Advances on Current Techniques and Methodologies in Milk Lipidomics
2021-02-21LiMohanRayhnigulAbdllaChenJialiLiuYimingZhangXiuminCaoXueyanYangMeiZhengYanandYueXiqing
Li Mo-han, Rayhnigul Abdlla, Chen Jia-li, Liu Yi-ming, Zhang Xiu-min, Cao Xue-yan, Yang Mei, Zheng Yan, and Yue Xi-qing*
1 College of Food Science, Shenyang Agricultural University, Shenyang 110866, China
2 Department of Foreign Languages, Shenyang Agricultural University, Shenyang 110866, China
3 Beijing Academy of Food Sciences, Beijing 100068, China
Abstract: Milk is a complex biological fluid containing lipids, proteins, carbohydrates and minerals, which are essential for infant growth. While the lipid portion constitutes only 3%-5% of the total milk composition, it accounts for over 50% of the infant's daily energy intake. The dominant portion (approximately 98%) is in the form of triacylglycerols and polar lipids, such as glycerophospholipids and sphingolipids, forming minor components. Recently, with the development of lipidomics, important progresses have been made in milk lipidomics, and the identification and quantification of several milk lipids at the group and molecular species level has become a reality, thereby providing useful information for the infant formula industry. In this review, an overview of the separation of the main components of milk lipids was presented, including glycerolipids, phospholipids and sphingolipids. The analytical methods and strategies for milk lipidomics, including gas chromatography-mass spectrometry (MS), capillary electrophoresis MS, nuclear magnetic resonance, matrix-assisted laser desorption ionization-MS, electrospray ionization-MS, shotgun lipidomics and liquid chromatography-MS, were reviewed. Additionally, the bioinformatics of lipidomics for milk lipid determination, including lipid classification, lipid databases and lipid analysis software, were investigated. This review would aid future investigations of the nutrition of milk lipids and refined researches on formula milk powder.
Key words: milk, lipidomics, triacylglycerols, glycerophospholipids, sphingolipids
Introduction
A comprehensive study of the roles and functions of lipids in molecular biology requires accurate identification and quantification of lipid species. The high structural diversity of lipids presents a significant challenge for lipidomics (Fahyet al., 2009). The first popular mass spectrometry (MS)-based lipidomics technique, namely, shotgun lipidomics, was introduced by Han and Gross in 2003. The method employs direct infusion to introduce lipids into a mass spectrometer combined with either precursor ion scanning or neutral loss scanning (Han and Gross, 2005). Later, a direct infusion approach with data-dependent acquisition using high-resolution MS was developed (Schwudkeet al., 2006). Hundreds of lipids from complex biological samples can be directly identified and quantified without any chromatographic separation. However, this method is limited by ion suppression and the lack of discrimination among isomeric lipid species (Köfeleret al., 2012). Alternatively, coupling of liquid chromatographic (LC) separation to datadependent MS/MS acquisition for lipidomics can effectively reduce matrix effects and resolve isobaric lipids (Sandraet al., 2010; Cajka and Fiehn, 2014). The acquired MS/MS spectra provide rigorous identification of lipids through a spectral match with a standard or predicted MS/MS spectral library (Ivanisevicet al., 2013; Cajka and Fiehn, 2017). Additionally, some large-scale predicted MS/MS spectral libraries are developed to facilitate lipid identification using LC-MS/MS, such as LipidBlast (Kindet al., 2013).
A wide range of possible compositions of milk fat gives rise to one of the most complex natural components in milk with thousands of molecular species (Liuet al., 2018). To comprehensively elucidate such complicated lipids in milk, reliable analytical methods must be employed. Lipidomics is an important part of metabolomics, which aims to characterize and quantify intact lipid molecules. In recent decades, with the rapid development of techniques, especially for MS, advances had been made in lipid detection and quantification. Over the past few decades, with the constant improvement of lipidomics, positive progress had been made in milk lipidomes (Benoitet al., 2010; Sokolet al., 2015; Garwolińskaet al., 2017; Tenget al., 2017; Chenet al., 2019, 2020; Daiet al., 2020). In addition, the fat used in infant formulas should be based on the characteristics of human milk fat. Thus, attention has also been paid to the comparison of milk lipidomes between humans and other domestic animals.
The aim of this review was to present an overview of the separation of the main components of milk lipids, analytical methods and strategies, and the bioinformatics of lipidomics for human milk lipid determination. This review mainly focused on human milk, while the lipidomics of other mammalian milk were not described in details, except for information on domestic animal milk for comparison. Problems to be solved and the importance of future perspectives were also discussed.
Separation of Main Components of Milk Lipids
Glycerolipids
Milk lipids contain 98%-99% triglycerides, 0.26%-0.80% phospholipids, 0.25%-0.34% sterols (mainly cholesterol), and traces of monoglycerides, diglycerides, free fatty acids and other lipids (Liuet al., 2018; Liet al., 2019, 2020a, 2020b). Analysis of triglycerides in milk fat first requires the extraction of triglycerides. The most efficient and convenient method at present is thin layer chromatography (TLC). Triglyceride extraction is achieved by TLC using silica gel as the sorbent and n-hexane, diethyl ether and acetic acid (or formic acid) as the mobile phase.
Reversed-phase high-performance LC (RP-HPLC) is the most commonly used liquid phase method in milk lipids. Triglycerides are weakly polar compounds that are hardly soluble in water and organic solvents can increase the solubility of triglycerides. A combination of widely used organic solvents is acetone and acetonitrile (Buchgraberet al., 2004). The most commonly used stationary phase is the octadecylbonded silica column (C18 or ODS). The peak of the triglycerides is analyzed by RP-HPLC to elute in order of the number of carbon atom equivalents.
Silver ion HPLC is the most efficient chromatographic method for isolating triglyceride isomers in milk fat. The triglyceride is separated, according to the degree of unsaturation of its constituent fatty acids, and the larger the number of double bonds in the triglyceride molecules is, the longer the retention time is. The order of separation of triglycerides is SSS, SSM, SMM, SSD, MMM, SMD, MMD, SDD, SST, MDD, SMT, MMT, DDD, SDT, MDT, DDT, STT, MTT, DTT and TTT, where S, M, D and T are saturated, monounsaturated and fatty acids containing two and three double bonds, respectively. The order of the peaks of the isolated triglyceride isomers is SMS, SSM, SMM, MSM, SDS, SSD, DSD, DDS, DMD and DDM (Buchgraberet al., 2004).
High-temperature gas chromatography (GC) analysis of triglycerides has high stability and good accuracy, but is only suitable for samples with high fatty acid saturation (Buchgraberet al., 2004; Ruiz-Sambláset al., 2013). Triglycerides with higher unsaturation may oxidatively cleave at high temperatures. The peak order of triglycerides is based on the total number of carbons (CN) from small to large peaks. Currently, two main types of non-polar and medium-polar fillers are used; the former can obtain different CN triglycerides and the latter can be specific to the single triglyceride species. A commonly used WCOT column material is fused silica coated with methylphenylsiloxane polymer, 25 mol · L-1×0.25 mmol · L-1. The order of separation of triglycerides is SSS, SSM, SSD, MMM, SDM, MMD, SDD, MDD, DDD and DDT (Buchgraberet al., 2004).
Supercritical fluid chromatography (SFC) is an important complement to GC and HPLC. The mobile phase is a liquefied gas (usually CO2) (Milneet al., 2006). SFC uses a GC-like column that separates and identifies triglyceride isomers, if modifiers are added for better resolution. The order of peaks of triglycerides is SMD, MMM, SDD, MMD, MMT, MDD, DDD, MDT, MTT, DDT and DTT (Buchgraberet al., 2004).
Phospholipids
Due to the wide variety of phospholipids and different lengths and unsaturation of unsaturated acyl chains of phospholipid molecules in different biological sources, there are theoretically more than 1 000 identifiable phospholipid molecules, thus, various types can be qualitatively and quantitatively analyzed in complex mixtures. Phospholipid molecules are technically difficult. At present, the main methods for extracting phospholipids are liquid-liquid extraction and solidphase extraction (SPE).
Liquid-liquid extraction involves placing the sample in a two-phase system that is incompatible with it. Typically, this system includes a lipophilic organic phase and a hydrophilic phase such that a highly lipophilic lipid will dissolve in the organic phase. The most classical method for extracting phospholipids by liquid-liquid extraction is the Folch method (Folchet al., 1957). The organic solvent used in this method is a chloroform : methanol mixture (v/v =2 : 1). Bligh and Dyer (2011) improved the method by adding water to the chloroform-methanol mixture to make the phase separation of the lipids faster. Usually, the Folch method is suitable for the extraction of lipids from all the tissue samples, while the Bligh-Dyer method is more suitable for the extraction of biological fluid samples (Schilleret al., 2004). In 2013, Chenet al. extracted lipids and their lipid metabolites simultaneously with methyl tert-butyl ether. This new method can achieve a comprehensive analysis of lipids and their metabolites. Due to the toxicity of chloroform and ether, some studies use n-hexane : isopropanol (V/V = 3 : 2) solvent, which is less toxic (Fuchset al., 2011), but the extraction rate of this method is relatively low. The lipid obtained by liquid-liquid extraction is mixed with a neutral ester and a free fatty acid in addition to phospholipids. To obtain a pure phospholipid, the acetone-treated extract is usually used to separate the phospholipid precipitate from other lipids. Gładkowskiet al. (2012) studied the extraction and purification methods of egg yolk phospholipids and found that the acetone temperature has a great influence on the purification of phospholipids and egg yolk phospholipids containing no cholesterol and triglycerides are obtained with acetone at -20℃.
Compared with liquid-liquid extraction, it is necessary to extract the total lipids, then separate phospholipids from the total lipids. SPE can specifically extract phospholipids, and the solvent consumption is small, making it a good method for phospholipid enrichment, especially suitable for the extraction of small amounts of phospholipids. The SPE cartridge used for SPE is usually filled with silica or silica gelmodified fillers (usually aminopropyl, dihydroxy, propylamino, cyanopropyl silica gel, etc.) (Mills and Goldhaber, 2010); commonly used organic solvents include methanol, chloroform and n-hexane. Pinkartet al. (1998) used an aminopropyl silica SPE column to separate lipid components, which can result in neutral lipids, PHA, polar lipids, sterol esters, triglycerides, sterols and diglycerides.
Sphingolipids
Among many methods, TLC is applied to the qualitative detection of glycoconjugates in the mid-1960s (Furukawaet al., 2013). TLC has the advantages of simple device structure, convenient operation steps, color development with high concentrations of acid and other corrosive color developer, high expansion rate and strong resolution, and can be carried out using a glycolipid-specific chromogenic reagent and antibody. After chromatographic separation, it is easy to eliminate the interference of non-glycolipid compounds and achieve specific detection. However, the disadvantage is that the separation effect of complex substances is not ideal. Therefore, highperformance TLC (HPTLC) has emerged. Compared to conventional TLC, the silica gel particles of the matrix (usually silica gel plate) used by HPTLC have a smaller particle radius and better particle size uniformity. HPTLC uses a multi-stage expansion technique to overcome the absorbance. The smear caused by small particles increases the concentration of the sample per unit area in the spot, thereby improving the detection sensitivity. HPTLC not only has higher precision and separation capabilities, but also has the advantage of smaller sample usage. In the past 10 years, with increased researches regarding shortening the separation time of TLC, improving detection sensitivity, enhancing separation efficiency, improving quantitative accuracy, and expanding the application range, TLC technology had made great progresses, but it remained difficult to carry out quantitative detection. This limits the widespread use of TLC technology. Therefore, a research method combining TLC/HPTLC with MS technology has been developed and can be directly combined by TLC and MS (TLC-MS) without further purification. The relative molecular mass of the test substance and its structural information are obtained (Leeet al., 2004).
By improving TLC-MS method, Parket al. (2014) first used liquid extraction surface analysis (LESATM) and TLC-MS technology platforms in combination with electrospray ionization-time-offlight MS. The chromatographic analysis of bovine brain acid glycosphingolipids is carried out and the condition parameters of TLC-MS are optimized. The results showed that TLC-MS analysis under different conditions not only accurately obtains the expression profile of bovine brain glycosphingolipids containing gangliosides and sulfatides, but also analyzes the neutral sugar sheaths in the samples. The structures of lipids and sphingomyelin are also mapped. The above technology platform are not only applied to the analyses on metabolites, peptides, blood and tissue samples (Jautz and Morlock, 2006; Morlock and Ueda, 2007; Kertesz and Berkel, 2010; Walworthet al., 2010, 2011), but also shows good characteristics of small sample sizes, small loss of analytes, high stability and ability to analyze complex and fragile samples.
Analytical Methods and Strategies for Lipidomics
Until now, there has been no consensus method or single analytical system that can characterize all the lipids. As a result, variously analytical platforms have been applied to the lipidomic analysis of biological materials. To increase the coverage of researches, the analytical methods of lipidomics are increasingly diversified. Currently, GC-MS, capillary electrophoresis MS (CE-MS), nuclear magnetic resonance (NMR), matrix-assisted laser desorption ionization-MS (MALDI-MS), electrospray ionizationmass spectrometer (ESI-MS), shotgun lipidomics and LC-MS are widely used.
Gas chromatograph-mass spectrometry (GCMS)
Jameset al. (1952) first applied GC to the analysis of fatty acids. Polar capillary columns, such as DBWax or INNOWax, are often used in GC analysis. The sensitivity of GC-MS has been greatly improved by connecting MS to GC as a GC detector. As an important tool for lipid analysis, it has good separation efficiency and is relatively economical. The preparation of samples includes preliminary separation, hydrolysis, derivatization or thermal decomposition of lipids. As GC-MS can only analyze volatile organic compounds, for non-volatile lipids, lipids must be hydrolyzed by phospholipase C before analysis. The hydrolyzed products include free fatty acids, watersoluble products (saponified part) and non-polar, unsaponified components. The hydrolyzed product is then trimethylmethane. Silylation or methyl esterification is used to improve their volatility, and further GCMS analysis is performed (Peterson and Cummings, 2006). Because the esterified fatty acids are determined in the actual analysis, the site information of the fatty acid chains of the sn-1 and sn-2 acyl groups of lipids is lost during the determination process. In addition, the sample derivatization process not only takes extra time, but also requires a large number of samples and is prone to sample changes. Because of these shortcomings, GC-MS has been widely used in the analysis of relatively simple fatty acids, which limits its further application in the determination of complex lipid molecules. GC is also used in earlier studies to directly characterize TAG (Fontechaet al., 1998; Goudjilet al., 2003; Gutiérrezet al., 2009) and for quantification of cholesterol in milk (Contariniet al., 2017; Liuet al., 2018).
Capillary electrophoresis-mass spectrometry (CE-MS)
CE-MS has high separation ability and sensitivity and can be applied to the separation and detection of biological macromolecules, including lipids. Some common separation modes of CE, including capillary zone electrophoresis (CZE), micellar electrokinetic chromatography and capillary electrochromatography, have been applied in CE-MS, among which CZE is the most widely used (Ramautaret al., 2009). The application of CE-MS in milk is very common, but most involve the detection of peptides and some drug residues, and there are few studies on lipid characterizations in milk (Santoset al., 2005; Wanget al., 2007; Blascoet al., 2009; Klampflet al., 2009). Gaoet al. (2007) established a CE-MS technology for a non-aqueous buffer system to increase the solubility of phospholipids. Using the optimized organic solvent, the phospholipids in mouse peritoneal extract are separated within 16 min. The separation of PC, PE and PI was analyzed in this study, which proved the feasibility of CE-MS in phospholipid analysis.
Nuclear magnetic resonance (NMR)
NMR is a fast and simple method for the determination of small amounts of samples. Unlike LC-MS, NMR is an effective quantitative method that does not require a large number of optimized parameters. However, it cannot be used directly for the qualitative analysis of complex mixtures. Another challenge for NMR is limited knowledge of the carbon shelf structure and obtaining similar maps from two substances (Sundekildeet al., 2013). However, from another perspective, NMR is very useful for identifying phospholipids by analyzing the structure of the head group. Overall, the sensitivity of NMR is not as good as that of MS. Estradaet al. (2010) used NMR and MALDI-MS to re-evaluate the phospholipid contents in the lens of adult lens cells and found that the human lens crystal membrane contains a considerable amount of an ether-based glycerophospholipid and its corresponding hemolytic molecule. Using 31P NMR spectroscopy, Garciaet al. (2012) identified and quantified phospholipids in milk from different species (human milk, cow milk, camel milk and mare milk) (Estradaet al., 2010). Because of its rapid and simple analysis method, NMR has become one of the most commonly used methods of lipidomics in milk, but its accuracy and sensitivity still need to be improved.
Matrix-assisted laser desorption ionizationmass spectrometry (MALDI-MS)
MALDI-MS method is also a relatively mature analytical method in lipidomic researches, mainly used for tissue and body fluid analysis as well asin situdetection and imaging analysis (Ejsinget al., 2009; Chughtai and Heeren, 2010). This technique is simple and sensitive and is therefore suitable for the rapid screening of polar and non-polar lipids (Giddenet al., 2007; Fuchs and Schiller, 2009). MALDI technology is a soft ionization method based on laser technology. It is commonly used to analyze protein molecules and is now successfully applied to lipid molecular analysis. This method allows the ionization of the molecule to be measured by mixing the sample with the matrix and transferring the laser energy through the matrix to the sample. MALDI ion source is typically used in conjunction with time-offlight MS (TOF-MS). The application of milk lipids in MALDI-TOF-MS is low (Picarielloet al., 2007; Tzompa-Sosaet al., 2018), but this method is fast, convenient, reproducible, enabling rapid, accurate and high-throughput, thus, the future use range will also be expanded. However, the comprehensive analysis of MALDI- MS is limited and is generally considered unreliable for quantitative analysis (Han, 2016; Liuet al., 2018). Although MALDI has high sensitivity and analytical flux, the protonated molecule [M+H]+of phospholipids is often not seen in a typical MALDITOF map. The ionization mode of MALDI is greatly affected by the matrix. It is necessary to find a suitable matrix to assist the ionization of samples in order to achieve better reproducibility and comparability. In addition, MALDI-TOF-MS is a good method for identifying fatty acid composition in certain lipid species. However, quantitative analyses of the two different types of phospholipids are very difficult (Schilleret al., 2004).
Electrospray ionization (ESI)-mass spectrometry
ESI is a soft ionization method widely used in lipid histology analysis. The method is to eject the eluent containing the analyte through the tip of a high-voltage needle. The charged droplet is heated to evaporate the solvent. Finally, the analyte molecule forms gas-phase ions. Fennet al. (1989) first applied ESI technology to the analysis of a large number of mixtures. According to different analytical methods, ESI-MS can be divided into the following three categories: ESI-MS direct quantitative method, ESI-MS/MS quantitative method and HPLC-ESI-MS quantitative method. ESI technology can ionize biomolecules without fragmentation, which is conducive to better analyze of sample mixtures. Moreover, ESI does not require derivatization of the analyte. It requires fewer samples, simplifies the analysis steps and has high monitoring efficiency. According to the differences in analytical methods, the methods of quantitative analyses of lipids by ESI-MS can be divided into three categories; namely, ESI-MS direct quantitative method, ESIMS/MS quantitative method and HPLC-ESI-MS quantitative method. Compared with other analytical methods, ESI-MS has the advantages of simple pretreatment, high resolution and easy automation. It is suitable for qualitative and quantitative studies of lipid mixtures, such as phospholipids and mixtures (Han and Gross, 1994; Brüggeret al., 1997).
Shotgun lipidomics
Shotgun lipidomics is developed by Han and Gross (2005) based on ESI technology and triple quadrupole tandem MS. ESI technology is suitable for the analysis of polar, thermodynamically stable and non-volatile molecules, which can meet the requirements of most lipid analyses. The ionization efficiency of the analyte under ESI is one of the important factors affecting the accuracy of the analysis. In addition, new electrospray methods, such as Nanospray, have greatly improved the sensitivity and throughput of the analysis (Herreroet al., 2007; Déglonet al., 2012). Triple quadrupole MS is a mass spectrometer in which three quadrupole mass analyzers are connected in series. Among them, the first quadrupole (Q1) and the third quadrupole (Q3) are usually used for mass separation, while the second quadrupole (Q2) is usually used as a collision cell for collision-activated dissociation (CAD). By adjusting the function of the three quadrupoles, triple quadrupole MS can achieve product ion scan, precursor ion scan (PIS), neutral loss scan (NLS) and selection functions, such as selected reaction monitoring and multiple reaction monitoring. Because the same type of lipids will have the same sub-ion fragments or loss of neutral mass in CAD, according to the mass spectrum fragmentation characteristics of lipids, the specific scanning function of the series triple quadrupole, PIS and NLS can be used to realize the specific scanning and identification of different lipids (Han, 2016). ESI-MS/MS-based shotgun lipidomics technology is widely used in lipidomics because of its high accuracy, sensitivity and reproducibility and lack of pretreatment required for complex samples. Given that creamy matrices contain thousands of lipid species and a large number of analogs and isoforms, chromatographic separation is considered the key to avoid ion suppression and distinguishing among heterogeneous species, and shotgun MS is rarely used to identify milk lipidomics (Sokolet al., 2015; Liuet al., 2018).
Liquid chromatography-mass spectrometry (LC-MS)
LC-MS is one of the most widely used analytical platforms for studying lipids in biological samples, including milk lipids. There are many LC techniques used for lipid separation, including silver-ion-LC, chiral LC, supercritical fluid LC, off-line 2-D LC, normal phase LC, RP-LC and hydrophilic interaction LC. MS analyzers mainly comprise Q-TOF, Q-Trap, ion trap, triple quadrupole (QQQ), Fourier transformion cyclotron resonance and linear trap quadrupole-Orbitrap (Laakso and Manninen, 1997; Gastaldiet al., 2011; Lísaet al., 2011; Köfeleret al., 2012; Lísa and Holčapek, 2013; Yamadaet al., 2013; Liet al., 2014; Zhouet al., 2014; Liuet al., 2015a, 2015b, 2017a, 2017b, 2017c, 2018). LC-MS can not only determine the species of milk lipids and fatty acid composition of each lipid species and in some cases fatty acid regiospecific position, but also provide quantitative information on lipid class and individual species (Tuet al., 2017; Xuanet al., 2018).
Lipids need to be extracted prior to LC-MS analysis. Lipid extraction methods invented by Bligh and Dyer (2011) and Folchet al. (1957) are currently the most widely used. Some antioxidants, such as butylated hydroxytoluene, can be added to avoid oxidation before lipid extraction (Rupasinghe, 2013). To improve the accuracy of LC-MS analysis, internal standards covering each lipid class are usually added before extraction (Cajka and Fiehn, 2014). Additionally, prior to LC-MS analysis, it is often desirable to remove the extraction solvent (chlorine form/methanol) by evaporation and the lipid is reconstructed in different solvent systems compatible with the flow of LC. For polar lipid analysis, this procedure is particularly important to avoid unstable peaks and splitting (Liuet al., 2018). Furthermore, to improve the quality of lipidomic analysis data and facilitate the identification of lipid species, sample pre-cleaning or pre-fractioning by SPE columns or TLC prior to LC-MS analysis are often used. Lipidomics researches cover a wide range of topics and none of the existing analytical methods can detect all the lipids. However, the combination of different analytical methods can better overcome the limitations of a single technology. In order to improve the research methods of lipidomics, traditional lipid biochemistry, lipidomics profiling and lipid bioinformatics are required (Robertset al., 2008).
Bioinformatics of Lipidomics
Lipid classification and lipid database
The original data obtained by MS need to be identified, normalized and quantified before statistical analysis and, ultimately, lipidomics information can be obtained. At present, many databases are used in the studies of lipidomics. Through these databases, lipid structure, MS information and classification can be queried. Due to the lack of generally accepted lipid classification rules, the establishment of a lipid database is based on the research scope of each research unit. At present, there are three main lipid databases: LMSD, lipid bank and LIPIDAT. These databases provide most of the information regarding lipids found online. The three databases and other websites related to lipid research are listed in Table 1.
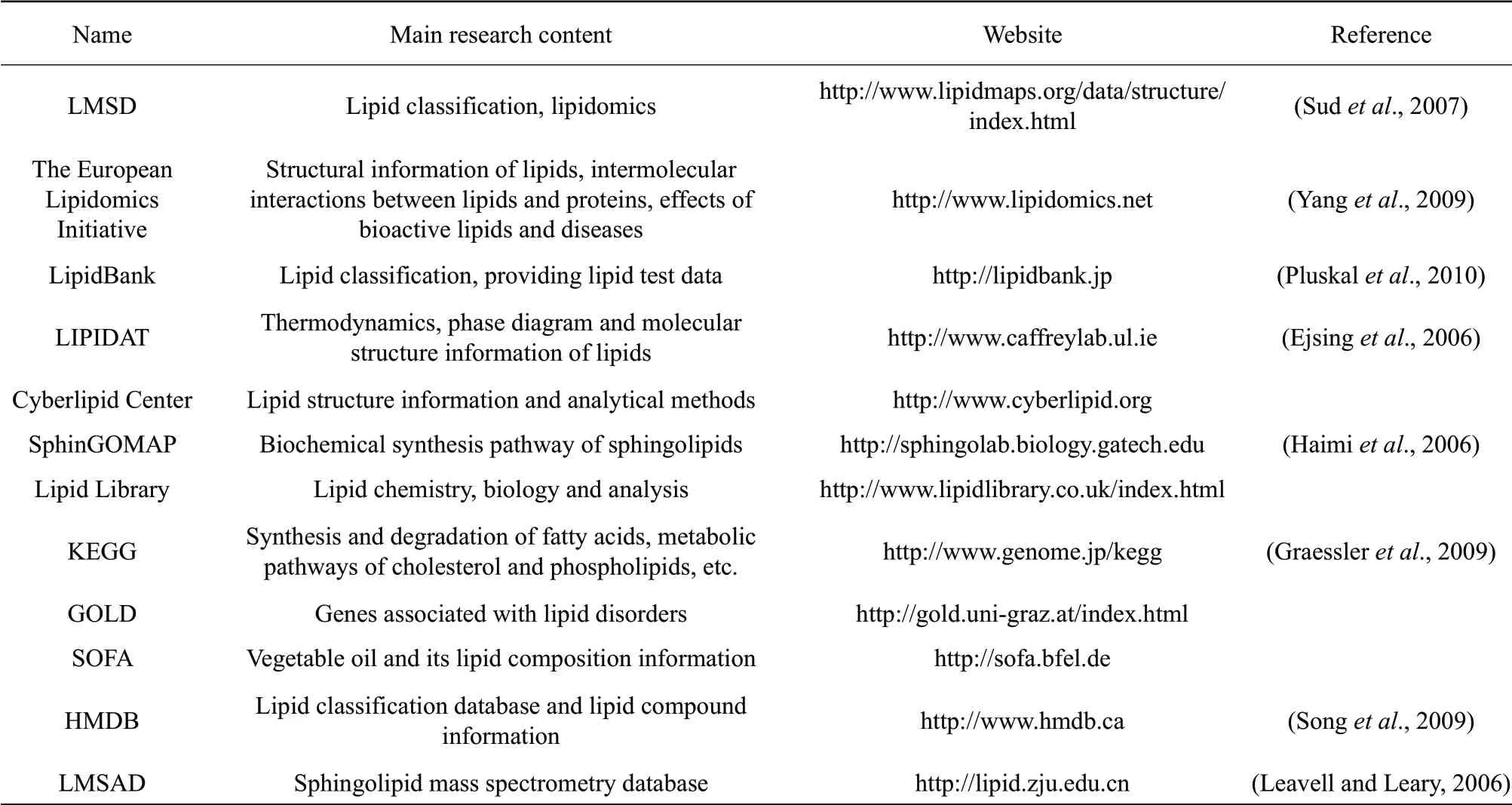
Table 1 Lipidomics databases and related websites
LMSD database was the result of the "Lipid Metabolism and Metabolic Pathway Research Strategy (LIPID MAPS)" project initiated by the USA in 2003. The database classifies lipids into eight categories as the followings: (1) fatty acyl; (2) glycerolipids; (3) glycerophospholipids; (4) sphingolipids; (5) cholesterol; (6) polyisoprene alcohol; (7) glycolipids; and(8) polyketone compounds. LMSD database identifies 1.68 million lipids and provides structural information on more than 10 000 lipids, including (1)-(5) lipids and cardiolipids, which have high levels in mammals. This structural information is mainly derived from the following four places: (1) LipidBank and LIPIDAT databases and other information bases; (2) core laboratories participating in LIPIDMAPS project and their collaborators; (3) LIPIDMAPS project experimentally identified lipids; and (4) derived from known corresponding lipids (Sudet al., 2007). The advantage of LIPIDMAPS system is the ease of database storage and management of biological information.
Lipid analysis software
In recent years, lipidomics analysis software based on MS data analysis had been continuously developed with various functions, such as data input, spectral filtering, peak detection, chromatographic alignment, standardization, visualization, multivariate statistical analysis and data output. With the rapid development of lipidomics, the functions of the lipidomics database and software will be continually improved. Lipid molecules are usually subjected to database search or software identification, and the internal standard method is usually used for the quantitative analysis of lipids. First, the collected MS data are subjected to isotope calibration, then the signal intensity of the internal standard and each test lipid in the mass spectrum is analyzed. Finally, the ratio is calculated according to the ratios of the two. The response sensitivity of different lipids in MS is different, thus, there are some bias in quantification using the internal standard method, but as most of the researches reflect the changes of lipids in a certain process of biological systems, such quantitative methods can roughly meet the research requirements. Lipid analysis software can be grouped into three categories as the followings: (1) freely available; (2) open resource; (3) purchased from commercial companies. Some commonly used lipidomic analysis softwares are listed in Table 2.
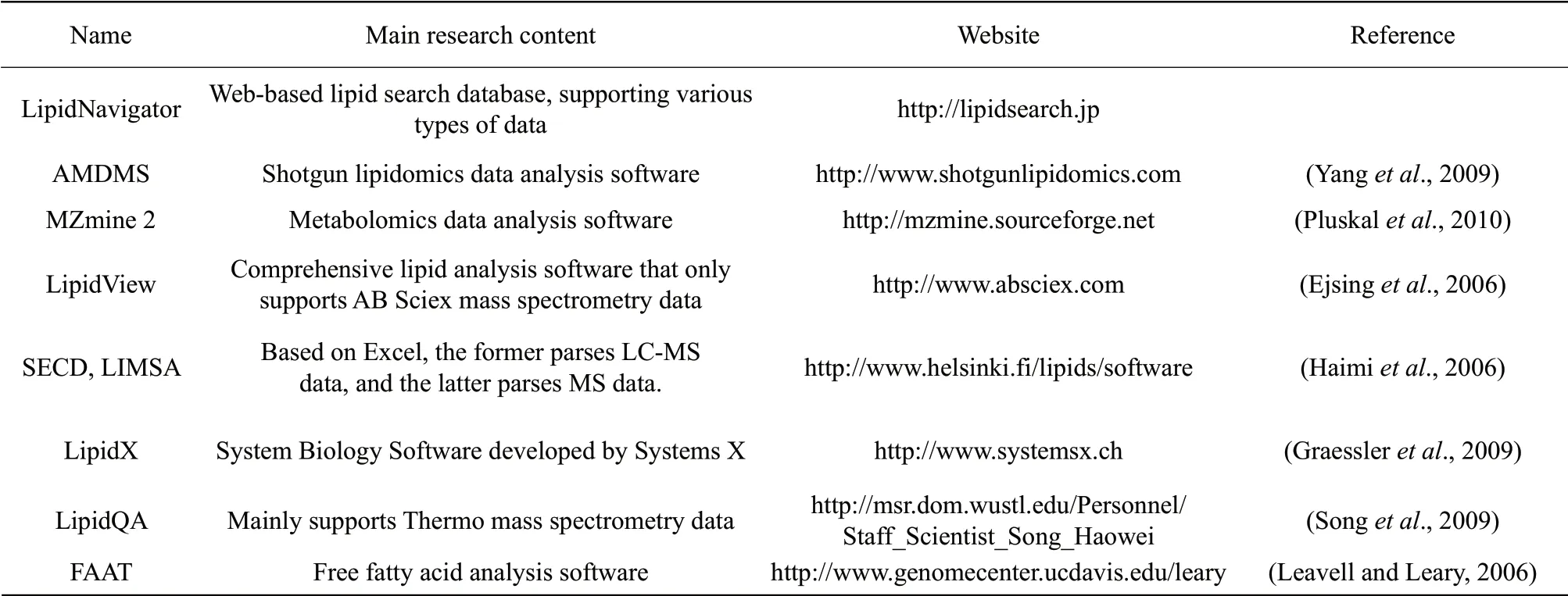
Table 2 Software of lipidomics and related websites
On the basis of qualitative and quantitative analyses of lipids, it is necessary to use bioinformatics to analyze the massive data obtained, especially the comprehensive data of lipid metabolites (Malavoltaet al., 2004). Bioinformatics analysis usually adopts multivariate statistical analysis based on pattern recognition, including unsupervised pattern recognition, such as principal component analysis and supervised pattern recognition and partial least squares (Songet al., 2009). Multivariate statistical analysis can be used to identify the association between lipid metabolites and physiological pheno-types and can effectively mine lipid biomarkers in mass data. Multivariate statistical analysis also includes the application of information technology to build metabolic networks, especially complex networks of lipids associated with specific proteins and genes. At present, application and bioinformatics mining with respect to milk fat are relatively scarce.
Conclusions
Milk has a unique composition and is an ideal food for newborn babies and infants. Increasing attention has been paid to the roles and characteristics of human milk. Studies focused on the elucidation of milk components have been performed. With the progresses of lipidomics, remarkable results have been achieved for lipids, including the identification and quantification of several humans and other mammalian milk lipids at the molecular species level, which provides useful information for the infant formula industry. However, there remain problems that must be solved, such as the complete identification of lipid species, especially for isomeric species in milk, the high-through analysis for milk lipidomes, the absolute quantification of lipids at the species level and the biological functions of human milk lipids at each molecular species. The progresses in chromatographic techniques as well as stationary phases in columns have greatly facilitated the analyses of TAGs in different types of samples. To elucidate the isomeric species and compositional isomers of TAGs and phospholipids as well as gain a better understanding of the low abundance lipids, multidimensional LC-MS should be developed for the exclusive separation of different kinds of lipids. For the complexity of lipids in human milk, finding sufficient standards for absolute quantification is not possible, thus, establishing an MS response algorithm for lipids of various structures might be helpful. Given the importance of infant formula to newborns and infants as well as milk and dairy products in human diet, building up the human milk lipidomes of mothers with different ethnicities, dietary habits and physical conditions, domestic animal lipidomes is urgently needed. In addition, there is a need to determine the biological functions of each molecular species of different kinds of milk lipids. With a variety of developing approaches in lipidomics, more useful results will be obtained in the future.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Effect of Foliar GA3 and ABA Applications on Reactive Oxygen Metabolism in Black Currants (Ribes nigrum L.) During Bud Paradormancy and Secondary Bud Burst
- Mining Heat Stress Associated Genes in Tomato Fruit (Solanum lycopersicum L.) Through RNA-seq
- Cloning of MYB2 Gene from Dryopteris fragrans and Its Response to ABA and Drought Stress
- NIRS Prediction of SOM, TN and TP in a Meadow in the Sanjiang Plain, China
- Screening of Feather Degrading Bacteria and Optimization of Fermentation Conditions from Poultry
- Isolation, Molecular and Phylogenetic Analysis of Porcine Encephalomyocarditis Virus Strain HLJ in China
