Complete Draft Genome Sequence of Cutibacterium (Propionibacterium) acnes Type Strain ATCC6919
2021-01-09HuiJingTangKaiWeiSiXiaoZhangHeLingZhuWenLiangZhengXiBaoZhangJingYaoLiangJianBoRuan
Hui-Jing Tang, Kai-Wei Si, Xiao Zhang, He-Ling Zhu, Wen-Liang Zheng, Xi-Bao Zhang,Jing-Yao Liang,∗, Jian-Bo Ruan,∗
1Department of Dermatology, Dongguan Hospital Affiliated with Jinan University Medical School, Dongguan, Guangdong 523905,China; 2Department of Dermatology, The Seventh Affiliated Hospital of Sun Yat-sem University, Shenzhen 510178, China; 3BGI Genomics, BGI-Shenzhen, Shenzhen 518083, China; 4Department of Dermatology, Guangzhou Institute of Dermatology,Guangzhou, Guangdong 510095, China.
Abstract
Keywords: ATCC6919, Cutibacterium acnes, resistance, whole-genome sequencing
Introduction
Cutibacterium (formerly Propionibacterium) acnes (C.acnes) is a commensal bacteria on human skin and an important pathogen in acne.1-2C. acnes type strain ATCC6919 (formerly DSM1897 and NCTC737) has been widely used as a reference susceptible strain in comparisons of antibiotic resistance and gene sequences.3-4The first draft genome sequence of ATCC6919 was published in 2014 using whole-genome shotgun sequencing with Illumina Miseq.5Recently, several bacterial genomes have been resequenced by next-generation sequencing technology; this has revealed a large number of errors and undetermined gap regions in the previously published draft genome sequencing, which have affected the accuracy of comparative genomics and protein annotation.6-7In the present study, we resequenced the ATCC6919 genome by high-throughput deep sequencing and reannotated the genome to provide a high-quality,complete whole-genome sequence.
Materials and methods
C.acnes ATCC6919 was purchased from the Guangdong Microbial Species Conservation Center, China, and was cultured on Luria-Bertani agar at 37°C. ATCC6919 was identified through the VITEK 2 system (bioMérieux,Marcy-l’Étoile, France). The antimicrobial susceptibility of ATCC6919 was tested using the agar dilution method containing 0.625-265mg/L of various antibiotics (sulfamethoxazole,erythromycin,azithromycin,clarithromycin,tetracycline, minocycline, metronidazole, trimethoprim,levofloxacin,ceftriaxone,and doxycycline),and the results were interpreted in accordance with the Clinical and Laboratory Standards Institute guidelines.8-9Genomic DNA was extracted and purified using Genomic-tip 100/G following the manufacturer’s protocol(Qiagen,Germantown,MD,USA).The genomic library was prepared using the standard Pacific Biosciences(PacBio,Menlo Park,CA,USA) protocol for 20-kb libraries. The genome was sequencedusingtheIlluminaHiseq4000platform(Illumina Inc.,San Diego,CA,USA)in combination with PacBio RSII single-molecule real-time sequencing (Pacific Biosciences).Thegenerated sequencereadsweredenovoassembledusing the HGAP3 protocol of single-molecule real-time analysis v.2.3.0,and were revised by Pilon v.1.22 software.10Genes were predicted with the National Center for Biotechnology Information(NCBI)prokaryotic genome annotation pipeline.11The tRNA and rRNA genes were predicted using tRNA scan-SE v1.3.1 and RNAmmer v1.2, respectively.The genome sequence was annotated using 15 databases,such as Gene Ontology,Kyoto Encyclopedia of Genes and Genomes,Cluster of Ortologous Groups of proteins,Nonsupervised Orthologous Groups (v.4.5), Swiss-Prot, Antibiotic Resistance Genes Database(v.1.1),and the Comprehensive Antibiotic Resistance Database(v.1.2.0).
Results
The sequencing reactions yielded 1,088,007,528 reads with sequencing depths of 100×. There were 462 Mb(PE150,insert size=270bp)of clean data in Illumine,and 1,083 Mb of filtered subreads with 108,813 subreads in PacBio. The draft genome sequence of ATCC6919 consisted of a single circular chromosome of 2,495,001 bp(one contig and an N50value of 2,495,001bp),with a GC content of 60.02% (Fig. 1A). In total,2,450 putative coding sequences(including 2,358 protein-coding sequences, 45 tRNA, 9 rRNA, and 1 sRNA) and 52 tandem repeats were detected. Of these 2,450 putative coding sequences, 2,437 (99.46%) were annotated by 15 relational databases. Furthermore, the gene (thy A) in the genome of the isolate was found in ATCC6919,which was associated with resistance to aminosalicylic acid in the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/).Antimicrobial susceptibility testing of ATCC6919 showed that this isolate was not sensitive to metronidazole and sulfamethoxazole,but was sensitive to all other tested antimicrobials (erythromycin, azithromycin, clarithromycin, tetracycline, minocycline, trimethoprim,levofloxacin,ceftriaxone,and doxycycline).Table 1 summarizes the genomic features of the new and previous sequences of ATCC6919.The average nucleotide identity of the new sequence of ATCC6919 was 99.91%identical to the previous sequence of ATCC6919 (Fig. 1B). The complete genome sequence of C.acnes strain ATCC6919 has been deposited at GenBank under the accession number CP023676.1 (https://www.ncbi.nlm.nih.gov/nuccore/1373293347/).
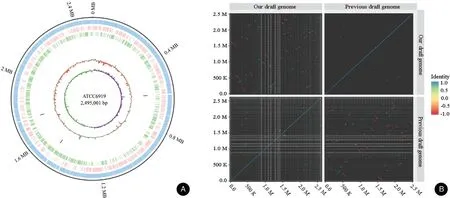
Figure 1. The sequence characteristics of ATCC6919 genome tested.(A)The Circos diagram of the ATCC6919 genome.The tracks from the outer to the inner rings represent the ATCC6919 genome size(in Mb;black),all anno gene(blue),forward stand gene(red),reverse stand gene(green),tRNA(light purple),rRNA(purple), sRNA(grey),GC(orange-green),and GC-skew(green-purple).(B)The average nucleotide identity values of the new and previous reported ATCC6919 genomes.
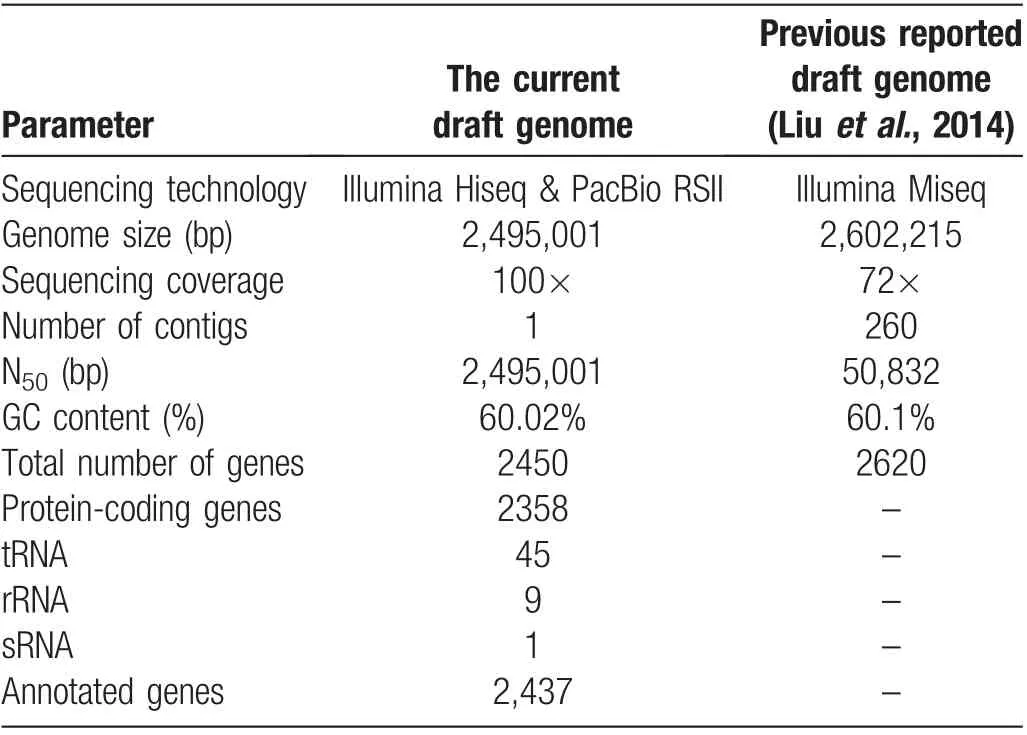
Table 1 Genomic features of the current and previous draft genome of ATCC6919.
Discussion
C.acnes play a critical role in the pathogenesis of acne,and is developing resistance to antimicrobials used for acne treatment, particularly through genomic variations.12-14The lack of a complete genome sequence of ATCC6919 has hindered comparative genomic studies evaluating the antimicrobial resistance of C. acnes. In the present study,we resequenced the genome of ATCC6919 using Pacific Biosciences sequencing technology and high-throughput sequence analysis platforms. Compared with the previously published ATCC6919 draft sequence acquired using second-generation sequencing technology,5the new genome sequence was assembled into only a single contig with a higher N50value and higher sequencing coverage,indicating better quality and accuracy. The new genome sequence of ATCC6919 is shorter than the previously published sequence; this may be because the previous genome sequence comprised a plasmid sequence and/or a large number of repetitive sequences. Indeed, the average nucleotide identity results showed that the new sequence identity was more than 99% comparable to that of the previous sequence, suggesting a very high degree of similarity between the two sequencing technologies.ATCC6919 has been used as an important reference in C. acnes studies globally.3,5,9,15-16The complete genome sequence of ATCC6919 will provide important insights into the genomic features, molecular evolution, epidemiology, and pathogenesis of C. acnes in future studies.
Acknowledgements
This study was supported by the Social Science and Technology Development Projects of Dongguan(Nos. 2013108101042 and 201950715025174), the Natural Science Foundation of Guangdong (Nos.2018A0303130011), and the Science and Technology Program of Guangzhou(No.201904010191).
杂志排行
国际皮肤性病学杂志的其它文章
- Instructions for Authors
- Platelet-Rich Plasma for Androgenetic Alopecia in Women: A Single-Center Case Series Study in Qatar
- Intracranial Abnormalities of Infantile Hemangiomas in the Head and Neck Regions:A Retrospective MRI Study
- Efficacy and Safety of Prednisolone Monotherapy Versus Prednisolone Plus Methotrexate in Erythema Nodosum Leprosum(Type 2 Lepra Reaction)
- The Past 70 Years in Control of Syphilis in China: Elimination and Responses to Resurgence
- Hair Matrix Cyst
