Generating a Hematopoietic Stem Cell Knowledge Graph for Scientific Knowledge Discovery
2021-02-12HUZhengyinLIULeileiCHENWenjieLIUChunjiangQIANLiSONGYibing
HU Zhengyin , LIU Leilei , CHEN Wenjie LIU Chunjiang , QIAN Li, , SONG Yibing
1.Chengdu Library and Information Centre, Chinese Academy of Sciences, Chengdu, Sichuan 610041, China
2.Department of Library, Information and Archives Management, School of Economics and Management, University of Chinese Academy of Sciences, Beijing 100190, China
3.National Science Library, Chinese Academy of Sciences, Beijing 100190, China
4.Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, Guangdong 510530, China
Abstract: [Objective]The hematopoietic stem cell (HSC) is one kind of the most effective stem cells for clinical treatments.It is of great significance to discover important knowledge entities, knowledge relations, and knowledge paths by literature mining for HSC knowledge discovery.Knowledge graph (KG), which represents knowledge entities and their relations with more details in a simple manner is widely used in scientific knowledge discovery (SKD).[Methods] This paper proposes a framework of generating KG using Subject-Predicate-Object (SPO) triples from literature, which includes six processes: literature retrieval, SPO extracting, SPO cleanup, SPO ranking, discovery pattern integrating, and graph building.Then, an HSC KG was constructed based on the Neo4j graph database following the framework.Finally, three kinds of SKD scenarios using HSC KG are introduced by empirical analysis.[Results] The results show that HSC KG has the advantages of “using graph data structure”, “integrating discovery patterns”, “fusing native graph mining algorithms”, and “easy to use”, which can effectively support deep open discovery, close discovery, and topic discovery in HSC.
Keywords: knowledge graph; SPO triple; scientific knowledge discovery; literature mining; hematopoietic stem cell
1 Introduction
Scientific and technological (S&T) literatures con- tains a large number of “credible, professional and standardized” scientific data, and information, which are important scientific research information resources[1].It will help effectively support data-driven scientific research by mining S&T literatures.However, with the overwhelming size and rapid growth of S&T literatures, it is a big challenge to efficiently and accurately mine and discover the new, useful, and potential knowledge from S&T literatures for scientific research, scientific management, and policymaking by understandable patterns in a credible way[1-2].SKD which derives from literature-based discovery (LBD), aims to alleviate these issues by combining natural language processing, text mining, semantic techniques, and scientometrics methods, and has become an important research area in biomedical informatics[1,3-4].
KG, officially proposed by Google in 2012[5], is essentially a kind of semantic network, which aims to describe the concepts, entities, events, and their relationships based on the graph model.In KG, the nodes are knowledge entities, the edges are relationships between them, and multiple connected nodes and edges form knowledge paths.As a kind of applicationoriented semantic technique, KG can effectively fuse heterogeneous data from various sources of information multi-dimensionally and fine-grained and enrich semantic relationships among them[4-6].KG which represents knowledge entities and their relations with more details in a simple manner and facilitates semantic retrieval, knowledge inference, knowledge path discovery, link prediction, and other knowledge discovery applications is widely used in SKD[4-6].
HSC is one kind of the most effective pluripotent adult tissue stem cell for clinical treatments with the earliest discovery and the longest research history[7].According to its biological characteristics of high selfrenewal, multi-lineage differentiation potential, and low immunogenic properties as well as migration and integration with the host tissue, HSC has been successfully used to cure many hematological malignancies, nonmalignant disorders, and some immune diseases, which may be more widely used in cancer treatment, cell therapy and other fields in the future[4,7].It is of great significance to discover important knowledge entities such as disorders, cells, genes, viruses, drugs, and pathogenic substances, latent knowledge relations between them and instructive knowledge paths among them by literature mining for HSC knowledge discovery.
The objective of this paper is to automatically gene- rate a HSC KG from S&T literatures to support HSC knowledge discovery.The rest of this paper is organized as follows.Section 2 briefly describes the related works about KG and SKD.Section 3 follows, presenting the framework of generating a KG using SPO triples from literatures in details.Three SKD scenarios using the HSC KG are given in Section 4, and Section 5 discusses the advantages of HSC KG and some limitations.Section 6 concludes this study and gives further possible applications of HSC KG.
2 Related works
In this section, we briefly introduce the construction of KG, SPO triples, and the corresponding tool SemRep[8]which is used to extract SPO triples from biomedical S&T literatures and three SKD scenarios.
2.1 Construction of KG and SPO triple
Generally, the construction process of KG could be divided into four parts: knowledge acquisition, know- ledge representation, knowledge storage, and knowledge inference[5-6].The knowledge acquisition aims to disco- ver and identify entities and relationships from unstruc- tured or semi-structured data, which includes relationship extraction, entity recognition and alignment, and know- ledge completion, etc.[5-6].Knowledge representation is a set of conceptual models and related techniques which are used to describe and model objectively existing knowledge with computer symbols to support knowledge com- puting and inferring[5,9].Knowledge storage is to save preprocessed and interrelated facts into a corresponding knowledge database according to a specific knowledge representation model[5,9].Knowledge inference refers to generalizing or discovering new facts from the existing knowledge[5,9].Knowledge databases such as Neo4j, TigerGraph, can realize knowledge storage and reasoning and hence continuously improve the KG.
Knowledge representation is the basis of the construction of a KG.There are some mature knowledge repre- sentation frameworks such as SPO triple, Resource Description Framework (RDF), and OWL.SPO triple, also called semantic predication, consists of a subject argument, an object argument, and the relation that binds them and helps to translate complicated free text into structured formats with rich semantic information[8].SemRep[8], a rule-based SPO triples extraction tool developed by the semantic knowledge representation (SKR) project of the National Library of Medicine (NLM), is wildly used to extract semantic predications from the content of biomedical S&T literatures.Elements of SPO triples are from the Unified Medical Language System (UMLS) knowledge ontology, and the subject and object of each SPO triple are concepts and terms in UMLS Metathesaurus, namely knowledge entities in KG; the predicate corresponds to relationship types in UMLS Semantic Network, namely relationships in KG[8,10].The relationships are further summarized to more than 50 basic semantic relations, which have become the semantic relations gold standard of biomedicine subjects[11].Taking the sentence “we used hemofiltration to treat a patient with digoxin overdose that was complicated by refractory hyperkalemia” as an example, three SPO triples (Hemofiltration - TREATS - Patients, Digoxin overdose - PROCESS_OF - Patients, Hyperkalemia - COMPLICATES - Digoxin overdose) are extracted by SemRep and one SPO triple (Hemofiltration - TREATS - Digoxin overdose) can be inferred by rules[8].
A SPO semantic network formed by a large number of semantic predications is considered a kind of domain knowledge graph[4,10].For example, SemMedDB[12], a large-scale biomedical repository of semantic predications extracted from the entire set of PubMed, is taken as a general knowledge graph for biomedical knowledge discovery.PubTator[13]is a Web-based KG system that provides automatic annotations of biomedical concepts such as genes, diseases, species, and mutations using cuttingedge machine learning and deep learning techniques.
In domain KG, Lamurias et al.[14]generated a tolerogenic cell therapy KG from PubMed to describe the relationships between cell and cytokine, which is used for tumor cell therapy knowledge discovery.Lion LBD[15], a literature-based discovery system for cancer biology generated a domain KG by mining the knowledge entities such as chemical, disease, mutation, gene, cancer, the hallmark, and the semantic relationship between them from S&T literature, which helps researchers to discover latent knowledge related to cancer from the literature efficiently.Fengfeng et al.[9]constructed a knowledge graph of intestinal cells from literatures, which can help medical researchers efficiently obtain domain knowledge.Hu et al.[16]developed a stem cell KG, which involves the effective integration of “multi-form, multi-granularity, multi-dimension” S&T literatures, S&T information, research data, and S&T service resources in stem cell and provides high-quality data support for SKD of stem cells.
2.2 Scientific Knowledge Discovery
Following LBD and scientometrics application scenarios, SKD can be divided into open discovery, close discovery, and research topic discovery.The first two methods are mainly for scientific research, while the latter is for scientific management.
Open discovery shown in Figure 1 (a) is a kind of divergent and exploratory knowledge discovery, which originates from an expansion of traditional literature retrieval demands.According to their own knowledge background or traditional literature retrieval, it is easy to find the knowledge nodes set B (b1, b2…) which is directly associated with a single knowledge node (a1) of interest for researchers.Open discovery helps to further discover some potential indirectly associated knowledge nodes set C (c1, c2…) and rank them according to importance, which is useful for the hypothesis generation in scientific research[4,15,17].
Close discovery shown in Figure 1 (b) is a kind of goal-oriented knowledge discovery, which is the most common one in scientific research.According to the scientific research aims, researchers hope to find the knowledge nodes set B (b1, b2…) which connects a given start node (a1) and end node (c1) directly or indirectly and the instructive knowledge paths among them.Close discovery helps to discover these knowledge patterns, which are often used to evaluate or test the scientific hypotheses[4,15,17].
Research topics are important analysis objects of bibliometric, and it is of great significance for mining these topics through S&T literatures for scientific management and policymaking.By discovering the topics of research focus, research frontiers, and the deve- lopment trends, it will provide a scientific basis and refe- rence for “Benchmarking and Catching up”.Furthermore, it is also helpful to subject key technology mining and technology foresight[18].

Fig.1 Illustration of closed and open discovery [4, 15]
3 Data and Method
3.1 Data
In this study, two global S&T literature databases - PubMed as the authoritative biomedical paper database and Derwent Innovation (DI) as the leading patent database, are chosen as the data sources.With the help of domain experts, the HSC literature retrieval strategies shown in Table 1 were made.After excluding some irrelevant literatures manually, 21,098 papers and 4,786 patents were eventually used for HSC KG.

Table 1 Search Policy and Results of HSC Literatures
3.2 HSC KG generating framework
We propose a framework for HSC KG generating (Figure 2), which includes literature retrieval, SPO extracting, SPO cleanup, SPO ranking, discovery pattern integrating, and graph building.Although the conclusive sen- tences of literature include essential knowledge units that synthesize its knowledge content[19], we hope to extract more different semantic types of SPO triples for a comprehensive KG.So we got SPO triples from the title and abstract fields of each paper and patent by SemRep after literature retrieval.We briefly introduce record and field selection for the HSC data, and then, go into the next “four” steps.
SPO Cleanup.The number of original SPO triples is huge, which contains a lot of noise, so it needs to be cleaned before use.Based on four basic principles pro- posed by Fiszman et al.[20], which are relevancy, connectivity, novelty, and saliency, the SPO

Fig.2 Framework for HSC KG generating
cleanup process includes three steps: entity cleanup, predicate cleanup, and SPO screening.(1) Entity cleanup.Firstly, according to the relevancy and novelty principles, some general or common academic/scientific Subjects and Objects concepts such as “Cells”, “Pati- ents”, “Disease” were removed.Then, the CUIs (UMLS Concept Unique Identifier) was chosen to align the concepts and used as the knowledge entity nodes in HSC KG.Afterward, the semantic types of entities such as “Cell”, “Gene or Genome”, “Disease or Syndrome”were used as the labels of these nodes[21].(2) Predicate cleanup.Some predicates such as “LOCATION_OF,” “ISA”, “PART_OF” that reflect hierarchy or posi-tion relationship and are meaningless for knowledge discovery were also removed.Then, the predicate gold standard[10]was chosen to align predicates and used as the edges between the knowledge entity nodes in HSC KG.(3) SPO Scree- ning.According to the connectivity and saliency principles, some very low frequency or isolated Subjects, Objects or SPO triples will be discarded.
SPO Ranking.This step calculates the weights of knowledge entities and relationships in HSC KG.The frequencies of concepts appearing in literature are used as the weight of knowledge entity nodes.In reference to the literature[22], we measure the strength of SPO triple using the Pearson correlation coefficient (Formula 1).

P (A, r) means the probability of occurrence for the concept A with Predicate r, P (r, B) means the probability of occurrence for Predicate r and the concept B, and P (A, r, B) means the probability of co-occurrence for the concept A and B with the Predicate r.
Discovery Patterns Integrating.The so-called kno- wledge discovery patterns refer to some conventional in- ference rules in KG.Hristovski et al.[23]proposed the know- ledge discovery patterns with semantic predications, and summarized a May_Treat pattern as follow:
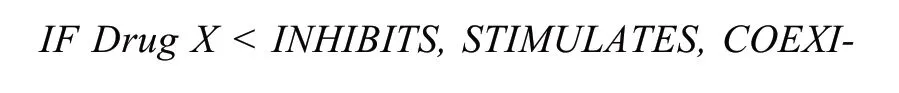
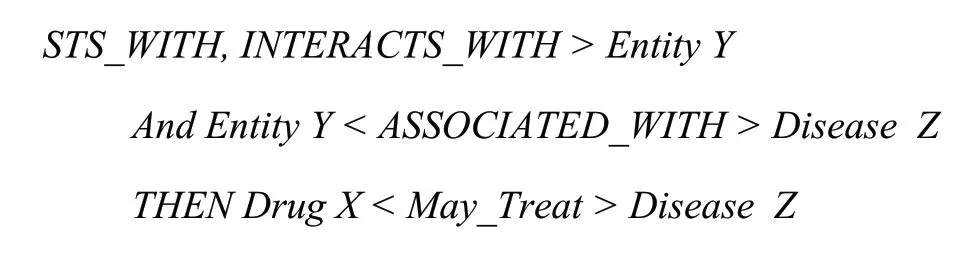
The May_Treat pattern concentrates on pharmacogenomics (relationship among drugs, genes, and disea- ses).The first line matches SPO with predicate INHIBITS, STIMULATES, COEXISTS_WITH, or INTERACTS_WITH, representing the corresponding action of one bioactive substance on another (X-Y relations).The second line matches SPO with predicate ASSOCIATED_WITH, representing association relations between a bioactive substance and a disease (Y-Z relations).The third line is the inference rule which means X may threaten Z (X-Z relations)[23].To discover subject know- ledge more effectively, some discovery patterns from literatures[23-25]were integrated into HSC KG (table 2).
Graph building.Neo4j, a popular graph database management system based on the native graph model, is used to build the KG from SPO triples.Neo4j provides a graph query language called Cypher to store and retrieve the patterns of nodes and relationships in the graph.Neo4j also contains a variety of graph mining algorithms such as centrality algorithms, pathfinding algorithms, link prediction algorithms, community detection algorithms which help discover the latest knowledge in KG effectively.For example, central algo- rithms can be used to find key nodes in KG, and pathfinding algorithms to discover the hidden association relationship and knowledge paths among the nodes, and community detection algorithms to discover topics[26].These algorithms can be easily called by Cypher.
4 SKD based on HSC KG
In this section, we briefly introduce the HSC KG.Then, three HSC knowledge discovery cases conducted by Cypher are given.
4.1 HSC KG
At present, there are 14,617 knowledge entities and 224,742 relationships in HSC KG.These knowledge entities were given 98 labels according to the UMLS semantic types[21].In the SKD scenarios of this paper, we focus on discovering important knowledge entities such as disorders, cells, genes, viruses, latent knowledge relations and instructive knowledge paths among them and HSC.So we choose “DISEASE, GENE, CELL, TIS- SUE, and VIRUS” as the key labels, which are shown with details in Table 3.And the relationships among them belong to 28 types in three categories — “knowledge entity-knowledge entity, knowledge entity- literature, SPO triple- literature” (Table 4).A part of HSC KG is shown in figure 3.More details about the HSC KG can be found at the website of http://biomed.kmcloud.ac.cn/hsc.
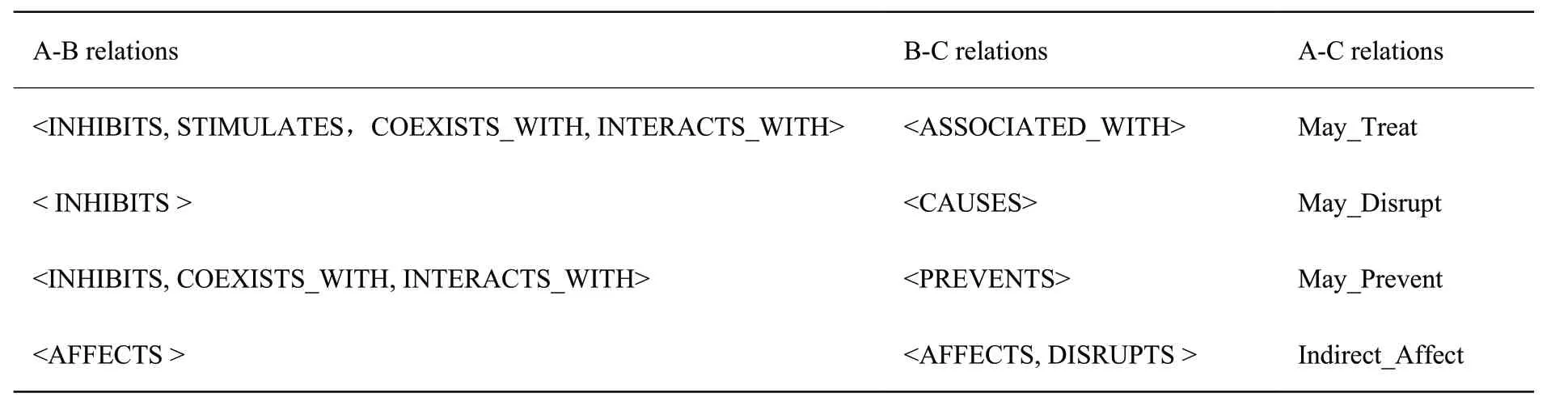
Table 2 Discovery Patterns with Predicate

Table 3 Knowledge entities in HSC KG

Table 4 Relationships in HSC KG
4.2 Open discovery case
Figure 3 shows some knowledge entities which have direct semantic relationships such as AFFECTS, INTERACTS_WITH with HSC nodes whose CUI pro- perty value is C0018956.Sometimes, it is more helpful to find those knowledge entities which have indirect semantic relationships with the HSC node.In this case, we show how to discover the knowledge entities which have indirect AFFECTS with HSC node by Cypher query on HSC KG.Following the Indirect_AFFECTS discovery pattern described in table 2, there are two steps to discover nodes that have Indirect_AFFECTS relationship with the HSC node.Firstly, we need to find the knowledge nodes set B (b1, b2 ...) which has AFFECTS relationship with the HSC node.Then, further to discover the knowledge nodes set C (c1, c2 ...) has AFFECTS or DISRUPTS relationship with each node in set B.The complete Cypher query statement for Indirect_AFFECTS discovery pattern of HSC is shown as follow:

Fig.3 Visualization of the part of HSC KG (focus on HSC node).Entities are shown as circles in different colors, such as blue CELL nodes, pink GENE nodes, green PROTEIN nodes, and more.Relationships among entities are represented by edges with arrows.
Excluding the knowledge entities with direct AFFE- CTS relationship with HSC, 169 knowledge entities with Indirect_AFFECTS relationship with HSC were discovered.By expert’s consultation, some important knowledge entities such as Bone remodeling, Tissue homeostasis, Natural regeneration, Degenerative abnormality, Cytokine production were picked out for further
analysis (Table 5).
4.3 Close discovery case
There is growing interest in the strategies for HSCbased therapy for HIV disease in academic circles[27].In 2019, Nature reported a case of a “London patient” who had no HIV virus replication in vivo for 18 months after bone marrow stem cell transplantation, which was praised as “the second cured AIDS patient in the world” and caused a sensation in the academic circles[28].In this case, we try to discover some heuristic knowledge paths (KP) which link the HSC and HIV in HSC KG by close discovery method.Here, the start node includes two closely related concepts of Hematopoietic stem cells (C0018956) and Hematopoietic Stem Cell Transplantation (C0242603), and the end node is HIV (C0019682).
Firstly, KP between HSC and HIV were extracted from HSC KG by path traversal algorithm.The short KP is generally public knowledge, and it is difficult to interpret the long KP.So KP whose length is between 2 to 4 were filtered with the following Cypher query statement 2.Then, following the Indirect_AFFECTS discovery pattern described in table 2, the specific knowledge paths containing AFFECTS relationship were filtered with Cypher query statement 3.

Some knowledge paths containing AFFECTS are shown in figure 4.Some interesting knowledge paths are picked out by experts as follows.KP 1 shows the following knowledge path: HSC differentiate immune cells which sense HIV invasion through receptors, and then they secrete antibodies to prevent HIV.KP 2 shows how the HSCT AFFECT HIV.During the HSCT, the donor HSC replaces receptor HSC (Graft acceptance) and the CCR5 gene mutate in immune cells (Gene Expre- ssion), which affects HIV infection.

Furthermore, a link prediction algorithm called Cypher (seen in query 4) is used to calculate the closeness score (4.79) between HSC and HIV.The score is relatively high, which means HSC and HIV will be more closely linked in the future.The result joins the academic opinions.

Fig.4 Specific knowledge paths between HSC and HIV

4.4 Research topic discovery case
The HSC KG is composed of thousands of nodes and edges, and the whole network becomes very complicated.Therefore, it is very difficult for experts to recognize the valuable research topics from the KG directly.In order to effectively find some valuable research topics, we try to mine topics that are clustered by the knowledge entities that HSC has AFFECT relationship with by subgraph generating and community detection approach in this case.
Firstly, a subgraph including these knowledge entities with which HSC has direct AFFECTS or Indirect_AFFECTS relationships with generated from HSC KG.The Cypher query for generating the subgraph was shown in Cypher query 5.The Neo4j Graph Data Science (GDS) library includes several community detection algorithms such as Label Propagation (LP), Louvain (LV), Modularity Optimization (MO), Triangle Count, and Weakly Connected Components, etc.[26].Then, LP, LV, and MO three community detection algorithms were chosen to mine communities from the subgraph.Experts were invited to review the different results and the MO algorithm was chosen to conduct community detection on the subgraph (Cypher query 6).Some important communities with which HSC has AFFECT relationship are shown in Figure 5.The community of cyan nodes focuses on leukemia, cancer-related genes, and carcinogenic factors; red community on the biological reaction and process of cell; gray community on the biological process and disease-related development process of HSC; green community on functions of HSC such as bone remodeling, immune response and diseases that can be treated by HSC; and yellow community on HSC related diseases as well as the pathogenesis.

Fig.5 Community detection of HSC subgraph by MO algorithm


5 Discussions
In the open discovery case, researcher starts from a problem to be solved, that is, what are the indirect effects of HSC? The computational complexity and the cognitive complexity of open discovery are both very high[29].HSC KG helps to efficiently and accurately discover those implicit knowledge entities by looking for a series of concepts that may be indirectly affected by HSC, and interpreting why this might happen.In the close discovery case, researchers have made a hypothesis (or preliminary experimental findings) that there should be associations between HSC and HIV.However, it is not clear what the specific associations are and how they occur.HSC KG helps to discover some heuristic knowledge paths between HSC and HIV by pathfinding and knowledge inference techniques.In the topic discovery case, researchers hope to find some valuable research topics about the effects of HSC.HSC KG helps to generate these topics by subgraph generating and community detection techniques.
From these SKD scenarios, we summarizes the advantages and limitations of HSC KG for SKD.Compared with other knowledge discoveries based on cooccurrence analysis and relational data model, the SKD based on HSC KG has the following advantages.(1) Graph data structure will better support SKD.HSC KG organizes data in the form of SPO triples, in which Subjects or Objects are nodes and semantic relationships edges.For open discovery, it is easy to get the knowledge entities directly or indirectly linked with the queried entity by Cypher query statements or query statements links.For close discovery, the knowledge paths or knowledge subgraphs composed of multiple SPO triples which connect the given start queried entity and end queried entity are also easy to be extracted by Cypher pathfinding query statements.(2) HSC KG supports knowledge inference by integrating discovery patterns.Discovery patterns are very important tools that help efficiently and accurately discover knowledge from a complex data network.And it is easy to add or update discovery patterns with Cypher statements, which increases the knowledge inference ability of HSC KG greatly.(3) HSC KG is managed by a graph database system that provides many advanced graph mining algorithms such as pathfinding, link prediction, and community detection, and so on.The data in HSC KG is easily called by these algorithms to support complex and deep SKD scenarios.
Compared with RDF, OWL, and other heavy KG techniques, SPO triple which is a kind of light but power- ful semantic knowledge representation model can more flexibly represent the knowledge entities and their relationships in S&T literature.And there is a set of mature tools and data set based on SPO triples such as SemRep, UMLS for biomedical KG construction.It doesn’t rely on manually labeled training data sets or complex machine learning models.Instead, it extracts SPO triples with normalized concepts and semantic relationships, which has the advantages of high accuracy, high effi- ciency, and easy interpretation.So it is easier to construct and update KG for SKD.
There are some limitations in HSC KG.First of all, the data of HSC KG are extracted from S&T literature which is increasing every day.How to update the HSC KG data timely and automatically is very important.Secondly, the interpretability of SKD results is relatively poor.Early LBD analyses, such as co-occurrence analysis of S&T terms, are easy to be interpreted and understood.However, SPO triples, knowledge path, and subgraph are relatively obscure and complex for domain experts who often find it difficult to understand the KG analysis process and think that the interpretability of results is also poor.In addition, it is difficult for domain experts to use HSC KG directly.At present, all analyses are conducted on the Neo4j system using the Cypher language and there is no user-friendly interface.
6 Conclusions and Future Directions
With the rapid rise of the data-intensive science paradigm, data-driven SKD will become a significant feature and important pathway of scientific research and is becoming the development direction of subject knowledge service[4,18].In order to meet the needs of deep SKD, this paper proposes a framework for generating KG using SPO triples from S&T literature for SKD.Following the framework, we construct an HSC KG based on the Neo4j graph database by five processes: literature retrieval, SPO extracting, SPO cleanup, SPO ranking, discovery pattern integrating, and graph building.Finally, three SKD scenarios based on the HSC KG were introduced.The results show the HSC KG has advantages of “using graph data structure”, “integrating discovery patterns”, “fusing native graph mining algo- rithms”, and “easy use”, which can effectively support deep open discovery, close discovery, and topic discovery in HSC.
However, there are still some challenges, such as enriching HSC KG data, increasing the interpretability of SKD results, and providing a friendly user interface to access the KG.In the future, we intend to enrich the HSC KG data, including extracting SPO triples from the clinical trials literature and integrating with some correlative linked data sets, etc.An intelligent question answering system is also a typical SKD application of HSC KG.Moreover, further research about how to improve the interpretability of the process and results of SKD based on HSC KG will be a hot and difficult topic in the future.
Acknowledgments
We appreciate the professional consultation provided by Dr.Wei Sixuan, Zhang Xuechun from the Institute of Zoology, Chinese Academy of Sciences, and Dr.Zhu Yanling from Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences.
