Identification of Typical Rice Diseases Based on Interleaved Attention Neural Network
2021-02-21WenXinJiaYinjiangandSuZhongbin
Wen Xin, Jia Yin-jiang, and Su Zhong-bin
College of Electrical and Information, Northeast Agricultural University, Harbin 150030, China
Abstract: Taking Jiuhong Modern Agriculture Demonstration Park of Heilongjiang Province as the base for rice disease image acquisition, a total of 841 images of the four different diseases, including rice blast, stripe leaf blight, red blight and bacterial brown spot, were obtained. In this study, an interleaved attention neural network (IANN) was proposed to realize the recognition of rice disease images and an interleaved group convolutions (IGC) network was introduced to reduce the number of convolutional parameters, which realized the information interaction between channels. Based on the convolutional block attention module (CBAM), attention was paid to the features of results of the primary group convolution in the cross-group convolution to improve the classification performance of the deep learning model. The results showed that the classification accuracy of IANN was 96.14%, which was 4.72% higher than that of the classical convolutional neural network (CNN). This study showed a new idea for the efficient training of neural networks in the case of small samples and provided a reference for the image recognition and diagnosis of rice and other crop diseases.
Key words: disease identification, convolutional neural network, interleaved attention neural network
Introduction
Rice is an important food crop in China, and its yield is of great significance to national food security. Rice easily infected by diseases during its growth, which directly affected the qualities and yields of rice. Rice diseases mostly occur in leaves, and the accurate identification of rice diseases is helpful for agricultural producers to implement controlling measures precisely. At present, the identification of rice diseases mainly relies on field identification by experienced agricultural technicians. This method has high professional requirements for agricultural technicians. It not only consumes a lot of manpower and material resources, but also has strong subjectivity and great error in the judgement results, which makes it difficult to achieve accurate identification of the diseases and easy to miss the best time to prevent and control the diseases. Therefore, it is of great significance to carry out researches on the identification method of typical rice diseases and realize the accurate diagnosis of rice diseases (Zhuet al., 2015; Houet al., 2015).
With the development of computer vision and artificial intelligence technology, artificial intelligence algorithms are widely used in agriculture (Lvet al., 2021; Maet al., 2021), medicine (Kiliarslanet al., 2020; Antropovaet al., 2016), industry (Zhang, 2020; Wanget al., 2020) and other fields. The use of an image to identify crop diseases has become a research hotspot in recent years. Zhuet al. (2021) used CNN-VGG16 and Resnet50 networks to identify common leaf diseases of typical crops, such as apples, corns, tomatoes and grapes. Ozguven and Adem (2019) proposed the Faster R-convolutional neural network (CNN) structure for automatic detection of leaf spot images in sugar beet. Huet al. (2019) proposed an improved deep CNN, which adds the module of multi-scale feature extraction to the rapid model of CIFAR10 to improve the ability of automatic extraction of image features of different tea diseases. Karthiket al. (2020) proposed two different depth architectures, which use successfully to detect the infection types of tomato disease leaves. However, all the above deep learning frameworks have disadvantages, such as huge amounts of parameters and low classification performance and huge amounts of data sets are needed to improve the classification performance of the models. However, in practical engineering applications, it is difficult to implement the acquisition of huge amounts of samples due to the heavy workload and high cost.
To sum up, in terms of data sources, most existing studies are based on data set PlantVillage, such as corn (Liuet al., 2021; Xuet al., 2020; Zhanget al., 2018), tomato (Renet al, 2020; Guoet al., 2019; Huet al., 2019), potato (Yanget al., 2020), etc., but there are no images of rice diseases in these data sets. Most of the existing research data on rice diseases comes from network collection, with few samples. In terms of processing methods, the traditional identification method of machine learning algorithm is cumbersome and its accuracy is low, while deep learning algorithm needs huge amounts of data sets to improve the classification performance of the model. However, in the actual engineering application, it is difficult to obtain huge amounts of sample data, leading to difficulty in training the parameters in the network. Based on the above deficiencies and limitations, this study proposed an interleaved attention neural network for rice disease image recognition based on the basic structure of deep learning network to reduce the number of model calculation parameters and enhance the classification performance of the model.
Materials and Methods
Image collection
Rice disease images were collected from June to August 2019, at Jiuhong Modern Agriculture Demonstration Park, Qing'an County, Heilongjiang Province. During the collection process, the infected leaves were first cut off with scissors, then the collected rice leaves were stored in a constant temperature and humidity incubator, and finally brought back to the laboratory for treatment. The camera of the HUAWEI Mate 9 mobile phone (eight megapixels, with a maximum resolution of 5 120×3 840) was used to take pictures of rice leaves. The camera's shooting direction was perpendicular to the plane of rice leaves, and all the backgrounds of leaf images were off-white desktop of the laboratory, and the light was even when the photo was taken. Finally, 841 pictures of the four diseases including rice blast, stripe leaf blight, red blight and bacterial brown spot were obtained, as shown in Fig. 1.
Data set establishment
The collected rice disease leaf images were manually annotated, according to the types of disease, and the classification labels and sample numbers of the disease images are shown in Table 1. With a total of 841 pictures of rice leaf diseases collected, 80% of the pictures of each category were randomly selected as the training set to establish the interleaved attention neural network (IANN) model, and the remaining 20% were used as the test set to test the classification performance of the model. In the end, 673 samples of the training set and 168 samples of the test set were included.

Table 1 Image labels and sample numbers of different diseases
After obtaining the disease images, the research objective and the main technical difficulties were established. During the experiment, the data set was divided into training set and test set, and then the data set was preprocessed. Then, the classification and recognition model of rice disease leaves was constructed. The classic CNN model was used to extract features and classify and recognize rice disease data sets, and its deficiencies and limitations were analyzed. Then, the collected data samples were input into the structure of the cross-attention network for optimization. Finally, the classification performance was verified on the test set, and the identification results of several methods were compared and analyzed in terms of performance and number of parameters. Fig. 2 is the technical roadmap of this paper.

Fig. 1 Pictures of rice leaf diseases
Data preprocessing
When the convolutional neural network was used to recognize images of rice infected leaves, the lesion images needed to be converted to a uniform size to facilitate feature extraction and pattern recognition. All the 841 original infected leaf images were uniformly converted to 256×256 pixels. Some of the corrected sample images are shown in Fig. 3.
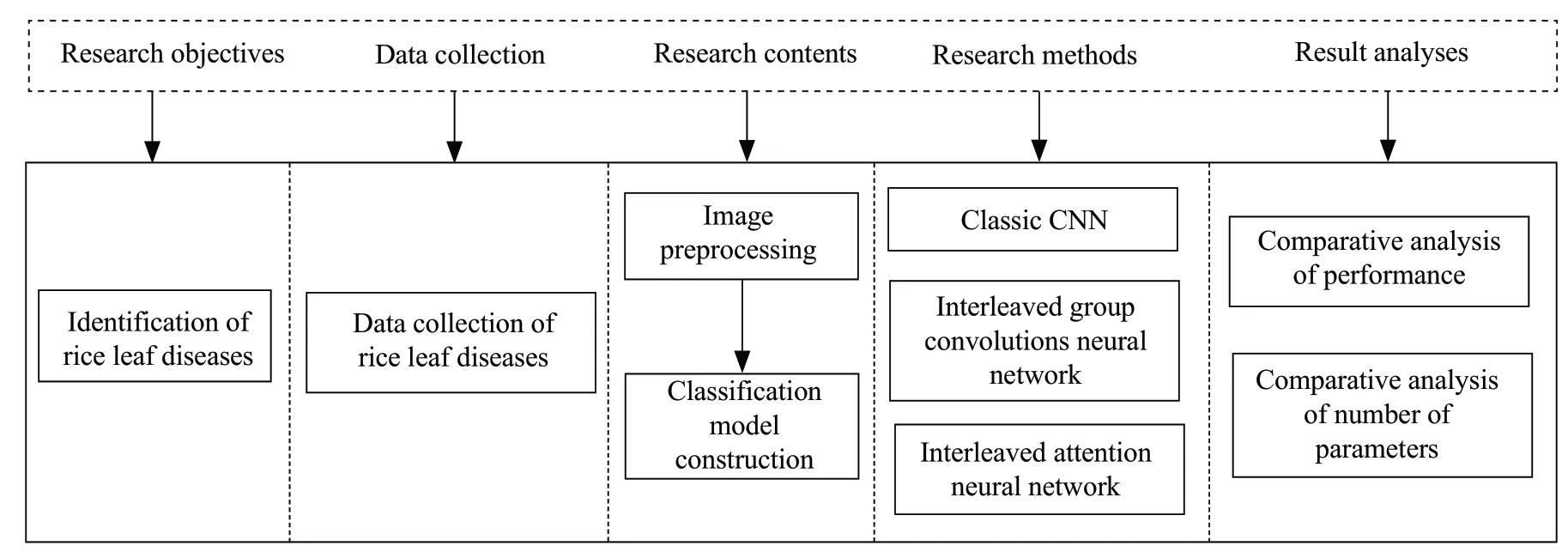
Fig. 2 Technical roadmap
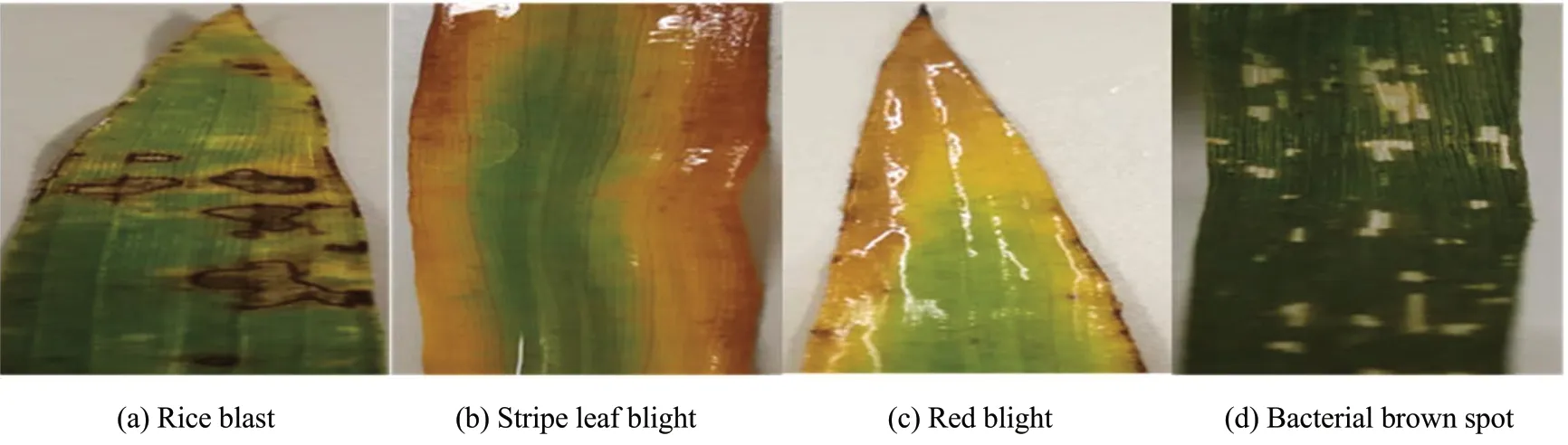
Fig. 3 Corrected image of rice leaf disease
Construction of Rice Leaf Disease Identification Model
Interleaved group convolutions
An interleaved group convolutions divided the input channel into several parts and performed regular convolutions on each partition (Zhanget al., 2017). Group convolution could be viewed as a regular convolution with sparse block to angle of convolution kernels, where each block corresponded to a partition of the channel and there was no connection between the partitions. The interleaved group convolution consisted of the two group convolutions, the primary group convolution and the subgroup convolution (Zhanget al., 2018). Firstly, the main group convolution was used to deal with the spatial correlation and the spatial convolution kernel was used to perform the conventional convolution on each channel partition. Then, the features were mixed using subgroup convolution, which consisted of cross-partition channels outputted by the first convolution set, and during that process the form of a 1×1 convolution kernel was simply computed in. Interleaved group convolutions (IGC) could effectively reduce the number of convolution parameters using grouping convolution (Huet al., 2018; Chollet, 2017).
Convolutional block attention module (CBAM)
Convolutional block attention module was an effective attention mechanism module, which consisted of channel attention module and spatial attention module (Wooet al., 2018). The main purpose of CBAM was to focus on the key features in the process of convolution, to make the convolution calculation more focused and targeted, thus the classification performance of the network could be improved.
Channel attention module: focusing on "what" mainly, channel attention could provide more valuable information and generates channel attention diagram by exploring the relationship between channels. In order to effectively calculate channel attention, maxpooling and average-pooling were introduced to aggregate the information inputting feature map. Then, they were sent to the multi-layer perceptual network to obtain two sets of vectors, which were merged into vectors featuring output through point multiplication operation. The formula for channel attention was as the followings:


In the formula,σwas the activation function of Sigmoid,W1∈C/r×Cndwere the weight ofMLP, andrwas the attenuation rate. In this paper,r=0.5.
Spatial attention module: spatial attention focused on which part was important, which was complementary to channel attention. The internal spatial relations between features were used to generate maps of spatial attention. Similarly, in order to calculate spatial attention, operations of average-pooling and max-pooling were applied on the channel axis to gather important information and obtain two feature identifiers, which were connected by operation of concat and sent to the convolutional layer to obtain the spatial attention map. The spatial attention formula was as the followings:
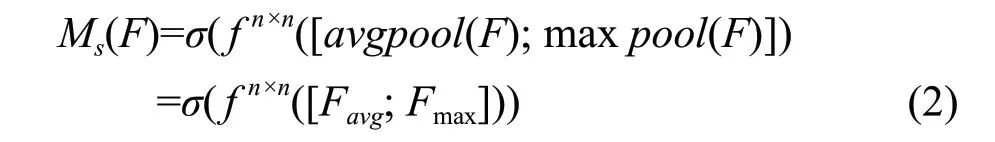
In the formula,f n×nrepresented the size of the convolution kernel inhe cnvolution operation wasn×n, and the size of the convolution kernel in this framework was 3×3, adding padding calculation.
Finally, for inputtedF∈RC×H×Wfeaturing 3D, CBAM was used to generate a 1D channel attention map and 2D spatial attention map. Combined with CBAM framework, the whole attention mechanism process was as the followings:

In the formula,ⓧdenoted element-wise multiplication. The attention value was transferred through point multiplication operation.Mcwas the map of channel attention, with size ofRC×1×1, andMswas the map of spatial attention, with size ofR1×H×W.F' was the output of attention through the channel, andF'' was the final output. CBAM module was only to extract the key features that affected the classification performance and would not change the number of feature channels.
Interleaved attention neural network framework
In order to realize the full information exchange in the process of CNN computing, the structure of IANN was further proposed based on no reduction of the classification performance and no increase in the amount of CNN computing. Firstly, after expanding the original convolution channel, all the channels were divided into groups, and each group contained the same number of channels. Then, the 3×3 convolution kernel was used to carry out intra-channel convolution with adding padding calculation. Next, the channel attention and spatial attention of CBAM module were used to focus on the key features. After key feature extraction, information interaction between channels was carried out. The information interaction here was completed by a point convolution operation. Finally, the information was restored to the input form of original data for further processing.
By using this framework structure, the key features that affected the classification performance were extracted, the convolution parameters were reduced, and information exchange was effectively completed, thus improving the classification performance of the network.
Evaluation index
The performance of the proposed model was evaluated through identifying accuracy and the index function expression was shown in Equation (4):
In the formula,Ncorrectwas the number of samples identified correctly in the data set andNtotalwas the total number of samples in the data set.
Calculation method of the number of parameters
The number of parameters in the calculation process of classical CNN could be expressed by formula (5):

In the formula, the size of the convolution kernel wasSand there wereLinput channels andMoutput channels.
In the interleaved attention module, the calculation of convolution parameters came from the calculation of the primary group convolution and the secondary group convolution in IGC, as well as the convolution calculation of spatial attention mechanism in CBAM module. The calculation of the number of convolution parameters in the interleaved attention module was shown in formula (6):
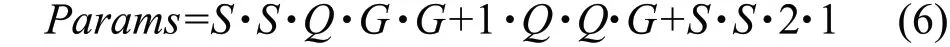
In the formula,Qrepresented the number of input channels divided into groups, andGrepresented the number of channels contained in each group. As a result of this grouping convolution, the number of feature channels before and after convolution remained constant.
Results
Experimental development environment
Hardware: i7-3770 CPU@ 3.40 GHz 3.40 GHz, 32 GB RAM, graphics card NVIDIA GeForce GTX 1080.
Software: Win10 operating system, Python 3.7 version, PyTorch 1.6.0 version, NumPy 1.19.2 version, Pandas 1.2.2 version.
Hyper parameter setting
In the training process of the deep learning model, after the pre-adjustment of the super parameters, the training parameters were determined as the followings: Adam optimizer with a learning rate of 0.01 was used to update the weight, deviation and convolution parameters. The size of the batch training was set as 32 and the number of iterations was set as 200. The batch normalization method (BN) was applied to keep the convolution kernel with the same parameter distribution. After the convolution calculation, the ReLU activation function was selected for nonlinear mapping, and a fully connected layer was designed, including four neurons. And the Softmax function was applied to the fully connected layer for the final classification and recognition. During the training process, L2 was used to regularize the loss function, and the regularization coefficient was 0.01. The loss value was calculated as shown in formula (7):
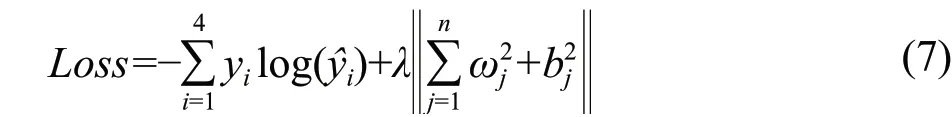
Where,yiwas the actual category of the picture, andwas the model prediction category.ωjandbjwere the weights and thresholds of the fully connected layer, respectively. In order to eliminate randomness, 10 tests were carried out, and the average accuracy rate obtained during the 10 tests was used as the final evaluation index.
Discussion
Accuracy analysis of classical CNN recognition
The rice disease data in this paper were identified based on the classical convolutional neural network. The classification performance of the classical CNN was mainly affected by the size of the convolutional kernel and the level of the network structure. Since there were only 841 images in the rice disease data set in this study, the deep network structure had many convolution parameters that needed to be trained. Therefore, a superficial CNN structure to recognize the four different kinds of disease images was used in this paper. In terms of the selection of volume set kernel size, a large convolution kernel would increase the convolutional receptivity field, making feature extraction too rough and failing to obtain important features that affected the performance of convolutional classification, which ultimately led to poor classification performance of the network. Therefore, the 7×7 convolution kernel was used in this paper. In this study, the influence of different shallow CNN structures on classification performance was discussed. With the increase in the number of C-P modules, the classification performance of CNN showed an overall upward trend. However, when the number of C-P modules was 5, the classification performance of the network decreased. When the structure was C-CP-C-P-C-P-FC, the classical CNN showed a better classification performance, with an accuracy rate of 91.42% on the test set.
In the framework of C-C-P-C-P-C-C-P-C-P-FC and the convolution kernel of 7×7, the parameter information involved in each layer of classical CNN is listed in detail in Table 2, including the size of convolution kernel, convolution step size, pooling step size, number of convolution kernel, characteristic input size, output size and activation function used by each layer.
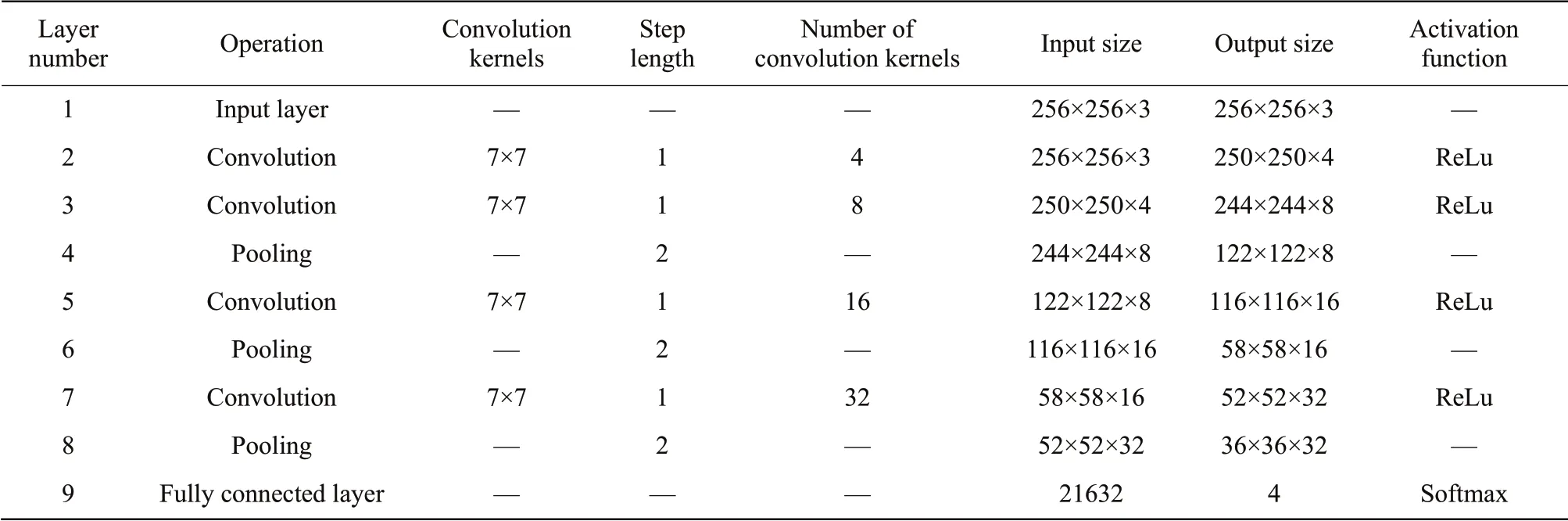
Table 2 Classic CNN frame parameter
Analysis of calculation quantity of classical CNN parameters
According to equation (5), the process of calculating the number of 7×7 convolution kernel parameters of C-C-P-C-P-C-C-P-FC except for the fully connected layer was as the followings:
(1) C: 3×4×7×7=588
(2) C: 4×8×7×7=1 568
(3) C: 8×16×7×7=6 272
(4) C: 16×32×7×7=25 088
Note:Pwas not involved in the calculation of the number of parameters.
The total number of parameters in the classical convolution frame was 588+1 568+6 272+25 088=33 516.
The classification accuracy of the rice disease data set based on the classic CNN framework only reached 91.42%, which was not enough to meet the actual demand. It could be seen that the process of convolution calculation involved a huge number of parameter calculation, which would increase the training difficulty of the model and reduce the classification performance of the model. These convolution parameters required a large amount of data for adequate training, but in practical engineering applications, it was difficult to obtain huge amounts of test samples. Therefore, it was very important to propose a lightweight CNN structure that could reduce the number of parameters in the calculation process.
Identification accuracy analysis of IANN
The original image set contained a total of 841 images. The deep network structure and the number of channels would increase the number of parameters of the model. Therefore, a 1×1 convolution kernel to expand the channel into 12 channels to extract the deep features without adding padding was used in this paper. Pooling was added only before the full connection layer for feature compression. The pooling size was 2×2, in the form of average pooling, and the step size was 2. C stood for convolution calculation, IG stood for IGC module (the main group convolution was calculated as a 3×3 convolution kernel, adding padding),Pstood for pooling, and FC stood for fully connected layer. The 12 channels were converted into a 3×4 group for IGC calculation. According to Table 3, added pooling operation after IGC module, with the number of IG-P modules increasing, the recognition accuracy of the network was continuously improved. When the network structure was C-IGP-IG-P-IG-P-FC, the best classification performance was obtained, and the classification accuracy of the test set was 91.52%. When IG-P module continued to be introduced, the accuracy tended to be stable and no longer increased.
The identification results of IGC network joining CBAM module were discussed. IGBS stood for IANN module, and the pooling method was the same as above. The identification results of IANN after the addition of the pooling module are shown in Table 3. With the increase in the number of IGBS-P modules, the classification accuracy of the network was on the rise. When the network structure was C-IGBS-P-IGBS-P-FC, the network obtained the best classification result. The classification accuracy on the test set reached 96.14%. The data transmission process of the C-IGBS-P-IGBS-P-FC network was as the followings: firstly, the 3-channel features were expanded into a 12-channel feature matrix by point convolution calculation, and the 12-channel features were divided into three groups with four channels in each group for IANN calculation. Then, the size of the feature matrix was halved by a pooling operation. Then, two IGBS and pooling operations were carried out to obtain the 28×28×12 feature form. Finally, the final feature was transformed into a 9 408 (28×28×12)×1 column vector as the input of the fully connected layer.
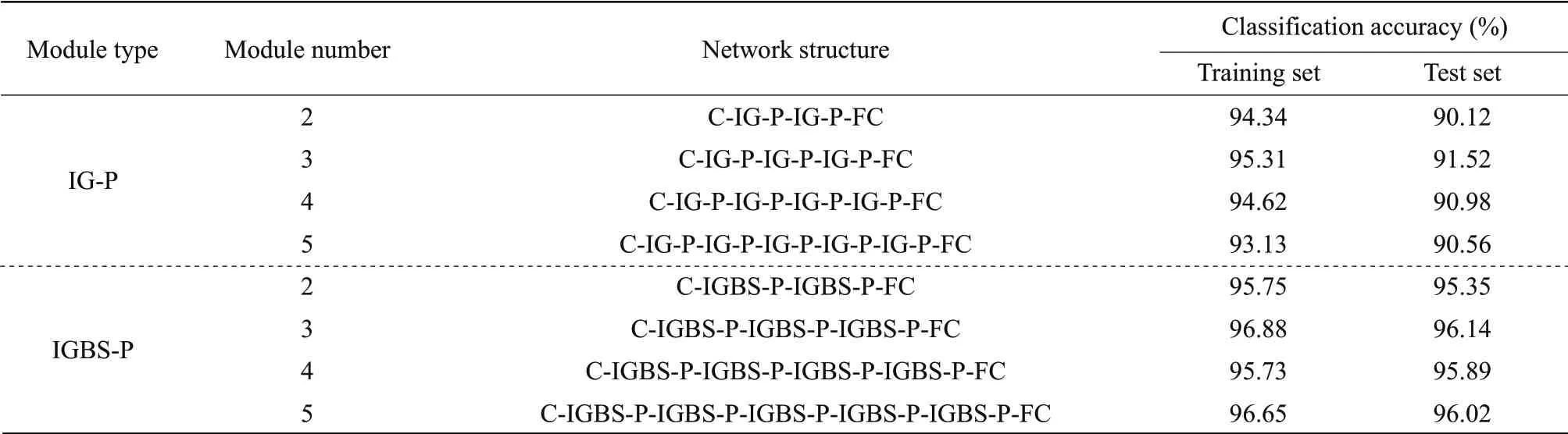
Table 3 IGC and IANN model recognition results
Comparing the network classification performance of IANN with that of the classic CNN structure and IGC structure, the size of the convolution kernel was 7×7 in the calculation process of the classic CNN structure. The classification and recognition results of the four kinds of rice disease images under multiple structures are shown in Table 4. It could be seen that the classification accuracy of IANN was 4.62% higher than that of IGC alone and 4.72% higher than that of the classic CNN. Compared with the classic CNN and IGC network, IANN had the best classification performance and the best recognition effect and successfully realized the accurate recognition of different rice disease images.
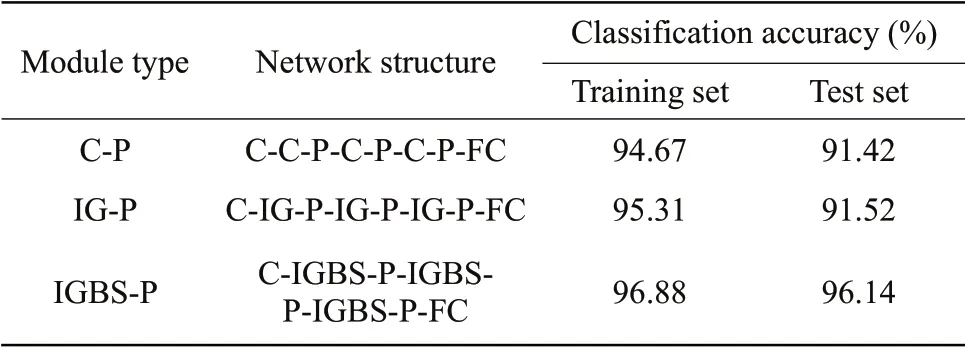
Table 4 Image recognition results of rice diseases under each structure
Analysis of calculation parameters of IANN
According to equation (6), the calculation process of C-IGBS-P-IGBS-P-IGBS-P-FC except for the fully connected layer was as the followings:
(1) C: 3×12×1×1=36
(2) IGBS: 7×7×3×4×4+1×3×3×4+7×7×2×1=2 486
(3) IGBS: 7×7×3×4×4+1×3×3×4+7×7×2×1=2 486
(7) IGBS: 7×7×3×4×4+1×3×3×4+7×7×2×1=2 486
Note:Pwas not involved in the calculation of the number of parameters.
The total number of parameters except for the fully connected layer under the interleaved attention frame was 36+2 486+2 486+2 486=7 494. Through the recognition of the four different rice disease images, the interleaved attention framework greatly improved the recognition accuracy and reduced the number of convolution calculation parameters except for the fully connected layer from 33 516 to 7 494, which greatly reduced the number of convolution calculation parameters in the process. Through experimental verification, the retained parameters could effectively represent the overall properties of the original image and effectively avoid the phenomenon of over-fitting of the model.
Conclusions
In order to solve the problem that rice suffering diseases in the process of growth could not be diagnosed and accurately identified in time, the deep learning technology to carry out the classification of rice disease images was used in this paper. Combined with the four different rice disease sample data in field collection, the convolutional neural network algorithm to extract and identify the features of the disease images was used in this paper. Furthermore, a rice disease identification and classification method based on IANN was proposed. In this paper, the classification performance and the number of parameters of the classical convolutional neural network framework and IANN framework were compared and analyzed. The main conclusions were as the followings:
A classic CNN structure C-C-P-C-P-C-P-FC with a convolution kernel size of 7×7 was designed. After iterative training, the framework achieved a classification accuracy of 91.42% on the test set. The classical CNN highly integrated the process of feature extraction and pattern recognition, making the classification process more intelligent.
Due to the large number of parameters involved in the classical CNN calculation process, the interleaved group convolution pattern to reduce the number of convolution parameters in the form of grouping convolution was introduced in this paper. According to the calculation and analysis of the number of parameters in this paper, it could be seen that the interleaved group convolution effectively reduced the number of calculated parameters.
In the process of classical CNN training, the convolution calculation process was all about the global convolution of the image without targeted calculation. Meanwhile, the convolution calculation between each channel was independent of each other, and there was no sufficient interaction after the calculation. Therefore, CNN calculation method of interleaved attention mechanism was designed in this paper, IGC was used to realize the information interaction between channels, and CBAM module was used to realize the important convolution under the attention mechanism. Combining CBAM module with IGC, based on reducing the number of convolution parameters, comprehensive attention was paid to the key features of the convolution process in channel and space, which significantly improved the classification performance of the network. Finally, classification result of 96.14% was obtained in the network structure under the framework of C-IGBS-P-IGBSP-FC.
In conclusion, IANN effectively reduced the number of convolution parameters, improved the classification performance, and successfully realized the accurate recognition of the four kinds of the rice disease images. This study showed a new idea for the efficient training of neural networks in the case of small samples and provided a reference for the image recognition and diagnosis of rice and other crop diseases.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Effect of Foliar GA3 and ABA Applications on Reactive Oxygen Metabolism in Black Currants (Ribes nigrum L.) During Bud Paradormancy and Secondary Bud Burst
- Mining Heat Stress Associated Genes in Tomato Fruit (Solanum lycopersicum L.) Through RNA-seq
- Advances on Current Techniques and Methodologies in Milk Lipidomics
- Cloning of MYB2 Gene from Dryopteris fragrans and Its Response to ABA and Drought Stress
- NIRS Prediction of SOM, TN and TP in a Meadow in the Sanjiang Plain, China
- Screening of Feather Degrading Bacteria and Optimization of Fermentation Conditions from Poultry
