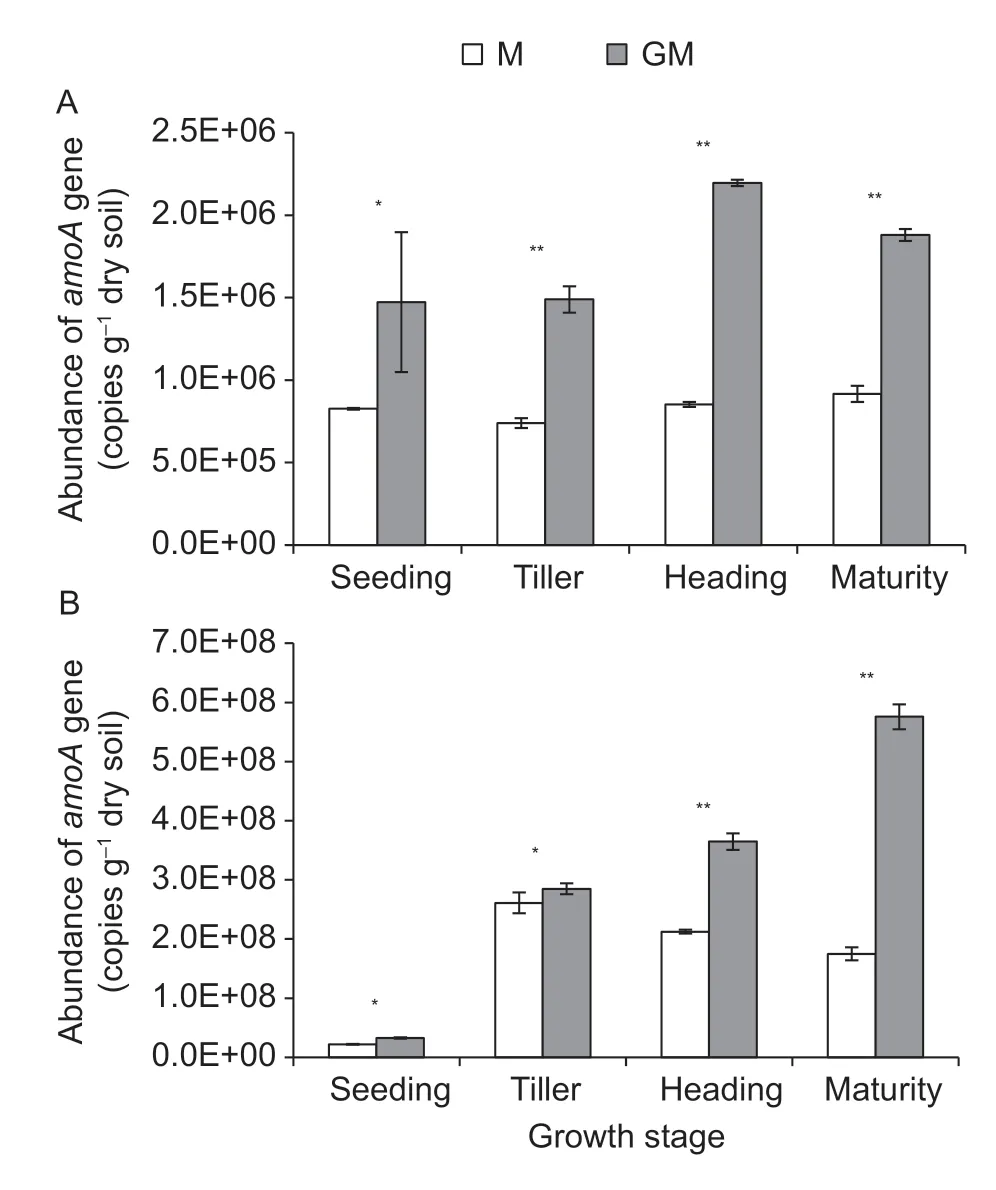
Fig.5 Abundance of amoA gene in the paddy soil with different rice lines during the growth period of rice. A and B indicate bacterial amoA gene and archaeal amoA gene,respectively.M,non-cry1Ab rice MH3301;GM,cry1Ab transgenic rice mfb-MH3301-1. *,P<0.05,**,P<0.01. Bar indicates SD (n=4).
3.3.Soil chemical properties in the paddy soils
Soil pH values in the paddy field cultivated withcry1Abtransgenic rice line GM were lower than those of non-cry1Abrice cultivar M (P<0.05) at the tiller,heading and maturity stages of rice,while no obvious differences in soil available N,P or K,or organic C were seen between cultivar M and line GM in the various growth periods of rice (Table 7). At the same time,soil available N,P or K decreased with the growth of rice. In the paddy soils with either cultivar M or line GM,differences in soil available K were statistically significant among the seeding,tiller,heading and maturity stages of rice (P<0.05) (Table 7).
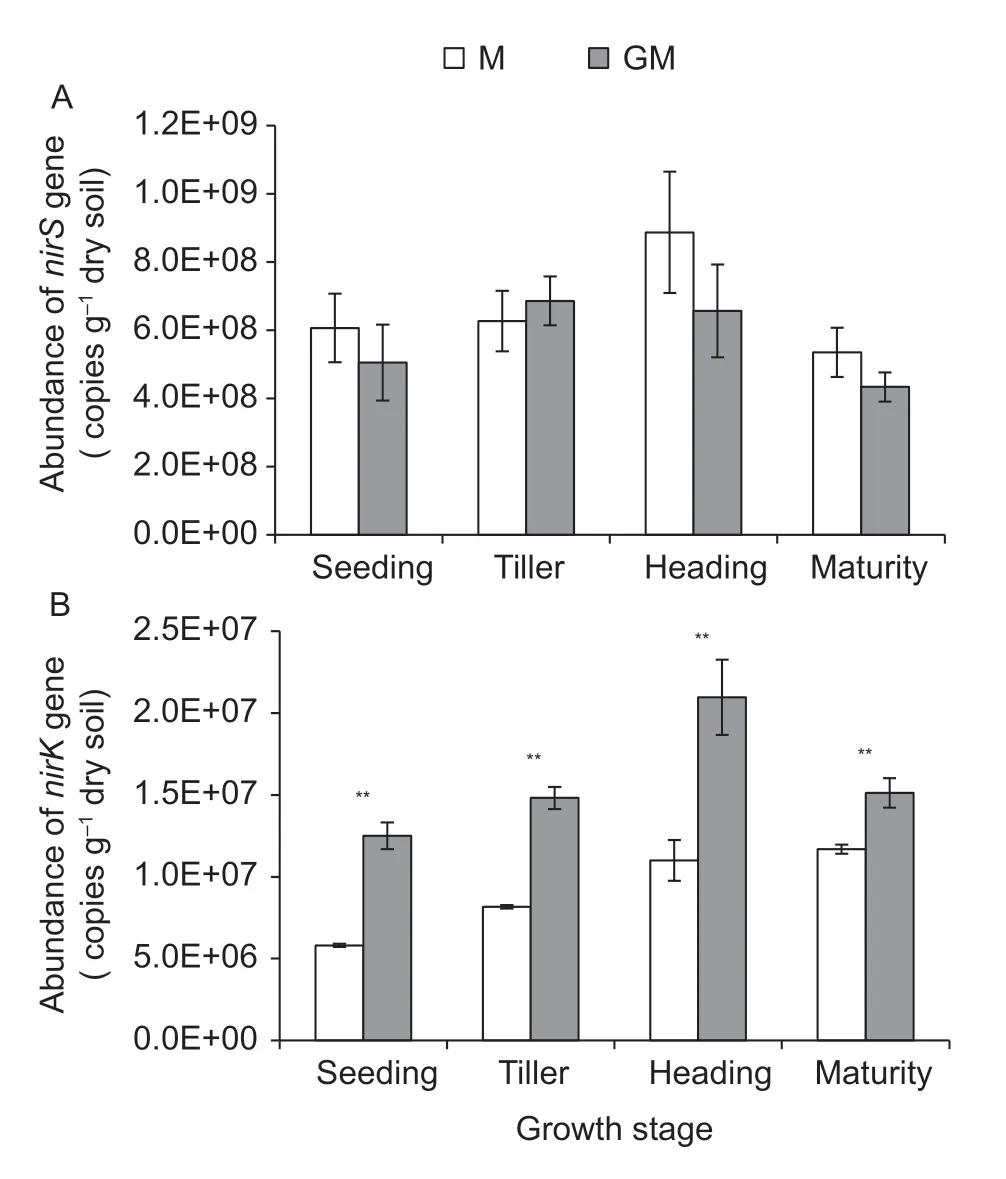
Fig.6 Abundance of denitrifying bacterial genes in the paddy soil with different rice lines during the growth period of rice.A and B indicate nirS gene and nirK gene,respectively. M,non-cry1Ab rice MH3301;GM,cry1Ab transgenic rice mfb-MH3301-1. **,P<0.01. Bar indicates SD (n=4).
Furthermore,the soil chemical properties were used as environmental factors for RDA of the bacterial communities.The results of RDA indicated statistically significant correlations between available N,available K and pH and the bacterial communities in the paddy soils,at significance levels of 0.01,0.01 and 0.05,respectively. Spearman correlations between environmental factors and microbial species are shown in Fig.7. The soil pH had statistically significant (P<0.05 or 0.01) negative correlations with the relative abundances of some species in the paddy soil,such asCandidatus_Koribacter,Acidibacter,Candidatus_Solibacter,Bryobacter,Geobacter,and others,which mostly belong to Acidobacteria. The available N and K had statistically significant (P<0.05 orP<0.01) positive correlations with the relative abundance ofClostridumin the paddy soil,while they had significant (P<0.05 orP<0.01) negative correlations with the relative abundances of some species such asMethylomonas,Sideroxydansand others.
3.4.Plant nutrition,agronomic and yield characters of the rice
The plant height of line GM was significantly (P<0.05)greater than that of cultivar M,and effective tiller number,seed setting rate,1 000-grain weight and yield per plot in line GM were markedly higher (P<0.05) than those in cultivar M (Table 8). There were no differences in plant nutrition of total N,P or K,or other agronomic characters such as grain number per panicle between line GM and cultivar M(Table 8).
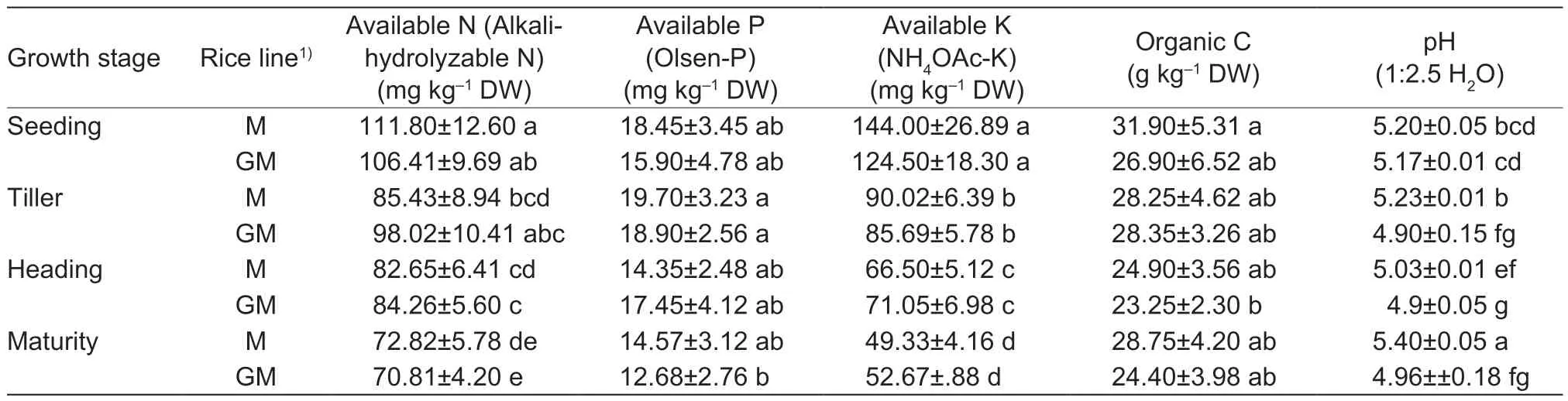
Table 7 Chemical characters in paddy soil with different rice lines
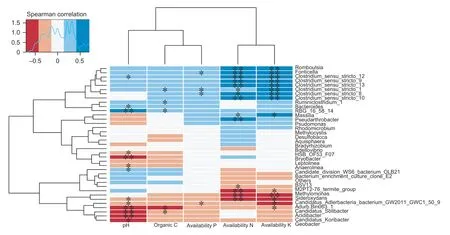
Fig.7 Spearman correlations between soil chemical properties and soil microbial species. Blue represents positive correlations,red stands for negative correlations,and the deeper the color is,the higher the correlation. *,P<0.05,**,P<0.01.
4.Discussion
Bt transgenic plants are genetically modified to express the insecticidal protein fromBacillusthuringiensis. The Btcry1Abgene has been transferred into plants of rice,cotton,corn,canola,tobacco and potato,which are then cultivated to resist lepidopteran pests (James 2011). The influence ofcry1Abtransgenic rice on the bacterial community in rice paddy has rarely been investigated by a high-throughput technique such as Illumina MiSeq. Our previous study showed that two events ofcry1Ac/cptitransgenic rice had no obvious effects on the diversity of bacterial or fungal communities in paddy soil during the growth period of rice,but it only used the detection method of PCR-DGGE (Songet al.2014). In this study,we focused mainly on using a highthroughput technique,Illumina MiSeq,to detect bacterial communities in soil planted withcry1Abtransgenic rice and its parental non-cry1Abrice.
In the experimental field site,cry1Abtransgenic rice mfb-MH3301-1 (GM) and the parental MH3301 (M) have been planted for five years. Data from the field experiment in the fifth year showed that some agronomic characteristics and yield,such as plant height,yield per plot and others,in line GM were better than those in the parental rice cultivar M. These findings indicated thatcry1Abtransgenic rice showed improved plant growth and increased yield in this study. However,the plant nutrition and most of the chemical characters in paddy soil withcry1Abtransgenic rice were similar to those of non-cry1Abrice,except for soil pH.The pH values were significantly lower in the paddy soil planted withcry1Abtransgenic rice at the tiller,heading and maturity stages of rice,with the greatest difference in soil pH between line GM and cultivar M at the maturity stage of rice. This indicates that for a period of at least five years the release ofcry1Abtransgenic rice has an influence on soil pH,promoting acidification of the soil,and the acidification was the most obvious at the maturity stage of the rice. The decrease in soil pH fromcry1Abtransgenic rice may be attributed to changes in the composition and amount of plant residues caused by thecry1Abtransgenic character (Doneganet al.1995).

We found that the diversity of soil bacterial communities of line GM was similar to that of cultivar M at any growth stage of rice,based on the majority of the alpha diversity indexes,such as Chao1,Shannon and Simpson. Nevertheless,the soil bacterial community compositions were different between line GM and cultivar M during the growth season of rice according to NMDS or PCoA with ANOSIM and ADONIS analyses. The results of this study indicated that plantingcry1Abtransgenic rice had a more obvious impact on the composition of soil bacterial communities than on their diversity,despite some diversity indexes of soil bacterial communities,such as PD_whole_tree and Observed_species,being different between line GM and cultivar M at some growth stages of rice. Otherwise,the growth period of rice had an obvious impact on the soil bacterial communities in this study.
The relative abundances of soil bacterial phyla were significantly different betweencry1Abtransgenic rice line GM and its control cultivar M at the different growth stages of rice. The relative abundance of Acidobacteria significantly increased in the paddy soil with line GM at the maturity stage of rice,which should be in accord with the marked decrease in soil pH of line GM.Acidobacteria is one of the highly diverse phyla of the domain bacteria,which inhabit a wide variety of terrestrial and aquatic habitats and are particularly abundant in acidic soils (Dedysh and Sinninghe 2018). Previous studies showed that an acidic soil environment was favorable for the growth of Acidobacteria (Claudiaet al.2002;Liuet al.2014). In this study,the decrease in soil pH of the field withcry1Abtransgenic rice coincided with the increase in the relative abundance of Acidobacteria. This result was consistent with previous studies which showed a negative correlation between the relative abundance of Acidobacteria and soil pH based on MiSeq sequencing (Wanget al.2019;Yanget al.2019). Furthermore,based on the present results of LEfSe,the specific bacterial phylum related to the line GM should be Acidobacteria,and the increase in the relative abundance of Acidobacteria played an active role in causing the differences in the compositions of soil bacterial communities between line GM and cultivar M.In addition,the relative abundance of Bacteroidetes was higher in the paddy soil with cultivar M than with line GM at the tiller stage of rice. It was also the specific bacterial phylum related to the cultivar M based on the results of LEfSe,and the enrichment of Bacteroidales,and Bacteroidetes_vadinHA17 contributed to the differences in soil bacterial communities between cultivar M and line GM. As to the decreases in the relative abundances of Proteobacteria and Nitrospirae in the paddy soil with line GM at the seeding and heading stages of rice,respectively,this correlation needs to be further tested in the field experiment.
Soil chemical and physical properties,such as soil nutrients,pH,moisture,temperature and ventilation,have well-known influences on the abundance and diversity of soil microbes (Yang and Crowley 2000;Marschneret al.2004). In this study,we also observed that soil pH and soil available N or K also had significant effects on the composition of microbial communities in paddy soil. In this study,the detection of the bacterial community by the Illumina MiSeq platform indicated that a change in the relative abundance of Acidobacteria,according to the variation of soil pH,was an important factor influencing the differences in bacterial communities in the paddy soils between cultivation withcry1Abtransgenic rice and its control cultivar.
The microbial functional genes in nitrification and denitrification were tested in this study,and they drive the transformation and utilization of nitrogen in the soil. Ammonia oxidation catalyzed by ammonia monooxygenase (amo) is known to be the first and rate-limiting step of nitrification (Caponeet al.1991). Ammonia oxidation is believed to be carried out by both ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA). The former belongs either to β-proteobacteria isolated from soils (Stephenet al.1996) or to γ-proteobacteria which has only been isolated from marine and brackish water (Woeseet al.1985),whereas the later belongs to Crenarchaeota (Könnekeet al.2005;Schleprtet al.2005).The functional genes ofnirSornirKare the main molecular markers of denitrification for the catalytic reaction of nitrite reduction (Zumftet al.1997;Brakeret al.2000).
We found that the release ofcry1Abtransgenic rice promoted an increase in the abundance of ammoniaoxidizing bacterialamoAgene,ammonia-oxidizing archaealamoAgene and denitrifying bacterialnirKgene in the paddy soil at any of the growth stages of rice. Our previous study showed thatcry1Ac/cptitransgenic rice had no obvious effect on the abundance of ammonia-oxidizing bacteria in paddy soil using the test of real-time PCR based only on the 16S rRNA gene (Songet al.2012). However,the test of real-time PCR based on the functional gene ofamoAin this study suggested that plantingcry1Abtransgenic rice did have an effect on the numbers of ammonia-oxidizing bacteria and archaea. A study using drop digital PCR based on theamoAgene also indicated that Bt+CpTItransgenic cotton can slow the rate of ammonia transformation by impacting the number of ammonia-oxidizing bacteria(Donget al.2014). At present,there are few reports on the effects of transgenic plants on the functional genes of soil denitrification,such as thenirKornirSgenes. In this study we firstly found that plantingcry1Abtransgenic rice had an effect on the abundance of thenirKgene. In addition,functional gene predictions based on 16S rRNA Illumina MiSeq data by using PICRUSt also showed that the abundance of functional genes of nitrogen metabolism was significantly increased in the paddy soil with line GM.The above results indicated thatcry1Abtransgenic rice may have an effect on the conversions of soil nitrogen in paddy fields,such as nitrification or denitrification. Moreover,it may influence nitrogen fertilizer effectiveness and the environmental problems caused by the loss of nitrogen or the release of N2O. Therefore,the diversity and composition of the functional microbes by deep sequencing of functional genes and GeoChip analysis are worth studying further.
5.Conclusion
The results of this study demonstrated that the planting ofcry1Abtransgenic rice could lead to a change in the bacterial community composition in paddy soil,at least in this fiveyear experiment. The increase in the relative abundance of Acidobacteria was in accord with the soil acidification in the field withcry1Abtransgenic rice. At the same time,the release ofcry1Abtransgenic rice could lead to an increase in the abundance of microbial functional genes which drive the conversion of nitrogen in soil,such as theamoAandnirKgenes.
Acknowledgements
The study was conducted under the support of the National Science and Technology Major Project of the Ministry of Science and Technology of China (2016ZX08001-001).
Declaration of competing interest
The authors declare that they have no conflict of interest.
杂志排行
Journal of Integrative Agriculture的其它文章
- Low glycemic index:The next target for rice production in China?
- Do cooperatives participation and technology adoption improve farmers’ welfare in China? A joint analysis accounting for selection bias
- Impacts of household income on beef at-home consumption:Evidence from urban China
- The water-saving potential of using micro-sprinkling irrigation for winter wheat production on the North China Plain
- Synergistic effect of Si and K in improving the growth,ion distribution and partitioning of Lolium perenne L.under saline-alkali stress
- Microbial community dynamics during composting of animal manures contaminated with arsenic,copper,and oxytetracycline
