Molecular Cloning, Expression and Characterization of Peroxisome Proliferators-Activated Receptors Gamma in the Sea Urchin (Strongylocentrotus intermedius)
2021-03-06QUANZijiaoHANLingshuCAOYueGAOPingpingLIUXiaoyuCHANGYaqingandDINGJun
QUAN Zijiao, HAN Lingshu, CAO Yue, GAO Pingping, LIU Xiaoyu,CHANG Yaqing, and DING Jun
Molecular Cloning, Expression and Characterization of Peroxisome Proliferators-Activated Receptors Gamma in the Sea Urchin ()
QUAN Zijiao#, HAN Lingshu#, CAO Yue, GAO Pingping, LIU Xiaoyu,CHANG Yaqing, and DING Jun*
,,,116023,
Peroxisome proliferators-activated receptor gamma () plays important regulatory roles in adipocyte differentiation. In this study, we cloned the full-length sequence of thegene and analyzed its expression profile in different developmental stagesand tissues of. The full-length cDNA ofcontains 2286 base pairs (bp)with a putative open reading frame of 1755bp, and the gene encodes encoding a polypeptide of 584 amino acid residues. The predicted molecular massof the protein is 67.27kDa, and its theoretical isoelectric point (pI) is 10.07. The protein contains conserved motifs, including an RRM (RNA recognition motif) domain.expression with the highest level was observed in the gonad, and the lowest was observed in the tube feet of. Time-course expression measurements at different developmental stages showed that the highest expression level ofis in the eggsand its weakest expression level isin the 32-cells stage. Knock-down ofby specific siRNA revealed thatexpression is significantly decreased in the gonads and intestines 48h post-transfection,indicating that theis a downstream target gene of.This finding suggests thatandhave positive regulatory effectsin regulating adipocyte differentiation. Changes in fatty acid levels in the gonads before and afterinterference were assessed, and decreased C18:2(trans, n−6) and C20:3(n−6) levels were observed 48h after siRNA transfection. The results showed the function ofin fatty acid anabolism,The data are helpful to improve the current understanding of the fatty acid synthesis pathways and regulatory mechanisms in. They also provide an experimental basis for improving fatty acid synthesis in sea urchins, which is important for cultivating sea urchins with high nutritional value.
;;gene cloning; siRNA
1 Introduction
Fatty acids (FAs) play important roles in biological processes, and are essential molecular components of all organisms. As fundamental components of phospholipids in cell membranes, fatty acids affect the composition of cell membranes and influence organism health. Fatty acids are divided into saturated fatty acids, monounsaturated fatty acids (UFAs) andpolyunsaturated fatty acids (PUFAs) depending on their degree of saturation. PUFAs are basic components of cell membranes that control cell configuration and permeability (Xiao., 2001). These fatty acids play essential roles in biological growth and lipid metabolism. Studies have shown that PUFA anabolism in fish is affected by external factors, such as nutrition and the environment, as well as genetic factors, which may influence the mRNA level of key enzymes in the highUFA synthesis pathway (Xie., 2013). Cook(2000) and Barberá(2011) studied the fatty acid composition of sea urchin gonadsand showed that three PUFAs mainly contribute to gonad composition, namely, eicosapentaenoic acid (C20:5n−3), arachidonic acid (C20:4n−6) and palmitoleic acid (C16:1n−7). PUFAs are also essential to sea urchin growth and development. Studies have shown that the growth rate of sea urchins rich in PUFAs is superior to that of control group (Ding, 2014).
Peroxisome proliferator-activated receptors () aremembers of the nuclear receptor superfamily that regulates the expression of target genes, particularly those asso- ciated withlipid metabolism (Zhu., 2000). Issemann(1990) discovered that this protein family is activated by fatty acid-like compounds called peroxisome pro- liferators; and the nuclear receptors came to be known as. The proteins can be divided into three structural type, namely, α, β (or δ), and γ, which are products of dif- ferent genes (Issemann, 1990; Dreyer, 1992; Kliewer, 1994; Zhu, 1995).Mammalianstrongly influences lipidmetabolism, cell proliferation, and inflammation.signalingpathways control lipid uptake, transport, storage, and disposal(Walczak and Tontonoz, 2002).also regulates the expression of gene networks required for cell proliferation, differentiation, morphogenesis, and metabo- lic homeostasis, thus highlighting its ability to activate li- pogenic genes and adipocyte differentiation. Given its in- teresting properties, the functions ofhave become an important research area in recent years.
Thegene sequences of several fish species, including(Ruyter.,1997),(Leaver.,1998),(Ibabe,2002),(Boukouvala.,2004),(Ibabe.,2004),(Batista-Pinto, 2005),(Mimeault.,2006),(Tsai.,2008), and(He.,2012), have been cloned and functionally identified. However, no reports ofiden- tification inhave yet been published
Uncoupling protein-2 () was discovered in 1997 as ahomologue of the brown-fat UCP (UCP1). Similar to UCP1, whichis able to dissipate caloric energy as heat by uncoupling mitochondrial respiration from ATP production,can uncouple respiration, at least(Fleury, 1997).is present in many tissues involved in fuel metabolism. Its expression is increased in fat and muscle in response to elevated circulating free fatty acidsfrom fasting and high-fat feeding (Medvedev., 2001).
Medvedev. (2001)proposed a role foras a mediator of physiological changes in, because thiazolidinediones increaseexpression in the cells of tissues related to fuel metabolism (Aubert, 1997). The-dependent pathway might play a role in mediatingthe response ofto fatty acids because treatment withagonists can increasemRNA levels bothand(Aubert, 1997; Camirand, 1998). However, the regulatory relationship between these two genes inremains unclear.
In 1989,was introduced from Japan by Dalian Ocean University and promoted in coastal areas,such as the Liaoning and Shandong Peninsulas, of China. Today, this species is one of the main sea urchin varieties cultivated in China (Chang., 2004). The only edible componentof the animal is the gonad, and its nutritional value is mainly reflected by the medicinal effects of its fatty acids (Zuo., 2016). These fatty acids are extremely complex and contribute to the growth and reproduction of the organism (Russell, 1998; Palacios., 2007; Wei., 2016). Many types of fatty acids occurin sea urchins, and their total amount outweighs those found in fish species from both seawater and freshwater environments. Sea urchins feature much higher amounts of UFAs compared with marine fish and shrimp (Wang., 2006). UFAs from gonads are efficacious for preventing certain cardiovascular and cerebrovascular diseases and have high nutritional and medicinal value. Previous studies showed that UFAs are closely related to the growth, development, and reproduction of sea urchins (Kang and Leaf, 1996; Tavazzi., 2008; Carboni., 2013).
is a fatty acid-related gene. To understand the function ofin, we used rapid amplification of cDNA ends (RACE) polymerase chain reaction (PCR) to obtain the full-length cDNA sequence encodingin this species. Then, we investigated the expression patterns ofusing fluorescence quantitative PCR and studied the relationship betweenand.
2 Materials and Methods
2.1 Animals, Rearing Conditions, and Sample Collection
The 2-year-oldsamples used in this study were bred at the Key Laboratory of Mariculture and Stock Enhancement in the North China Sea, Ministry of Agriculture and Rural Affairs, China. The healthy sea urchins were homogeneous in size with shell heights of (29.11±2.10)mm, shell lengths of (50.13±3.46)mm and weights of (41.53±5.05)g.
Tube feet, peristomial membrane, Aristotle’s lantern, coelomocyte tissues, gonads, and intestines were dissected from nine sea urchins. Samples from the different developmental stages were also collected as follows.Three fe- males and three males were induced to spawn by injection of 1mLof KCl (40μL per gram of body mass) into the coelom via the peristomial membrane (Kelly., 2000). The fertilized ovum develops from the multicellstage to the blastocyst stage through cleavage, enters the gastrula stage after blastocyst rotation, and then develops to the planktonic stage. The planktonic larvae are divided into two stages,including prismatic larvae and long-winged larvae. In the late stage of the long-winged larvae, a larva’s wrist gradually disappears, and it eventually becomes a juvenile sea urchin after a series of metamorphic transformations. Samples were pooled and passed through a 400μm sieve to remove impurities, taken up by pipettes, and placed in a 1.5mL centrifuge tube. All tissue and developmental stage samples were frozen in liquid nitrogen immediatelyand stored at −80℃ forRNA extraction and fatty acid ana- lyses.
2.2 Cloning of a Partial PPARγ cDNA Fragment
Total RNA was extracted from the intestines of threeindividuals using Trizol reagent (Invitrogen) and digested by the RNase-free DNase I (TaKaRa, China) to remove any possible genomic DNA contaminants. First- strand cDNA was synthesized with a reverse transcription system (TaKaRa) according to the manufacturer’s proto- col. A pair of degenerate primers,-F/-R (Table 1), was designed based on the conserved regions ofsequences from other species and used for PCR under following conditions: initial denaturation at 94℃ for 5min, followed by 35 cycles of94℃ for 30s, 60℃ for 30s, and 72℃ for 45s, and a final extension at 72℃ for 10min. The PCR product was cloned into the pMD18-T simple vector (TaKaRa) and sequenced.

Table 1 Primers used in this study
2.3 Cloning the Full-Length cDNA of PPARγ Using Rapid-Amplification of cDNA Ends (RACE) PCR
The partial cDNA sequences ofwere obtained from our results of cloning of a partialcDNA frag- ment, and the gene-specific primers were designed ac- cording to these cDNA sequences by Primer Premier 5.0. The nucleotide sequences of all of the primers used in this study are shown in Table 1, and the primers were obtained from Sangon Biotech (Shanghai, China). RACE products were extracted and purified using the Quick Gel Extraction Kit (Transgen Biotech, China). The purified products were subcloned into PEASY®-1 Cloning Vector (Transgen Biotech, China) and immediately transformed into Trans1-T1 phage-resistant chemically competent cells (Transgen Biotech, China). Positive recombinants were identified by colony PCR using M13 primers (Transgen Biotech, China) and finally sequenced at Sangon (Shanghai, China) for se- quence confirmation.
2.4 Bioinformatics Analysis
The nucleotide sequence of the full-lengthcDNA sequence was analyzed using the Basic Local Alignment Search Tool (BLAST) program at the National Center for Biotechnology Information (https:// www.ncbi.nlm.nih.gov/blast/), and the deduced AAsequence was determinedusing the Expert Protein Analysis System (http://www.expasy.org/). The open reading frame (ORF) was identi- fied with ORF Finder (https://www.ncbi.nlm.nih.gov/or- ffinder/). The physical and chemical parameters of the de- duced protein were computed using the ProtParam tool (http://web.expasy.org/protparam/). The computed parame- ters also included the molecular weight, theoretical iso- electric point (pI), and grand average of hydropathicity (GRAVY). The presence and location of the signal peptide cleavage sites in the AA sequence were predicted by SignalP 4.1 Server (http://www. cbs.dtu.dk/services/Sig- nalP/). The functional domains and important sites of the protein were predicted by InterPro (http://www.ebi.ac.uk/interpro/). Multiple sequence alignment of the deduced AA sequences derived fromwas performed using the ClustalW program (http://www.ch.embnet.org/software/ClustalW.html). A phylogenetic tree was constructed according to the AA sequences of the selectedusing the neighbor-joining (NJ) method in MEGA 6.0 software.
2.5 Real-Time PCR Analyses of PPARγExpression
The relative expression levels ofmRNA transcripts in differenttissues and at different developmental stages (., eggs, 2 cells, 16 cells, 32 cells, 64 cells, blastula, gastrula, four-armedarm larvae, and six-armedlarvae) were analyzed by quantitative real-time PCR (qRT-PCR), which was conducted on an Applied Biosystems7500 real-time PCR system (Foster City, CA, USA) following the manufacturer’s instructions on the use of SYBR Premix Ex Taq(SYBR PrimeScript™ RT-PCR Kit II, Ta- KaRa, Japan). TherRNA gene was used as the internal reference gene. Amplification was performed in a total volume of 20μL containing 2μL of 1:5 diluted original cDNA, 10µL of 2×SYBR Green Master mix (TaKaRa, Ja- pan), 0.4μL of ROX Reference DyeII, 6μL of PCR-grade water and 0.8μL (10mmolL−1) of each primer. The reaction was performed as follows: 40 cycles of 94℃ for 5min, 94℃ for 30s, 60℃ for 30s, and 72℃ for 30s. The final extension step was conducted at 72℃ for 5min. Three independent biological replicates and three technical re- petitions of each group were carried out. At the end of the PCR reactions, melting curve analysis of the amplification products confirmed that a single PCR product was present in these reactions. The relative expression levels of the target gene were calculated by the 2−ΔΔCTmethod described by Livak and Schmittgen(2002).
2.6 RNAi in S. intermedius and Identification of a Relationship Between PPARγ and UCP2
Small interfering RNAs (siRNAs) targeting(Ta- ble 1) were designed and synthesized by GenePharma (Shanghai, China). Six healthy sea urchins with consistent growth, an average shell diameter of (43.25±1.19)mm and an average wet weight of (25.36±2.25)g were randomly divided into the negative control and experimental groups with three individuals in each group. The first group was pipetted with 5μLof NC (negative control), 5μL of Lipo6000TM transfection reagent (Beyotime, Shang- hai), and 40μL PBS buffer; the second group was pipetted with 5μLof siRNA, 5μLof Lipo6000TMtransfection reagent (Beyotime), and 40μL of PBS buffer. These com- ponents were mixed evenly, allowed to stand for 5min, and then injected into the body cavity through the sea urchin’s mouth. After 48h, total RNA was extracted from the coelomocyte tissues, intestines, and gonads and reverse-transcribed. Finally, we detected the relative expression le-vels ofandgenes using qRT-PCR and detected changes of fatty acidcomposition before and after interference by capillary gas chromatography.
2.7 Statistical Analysis
All data were expressed as mean±SD (standard deviation). All analyses were performed using one-way ANOVA (Analysis of variance) with SPSS software (version 16.0). Thevalue was adjusted for multiple tests using a falsediscovery rate (Benjamini-Hochberg),and<0.05 was considered statistically significant
3 Results
3.1 Sequence Analysis of PPARγ
The full-length sequence ofcomprised 2286 base pairs (bp), including a 5’ untranslated region (UTR) of 145bp, a 3’ UTR of 386bp and a putative ORFof 1755bp, The gene encoded a protein with 584 AA residues. The predicted molecular mass ofis 67.27kDa, and its theoretical pI is 10.07. The protein sequence included an RRM (RNA recognition motif) domain (Fig.1).
3.2 Analysis of the Amino Acid Composition of PPARγ
The AA hydrophilicity/hydrophobicity of theen- zyme was analyzed using the online ProtScale database, which showed that the aliphatic index is 30.97, the maxi- mum hydrophilic coefficient is 1.389, the minimum hydrophilic coefficient is −3.500, and the GRAVY is 0.835. These data suggest the protein is hydrophobic in nature.
3.3 Multiple Alignment and Phylogenetic Analyses of PPARγ
The deduced AA sequence ofinwas analyzed using BLAST. The results showed thatshares high homology withfrom other vertebrates and invertebrates, including(97%),(50%),(47%),(52%),(46%), and(53%). We cons- tructed a phylogenetic tree using the NJalgorithm to reveal the phylogeny of, and the results are shown in Figs.2 and 3.
3.4 Tissue Expression of PPARγ
Tissue distributions ofmRNA were detected by qRT-PCR. Thegene was expressed in all tissues tested. Expression levels were the highest in the gonads and the lowest in the tube feet. The coelomocyte tissues also showed lowexpression levels (Fig.4).
3.5 Differential Expression of PPARγ at Different Developmental Stages
To understand the role ofat different developmental stages of sea urchin and the synthesis of fatty acids during development, we examinedexpression in the egg, 2-cells, 16-cells, 32-cells, blastula, gastrula, four-armed larva and six-armed larva stages of.was expressed at all nine stages of development. The relative expression level ofwas highest during the egg stage and lowest in the 32-cell stage.expression initially decreased and then increasedover the entire developmental process of sea urchin (Fig.5).

Fig.1 Nucleotide and deduced amino acid (AA) sequences of PPARγ. The initiation codon is shown in italics. The asterisk (*) in dicates a terminal codon. RRM (RNA recognition motif) combined with its domain is shown in the shaded area.

Fig.2 ClustalX alignment of S. intermediusPPARγ. Amino acid (AA) sequences.The shaded regions indicate identical residues. Other conserved but dissimilar AAs are shaded in gray.
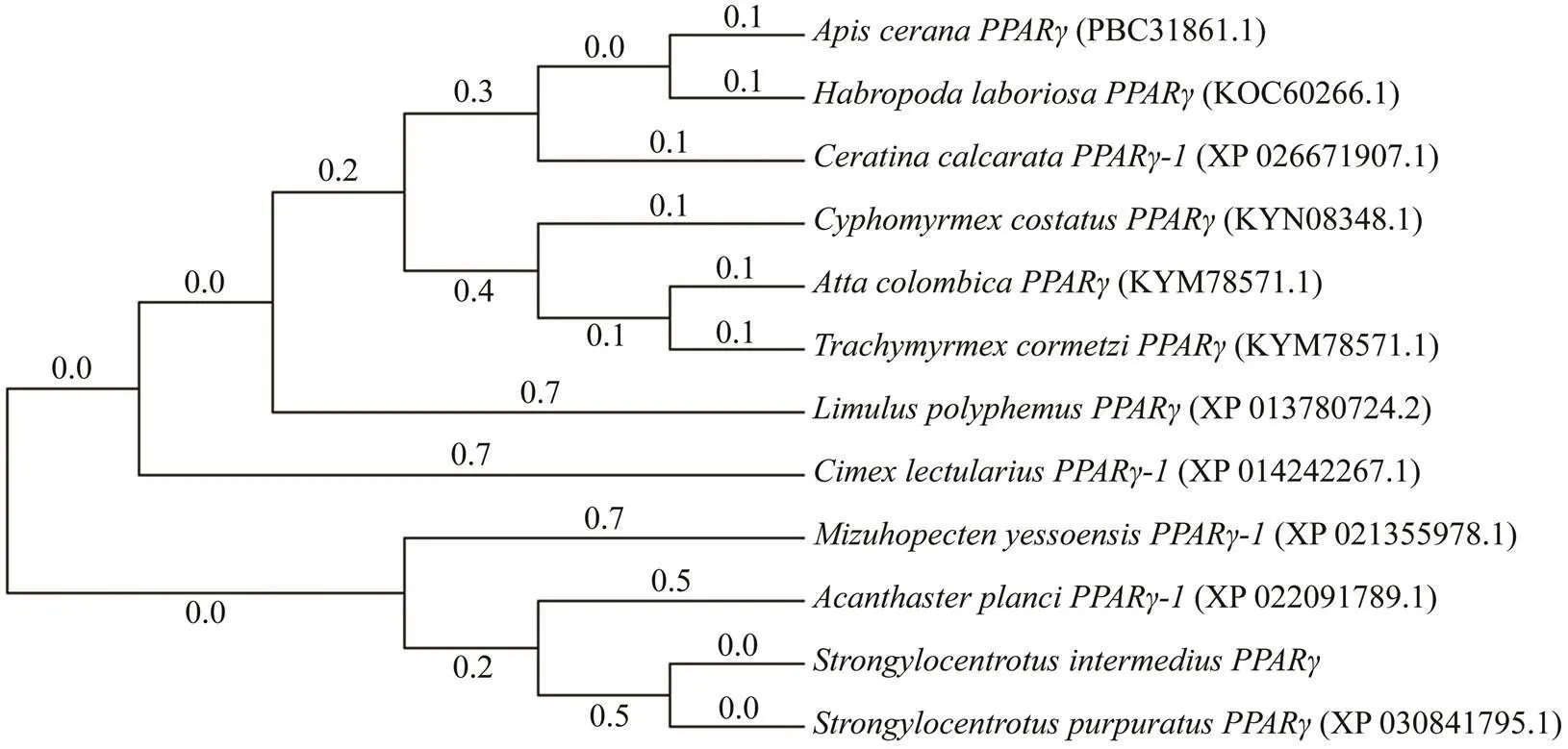
Fig.3 Consensus neighbor-joining (NJ) tree based on the amino acid sequences of PPARγ genes from other species. The evolutionary historywas inferred using the NJ method. Evolutionary analyses were conducted in MEGA7.0.

Fig.4 Relative expression levels of PPARγ in different tissues. Each vertical bar represents the mean±SD (n=3).18SrRNA was used as the reference gene. Different Roman letters above the bars provided indicate significant differences in different tissues at P<0.05.
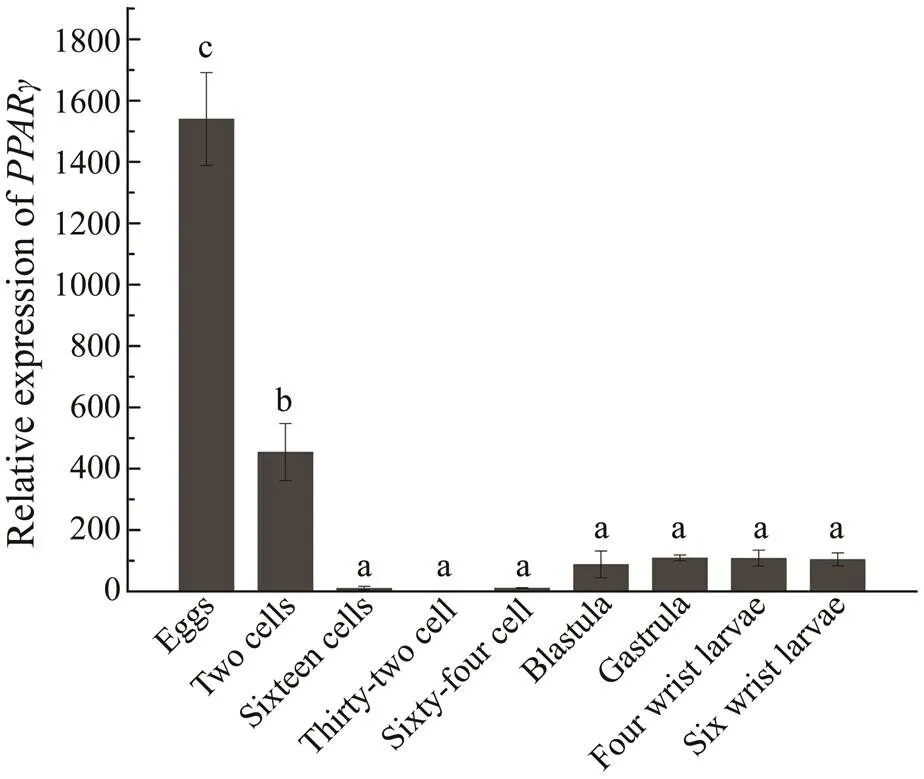
Fig.5 Relative expressionof PPARγ in S. intermedius at the different developmental stages. 18S rRNA was used as the reference gene. Different Roman letters above the bars provided indicate significant differences in different tissues at P<0.05.
3.6 Relationship Between PPARγ and UCP2
siRNA experiments were performed to understand the regulatory relationship betweenand. Compared with that in the negative control, 48h of trans- fection ofsiRNA reducedexpression in the coelomocyte tissue, intestines and gonads by 43.17%, 16.16%, 83.56%, respectively. Compared with that in the negative control, 48h of transfection ofsiRNA reduced the expression ofmRNA in the coelomocyte tissue, in- testines, and gonads by 5.96%, 16.29%, and 33.75%, respectively (Fig.6).

Fig.6 (A) PPARγ mRNA levels of RNAi-PPARγ. (B) UCP2 mRNA levels of RNAi-PPARγ. Values are presented as mean±SD (n=3). Asterisks indicate significant differences,P<0.05.
3.7 Changes in Fatty Acid Levels Before and After PPARγ siRNA Interference
To determine the role ofin fatty acid synthesis, we examined the changes of fatty acid levels in the gonads of sea urchins, before and aftersiRNA inter- ference. The results showed that C4:0, C18:2(trans, n−6) and C20:3(n−6) levels were decreased aftersiRNA interference (Fig.7).

Fig.7 Changes in fatty acid contents before and afterPPARγ siRNA interference (n=3). Asterisks indicate significant differences,P<0.05.
4 Discussion
Sea urchin is a delicacy in many countries, including China, Japan, and France. Essential nutrients such as li-pids and PUFAs are stored in the gonads. They not only determine the nutritional value of this delicacybut also en-sure the normal growth and reproduction of sea urchins in culture (Han., 2019).gonads are rich in fatty acids, especially UFAs.can activate lipogenic genes and adipocyte differentiation. In this study, we characterized and analyzed the expression of acDNA in. The full-length cDNA of, which includes 1755bp was obtained. Phylogenetic ana- lysis revealed thatandareclustered together, thus indicatingthat these two species are closely related and have a long relationship with other vertebrates. Similarities in the sequence, conserved domains, and phylogenetic analysesof these species provide supporting evidence that the cloned geneis indeed.
Many studies have investigated the relationship between the tissue expression of fishand their functions. Tsai(2008) cloned,,andfrom, and found thatis expressed in all tis- sues, particularly in visceral fat deposits. He(2012) cloned the threegenes fromand determined thatis highly expressed in the liver and mi- cro-expressed in other tissues. Because liver isone of the most important organs regulating glucose and lipid metabolism, these results confirm thatis an important transcriptional regulator of glucose andlipid metabolism. In this study, qRT-PCR analysis showed thatis expressed in all tissues of,especially in the gonads, followed by the intestines and Aristotle’s lantern. The high expressionofobserved in the gonads agrees with the hypothesis that the UFA content of the gonads of sea urchins is high (Chang., 2004). Our data partly agree with the tissueexpression patterns ofmRNA determined in the loach, where the highest levels was found in the brain, followed by the male gonads and then the female gonads (Cui.,2018). During gonadal development in sea urchin, fatty acids contribute to the accumulation of ovarian lipids before spawn-ing to ensure nutrient accumulation of yolkand maintain normal embryo development and early larvae development. Unlike other animals, both female and male sea urchins can accumulate high levels of yolk protein, which can reach 10%–15% of their total protein content (Chang, 2014). Yolk protein is a lipoproteinwith high lipid contents and it is also an important nutrient and energy source for larval de- velopment (Chang., 2004). In the loach, the expressions ofandcoincide withregulation,implying thatmay be involved in long-chain LC-PUFA biosynthetic pathways (Cui., 2018).
The normal accumulation of total lipids, triglycerides, phospholipids, and UFAs in aquatic animals is critical for gonadal and embryonic development, as well as early lar- val growth (Palacios., 2007; Barberá., 2011). In previous studies, the utilization of different fatty acids in embryos varied with the developmental stage (Yao and Zhao, 2006). During the early development of the sting-ray embryo, for example, PUFAs are present at higher pro-portions in the blastocyst stage, the gastrula stage, and during organogenesis, indicating increased PUFA levels are involved in metabolism,decomposition, and energy preparation for embryonic development in the yolk during these periods (Yao., 2009). Inthe present study,was expressed during all developmental stages of sea urchin.expression was the highest in the eggs because the protein is directly involved in the maturation of germ cells in the testes and oocytes (Huang, 2008). During the transition period from eggs to 32-cells embryos,expression gradually decreases because of cell division.expression gradually increases from the 32-cells stage to the gastrula stages because the sea urchin differentiates out of the intestine in the latter stage and ingests nutrients outside this organ.After the gastrula stage,expression stabilizes. Because rich UFAsare available in the food, marine animals are not required to synthesize their own UFAs; however, lower invertebrates appear to retain ancapacity forUFA synthesis (Zuo., 2016). The results indicate thatexpression is closely related toUFA synthesis during the early developmental stages of sea urchin. This inference provides a basis for investigating fatty acid synthesis at the different developmental stages of sea urchins.
is an important mitochondrial inner membrane protein associated with glucose and lipid metabolism (Zhou., 2016). Adipose-specific increases in expression ofhave been observed in obesity-resistant mouse strains in response to high-fat diets (Fleury, 1997). Fasting-in-duced increases of circulating free fatty acidslead to astimulation ofexpression in adipose tissue and muscle,whereas subsequent refeeding suppressesex-pression (Boss, 1997; Samec, 1998). These research findings have provide compelling evidence of the involve-ment of fatty acids inregulation, though the molecular mechanisms still unknown. It is possible thatmediates its response to fatty acids because some fattyacids through a-dependent pathway,considering some fatty acids can specifically bind, that acts as a natural ligand for this transcription factor (Forman., 1997; Kliewer., 1997). To understand the relationshipbetweenand, we targeted the siRNA ofand detectedandexpression using qRT-PCR. The results showed thatexpression inwas significantly down-regulated after 48h of interference (<0.05); and the interference efficiency was the highest in the gonads followed by the intestines. Similar to the resultsof, the expression ofalso significantly decreased in the gonads and intestines. This result may be due to the fact thatactivatesindirectly by altering the activity or expression of other transcription factors that bind to thepromoter (Medvedev.,2001). Thus,we speculate thatis located downstream ofand a positive regulatory relationship exists between these two genes.
The gonads are important tissues for reproduction and energy storage in sea urchins. Fat is an important source of energy and fatty acids for marine animals (Tong., 1998; Zuo., 2016). To determine the role ofin fatty acid synthesis, we examined the changes of fatty acid levels in the gonads ofbefore and aftersiRNA interference.was sufficiently inhi- bited after 48 h of interference while fatty acid C18:2(trans, n−6) and C20:3(n−6) synthesiscontents also decreased. Socan promotes fatty acid synthesis and affect fatty acid metabolism, which might be realized through the re- gulation of.
In conclusion,cDNA was cloned fromfor the first time, and bioinformatics analysis was conducted on the obtained sequences. The tissue distribution and time-course expression ofat different developmental stageswere detected, while the highest expression level ofwas observed in the gonads. Our results showed thatexpression gradually increases and then decreases during the embryonic development.andhave a positive regulatory effect.expression can directly affect C18:2(trans, n−6) and C20:3(n−6) levels. Future studies should elucidate what kinds of PUFAs are regulated by, which is helpful to de- termine the categories and functions of key enzymes involved in PUFA biosynthesis and reveal the enzymatic me-chanisms of nutritional regulation in.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31772849), the Liaoning Higher School Innovation Team Support Program (No. LT2019003), and the Doctoral Startup Foundation of Liao- ning Province (No. 20170520095).
Aubert, J., Champigny, O., Saint-Marc P., Negrel R., Collins S., Ricquier, D., and Ailhaud G.,1997. Up-regulation ofgene expression by PPAR agonists in preadipose and adipose cells.,238(2):606-11.
Barberá, C., Fernández-Jover, D., López Jiménez, J. A., GonzálezSilvera, D., Hinz, H., and Moranta, J.,2011. Trophic ecology of the sea urchinelucidated from gonad fatty acids composition analysis., 71(4):235-246.
Batista-Pinto, C., Rodrigues, P., Rocha, E., and Lobo-da-Cunha, A., 2005. Identification and organ expression of peroxisome proliferator activated receptors in brown trout ()., 1731 (2): 88-94.
Boss, O., Samec, S., Dulloo, A., Seydoux, J., Muzzin, P., and Gia- cobino, J. P., 1997. Tissue-dependent upregulation of rat uncoupling protein-2 expression in response to fasting or cold., 412(1):111-4.
Boukouvala, E., Antonopoulou, E., Favre-Krey, L., Diez, A., Bau- tista, J. M., Leaver, M. J., Tocher, D. R., and Krey, G., 2004. Molecular characterization of three peroxisome proliferator-activated receptors from the sea bass ()., 39(11): 1085-1092.
Camirand, A., Marie, V., Rabelo, R., and Silva, J. E., 1998. Thiazolidinediones stimulate uncoupling protein-2 expression in cell lines representing white and brown adipose tissues and skeletal muscle., 139(1):428-431.
Carboni, S., Hughes, A.D., Atack, T., Tocher, D. R.,and Migaud, H., 2013. Fatty acid profiles during gametogenesis in sea urchin (): Effects of dietary inputs on gonad, egg and embryo profiles., 164 (2):376-382.
Chang, Y. Q., Ding, J.,and Song, J., 2004.. Ocean Press, Beijing, 364pp (in Chinese).
Chen, W. C., Yang, C. C., Sheu, H. M., Seltmann, H., and Zou- boulis, C. C., 2003. Expression of peroxisome proliferator activated receptor and CCAAT/enhancer binding protein transcription factors in cultured human sebocytes.,121(3):441-447
Cook, E.J., Bell, M.V., Black, K.D., and Kelly, M. S., 2000. Fatty acid compositions of gonadal material and diets of the sea urchin,: Trophic and nutritional im- plications., 255 (2): 261.
Cui, Y., Liang, X., Cao, X. J.,and Gao, J., 2018. Molecular cha- racterization of peroxisome proliferator activated receptor gam- ma () in loachand its potential roles in fatty acid metabolism., 66:205-211.
Ding, Y. L., 2014. Construction and preliminary study of sea ur-chin family with different tube foot color and high unsaturated fatty acid. PhD thesis. Dalian Ocean University (in Chi- nese).
Di-Poi, N., Michalik, L., Desvergen, B., and Wahli, W., 2004. Functions of peroxisome proliferator activated receptors () in skin homeostasis.,39(11):1093-1099.
Downie, M. M. T., Sanders, D., Maier, L., Stock, D.M., and Kea- ley, T., 2004.Peroxisome proliferator-activated receptor and farnesoid X receptor ligands differentially regulate sebaceous differentiation in human sebaceous gland organ cultures., 151(4):766-775.
Dreyer, C., Krey, G., Keller, H., Givel, F., Helftenbein, G., andWalter W., 1992. Control of the peroxisomal β-oxidation path-way by a novel family of nuclear hormone receptors., 68(5):879-887.
Fleury, C., Neverova, M., Collins, S., Raimbault, S., Champigny, O., Levi-Meyrueis, C., Bouillaud, F., Seldin, M. F., Surwit, R.S.,Ricquier, D.,and Warden, C. H., 1997. Uncoupling protein-2: A novel gene linked to obesity and hyperinsulinemia., 15(3):269-272.
Forman, B. M., Chen, J., and Evans, R. M., 1997.Hypolipide- mic drugs, polyunsaturated fatty acids, and eicosanoids are li- gands for peroxisome proliferator-activated receptors α and δ., 94: 4312-4317
Göttlicher, M., Widmark, E., Li, Q., and Gustafsson, J. A., 1992. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor., 89(10):4653-4657.
Han, L. S., Ding, J., Wang, H., Zuo, R. T., Quan, Z. J., Fan, Z. H., Liu, Q. D., and Chang, Y. Q., 2019.Molecular characterization and expression of).,705:133-141.
He, S., Liang, X. F., Qu, C. M., Huang,W., Shen, D., Zhang, W. B., and Mai, K. S., 2012. Identification, organ expression and ligand-dependent expression levels of peroxisome proliferator activated receptors in grass carp ().,155(2):381-388.
Huang, J. C., 2008. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos., 2008:732303, DOI: 10.1155/2008/732303.
Ibabe, A., Grabenbauer, M., Baumgart, E., Fahimi, H. D.,and Ca- jaravillel, M. P., 2001. Expression of peroxisome proliferator-activated receptors (PPARs) in zebrafish ().,118(3):231-239.
Ibabe, A., Grabenbauer, M., Baumgart, E., Völkl, A., Fahimi, H. D., and Cajaraville, M. P., 2004. Expression of peroxisome proliferator-activated receptors in the liver of gray mullet ().,106(1):11-19.
Issemann, I., and Green, S., 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators., 347:645-650.
Juge-Aubry, C. E., Gorla-Bajszczak, A., Pernin, A., Lemberger, T.,Wahli, W., Burger, A. G.,and Meier, C. A., 1995.Peroxisome proliferator-activated receptor mediates cross-talk with thyroid hormone receptor by competition for retinoid X receptor. Possible role of a leucine zipper-like heptad repeat., 270(30):18117-18122.
Kang, J. X.,and Leaf, A., 1996. The cardiac antiarrhythmic ef- fects of polyunsaturated fatty acid., 31: S41-S44.
Karen, L. H., Bridget, M. C.,and Pamela, J. S., 2002. Peroxi- some proliferator-activated receptor gamma () and its ligands: A review.,22: 1-23.
Kelly, M. S., Hunter, A. J., Claire, L. S., and McKenzie, J.D., 2000. Morphology and survivorship of larval(Gmelin) (Echinodermata: Echinoidea) in response to vary- ing food quantity and quality., 183(3-4):223-240.
Kim, M. J., Deplewski, D., Ciletti, N., Michel, S., and Rosenfield, R. L., 2001. Limited cooperation between peroxisome proliferator-activated receptors and retinoid X receptor agonists in sebocyte growth and development.,74(3):362-369.
Kliewer, S. A., Forman, B. M., Blumberg, B., Ong, E. S., Bor- gmeyer, U., Mangelsdorf, D. J., Umesono, K.,and Evans, R. M.,1994.Differential expression and activation of a family of mu-rine peroxisome proliferator-activated receptors., 91(15):7355-9.
Kliewer, S. A.,Sundseth, S. S.,Jones, S. A.,Brown, P. J.,Wisely, G. B.,Koble, C. S.,Devchand, P.,Wahli, W.,Willson, T. M.,Lenhard, J. M., and Lehmann, J. M.,1997. Fatty acids and ei- cosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ., 94: 4318-4323.
Lapsys, N. M., Kriketos, A. D., Limfraser, M., Poynten, A. M., Lowy, A., Furler, S. M., Chisholm, D. J., and Cooney, G. J., 2000.Expression of genes involved in lipid metabolism correlate with peroxisome proliferator-activated receptor gamma expression in human skeletal muscle., 85(11):4293-4297.
Leaver, M. J., Wright, J.,and George, S. G., 1998. A peroxiso- mal proliferator-activated receptor gene from the marine flatfish, the plaice ()., 46(1-5): 75-79.
Li, S., Gul, Y., Wang, W., Qian, X. Q., and Zhao, Y. H., 2013., an important gene related to lipid metabolism and im- munity in: Cloning, characteriza- tion and transcription analysis by GeNorm., 512(2):321- 330.
Livak, K.J., and Schmittgen, T.D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCTmethod., 25: 402-408.
Medvedev, A. V., Snedden,S. K., Raimbault, S., Ricquier, D., andCollins, S.,2001. Transcriptional regulation of the mouse un- coupling protein-2 gene., 276(14):10817-10823.
Mimeault, C., Trudeau, V. L.,and Moon, T. W., 2006. Waterborne gemfibrozil challenges the hepatic antioxidant defense system and down-regulates peroxisome proliferator activated receptor beta (PPARbeta) mRNA levels in male goldfish ().,228(2-3):140-150.
Muller, E., Drori, S., Aiyer, A., Yie, J.,Sarraf, P.,Chen, H.,Hauser, S.,Rosen, E. D.,Ge, K.,Roeder, R. G., and Spiegelman, B. M., 2002. Genetic analysis of adipogenesis through peroxi- some proliferator activated receptor γ isoforms., 277(44):41925-41930.
Palacios, E., Racotta, I.S., Arjona, O., Marty, Y., Le Coz, J.R., Moal, J., and Samain, J.F., 2007. Lipid composition of the Pa-cific lion-paw scallop,, in relation to ga- metogenesis., 266(1-4): 266-273.
Ren, D., Collingwood, T. N., Rebar, E. J., Wolffe, A. P., Camp, H. S.,Ren, D., Collingwood, T. N., Rebar, E. J.,Wolffe, A. P.,and Camp, H. S., 2002.knockdown by engineered transcription fators: Exogenousbut notreactivates adipogenesis., 16 (1): 27-32.
Rosenfield, R. L., Deplewski, D., Kentsis, A., and Ciletti, N.,1998. Mechanisms of androgen induction of sebocyte differentiation., 196(1):43-46.
Russell, M.P., 1998. Resource allocation plasticity in sea urchins: Rapid, diet induced, phenotypic changes in the green sea urchin,(Müller)., 220 (1): 1-14.
Ruyter, B, Andersen, O., Dehli, A, Farrants, A.O., Gjøen, T., and Thomassen, M. S., 1997. Peroxisome proliferator activated re-ceptors in Atlantic salmon (): Effects on PPAR transcription and acyl-CoA oxidase activity in hepatocytes by peroxisome proliferators and fatty acids.–, 1348(3):331-338.
Samec, S., Seydoux, J.,and Dulloo, A. G., 1998. Role of UCP ho-mologues in skeletal muscles and brown adipose tissue: Mediators of thermogenesis or regulators of lipids as fuel substrate., 12(9):715.
Singh, R., Artaza, J. N., Taylor, W. E., Gonzalez-Cadavid, N. F., and Bhasin, S., 2003. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway., 144(11): 5081-5088
Tavazzi, L., Maggioni, A.P.,Marchioli, R., Barlera, S., Franzosi,M. G., Latini, R., Lucci, D., Nicolosi, G. L., Porcu, M., and Tognoni, G., 2008. Effect of n-3 polyunsaturated fatty acids in patients with chronic heart failure (the GISSI-HF trial): Aran- domised, double-blind, placebo-controlled trial.,372 (9645): 1223-1230.
Tong, S. Y., Chen, W., Yu, X.,and Liu, H. L., 1998. Study on li- pid and fatty acids composition of three kinds of Echinoidea’s gonad., 22(03):247-252 (in Chi-nese with English abstract).
Tontonoz, P., Hu, E.,and Spiegelman, B. M., 1994.Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-ac- tivated transcription factor., 79(7):1147-1156.
Tsai, M. L., Chen, H. Y., Tseng, M. C., and Chang, R. C., 2008. Cloning of peroxisome proliferators activated receptors in the cobia () and their expression at diffe- rent life-cycle stages under cage aquaculture., 425(1-2):69-78.
Walczak, R., and Tontonoz, P., 2002. PPARadigms and PPARadoxes: Expanding roles for PPARgamma in the control of li- pid metabolism., 43(2):177.
Wang, L. M., Wei, C.,and Ying, S., 2006. Analysis of general nu-tritional components and fatty acid composition of hybrid sea urchins and amphibians., 21 (3): 255-258 (in Chinese with English abstract).
Wei, J., Zhao, C., Zhang, L., Yang, L. M., Zuo, R. T., Hou, S. Q., and Chang, Y. Q., 2016. Effects of short-term continuous and intermittent feeding regimes on food consumption, growth, go- nad production and quality of sea urchinfed a formulated feed., 97(2):359-367.
Xiao, Y. F., Ke, Q.,and Wang, S. Y., 2001. Single point mutations affect fatty acid block of human myocardial sodium channel alpha subunit Na+channels., 98(6):3606-3611.
Xie, D. Z., Wang, S. Q., You, C. H., Chen, F., Zhang, Q. H, and Li, Y. Y., 2013. Influencing factors and mechanisms on HUFA bio-synthesis in teleosts., 20(2):456-466.
Yao, J. J., and Zhao, Y. L., 2006. Lipid changes during the embryonic development of., 4: 103-109 (in Chinese with English abstract).
Yao, J. J., Zhao, Y. L., Li, C., He, D.,and Hu, C. Y., 2009. Study on the changes of fatty acids in the early development of yellow catfish.,28 (11): 644-647 (in Chinese with English abstract).
Zhou, M. C., Yu, P., Sun, Q.,and Li, Y. X., 2016. Expression pro-filing analysis: Uncoupling protein 2 deficiency improves he- patic glucose, lipid profiles and insulin sensitivity in high-fat diet-fed mice by modulating expression of genes in peroxi- some proliferator-activated receptor signaling pathway., 7(2):179-189.
Zhu, Y.,2000. Isolation and characterization of peroxisome pro- liferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR., 275(18):13510-13516.
Zhu, Y., Qi, C., Korenberg, J. R., Chen, X. N.,Noya, D.,Rao,M. S., and Reddy, J. K., 1995. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR ga- mma) gene: Alternative promoter use and different splicing yield two mPPAR gamma isoforms., 92(17):7921-7925.
Zuo, R. T., Hou, C. Q., Chang, Y. Q., Ding, J., Song, J., Zhao, C.,and Zhang, W. J., 2016. Progress in nutritional physiology of sea urchin., 31 (4): 463- 468.
March 6, 2020;
April 29, 2020;
October 16, 2020
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
#The two authors contributed equally to this work.
. E-mail: dingjun19731119@hotmail.com
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Characteristics of Atmospheric Rivers over the East Asia in Middle Summers from 2001 to 2016
- Facile Synthesis of Fe/Cr-Codoped ZnO Nanoparticles with Excellent Adsorption Performance for Various Pollutants
- Experimental Investigation of Wave Load and Run-up on the Composite Bucket Foundation Influenced by Regular Waves
- Development of a Microfluidics-Based Quantitative Real-Time PCR to Rapidly Identify Photobacterium damselae subsp. damselae with Different Pathogenicity by Detecting the Presence of mcp or dly Gene
- In vitro Antioxidant Effects of Porphyra haitanensis Peptides on H2O2-Induced Damage in HepG2 Cells
- Otolith Shape Analysis as a Tool to Identify Two Pacific Saury (Cololabis saira) Groups from a Mixed Stock in the High-Seas Fishing Ground
