Effects of viral infection and microbial diversity on patients with sepsis: A retrospective study based on metagenomic next-generation sequencing
2021-01-06LiweiDuanJinlongQuJianWanYonghuaXuYiShanLixueWuJinhaoZhengWeiweiJiangQitongChenYanZhuJianZhouWenboYuLeiPeiXiSongWenfangLiZhaofenLin
Li-wei Duan, Jin-long Qu, Jian Wan, Yong-hua Xu, Yi Shan, Li-xue Wu, Jin-hao Zheng, Wei-wei Jiang,Qi-tong Chen, Yan Zhu, Jian Zhou, Wen-bo Yu, Lei Pei, Xi Song, Wen-fang Li, Zhao-fen Lin
1 Department of Emergency and Critical Care Medicine, Changzheng Hospital, the Second Military Medical University,Shanghai 200003, China
2 Department of Emergency and Critical Care Medicine, Pudong New Area People’s Hospital, Shanghai 201299, China
Corresponding Author: Wen-fang Li, Email: chzhedlwf@163.com; Zhao-fen Lin, Email: linzhaofen@smmu.edu.cn
KEYWORDS: Sepsis; Metagenomic next-generation sequencing; Viral infections; Bacterial infections; Microbial diversity
INTRODUCTION
Sepsis has been ranked as one of the major public health concerns in intensive care units (ICUs) worldwide,causing an increasing number of deaths each year.[1]Early identification of responsible pathogens and prompt administration of specific antibiotics are crucial steps for patients’ prognosis.[2,3]Early antimicrobial treatment is recommended in current clinical instructions, specifically within one hour after the diagnosis of sepsis.[4]However,the majority of current therapies are empirical. Blood culture has been the gold standard for sepsis diagnosis,whereas obtaining final results of blood cultures can be time-consuming and some pathogenic microsomes are difficult to be culturedin vitro.[5]Additionally, prior usage of broad-spectrum antibiotics may confound the real specific diagnosis.[6,7]Polymerase chain reaction (PCR)-based techniques have been introduced for pathogen detection in recent decades.[8]Nevertheless, these techniques are limited by the collection of tissues and the lack of quantitative measurement of microorganisms load, and are also unable to detect the cause of antibiotic resistance.[9]
Despite the limitations, the techniques mentioned above have improved the accuracy and speed of clinical diagnoses of causative pathogens. However, some clinical concerns remain to be addressed. In our clinical practice, we found that many patients with bacterialinfected sepsis did not acquire satisfactory improvement regarding survival rate and ICU scores, even those treated by targeted antibiotic therapy based on blood culture.Traen et al[10]found that ICU patients with positive herpes simplex virus (HSV) type 1 (HSV-1) could acquire lower ICU mortality after the prophylactic use of acyclovir. A different study conducted by Schuierer et al[11]reported that in patients with ventilator-associated pneumonia and antibiotic treatment failure, acyclovir treatment was associated with significantly longer survival time in the ICU and improved circulatory and pulmonary functions.We hypothesized that concomitant viral infection could affect the survival of patients with sepsis.
Next-generation sequencing (NGS) has recently been proposed for critically ill patients who suffer from bloodstream infections.[12]NGS allows for an unbiased analysis of the bloodstream, including prompt diagnosis,stable results, and quantitative score.[13]This technology could be used in medical microbiology laboratories and useful for infection prevention measures.
Therefore, this study aims to investigate the performance of an NGS-based diagnostic technique for the identification of potential bacterial and viral infections, and effects of concomitant viral infection on the survival rate of sepsis patients in the ICU.
METHODS
Study design
This present study was approved by the Ethics Committee Board of our institution. Data were collected from all ICU patients who were admitted to our institution from February 1, 2018 to June 30, 2019.
The inclusion criteria for patients were as follows: (1)complete blood culture results and medical records; (2)aged over 18 years; (3) body temperature over 38 °C or below 36 °C; (4) simultaneously combined with one of the following items: (a) the definite invasive sites or migration foci; (b) systemic toxemic symptoms but no obvious infection foci; (c) unexplained rashes or bleeding spots, hepatosplenomegaly, or increased blood neutrophil counts; (d) systolic pressure less than 90 mmHg (1 mmHg=0.133 kPa) or decreased over 40 mmHg; (e) the diagnosis of pathogenic microorganisms via blood culture (if coagulase-negativeStaphylococcus,Propionibacterium, and other common skin contaminants exist, the blood sample collection would be conducted at a different time point) or identification of antigen of responsible pathogens before recruitment.
The exclusion criteria for patients were as follows:(1) pregnant or breast feeding; (2) malignant tumor,blood-related diseases, or HIV infection; (3) severe organ dysfunction; and (4) chronic infection, such as chronic inf lammatory bowel disease.
Thirteen patients were excluded from this study,including two patients because of poor sample quality and 11 because of incomplete medical records. Finally,a total of 74 patients were enrolled, and all the patients signed an informed consent form. If the patient was incapable of giving consent owing to the underlying severe infection, informed consent could be given by the patient’s legal guardian until the patients were informed.
Sample preparation and processing
A total of 118 venous blood samples (5 mL each)were collected in cell-free DNA (cfDNA) tubes(PAXgene Blood ccfDNA Tube, Feldbachstrasse 8634 Hombrechtikon, Switzerland) from the 74 patients with the blood infection. Blood samples were stored at 4 °C before plasma separation. Samples were collected when the patient’s body temperature was higher than 38 °C or below 36 °C. The tubes were spun down at 1,600gfor 10 minutes at room temperature for plasma preparation.The cfDNA was extracted from plasma using QIAamp circulating nucleic acid kit (Qiagen, Valencia, USA)following the manufacturer’s instructions. The extracted DNA specimens were used to construct DNA libraries.
Sequencing
NGS libraries were prepared for sequencing.Negative controls (buffer only instead of plasma) and positive controls (healthy plasma spiked with a known mixture of microbial DNA fragments) were processed alongside patient samples in each batch. The quality of the DNA libraries was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA) to measure the adapters before sequencing. High throughput sequencing (MedcareDx Bio-Tech, Shanghai, China)was performed on the cfDNA sample using an Illumina NextSeq550Dx platform. On average, twenty-five million sequence reads were acquired for each sample.
Reference database and quality control
Reference genomes for Homo sapiens (National Center for Biotechnology Information [NCBI], National Library of Medicine [NLM]), and microorganisms(bacteria, viruses, fungi/molds, and other eukaryotic pathogens) were retrieved from NCBI ftp site (ftp://ftp.ncbi.nlm.nih.gov/genomes/). After elimination of the taxonomic mislabeling and sequence contamination in the reference genomes, the curated sequences were assembled into a single Fasta file and used to generate our microorganism reference database, which contained genomic sequences from 6,030 bacteria, 3,551 viruses, 185 fungi, and 87 parasites.
Qualified sequencing data of each clinical sample were generated by removing adapter contamination,low-quality reads, duplicates, and short reads (length<35 bp) from the raw sequencing data, followed by excluding human host sequences mapped to human reference (GRCh38.p12) with the Burrows-Wheeler Alignment tool.[14]After removing low-complexity reads,the unmapped sequencing reads in each sample were retained for further analysis.
Determination of pathogens
To identify the pathogenic sequences, the remaining unmapped sequences were aligned to the curated microorganism reference database as described above.The strictly mapped sequencing reads were classif ied into bacterial, viral, fungal, and parasitic at the species level.Considering the confounding factors, such as the number of sequencing reads, the genome size, and coverage rate,the quantity for each microbe identified was expressed as the normalized number (NN) of DNA sequencing reads in plasma in terms of Langelier’s study.[15]Species with NN less than three were removed, whereas species with NN greater than ten were reported. For those species with NN between three and ten, the Basic Local Alignment Search Tool for nucleotide was implemented in the nucleotide database to verify the identification accuracy, and the verif ied species were reported.[16]
Statistical analysis
Statistical calculations were performed in R (version 3.5.0) with the following packages: exactRankTests,survival, survminer, ggplot2, and PMCMRplus.[12]Continuous data were presented as mean and standard deviation and were compared using Student’st-test.Categorical data were compared using Fisher’s exact test or Chi-square test. Kaplan-Meier plots were used to visualize survival curves for a 90-day follow-up, where were compared using the log-rank test. We then applied univariate and multivariate Cox proportional hazards models to identify independent prognostic factors for 90-day mortality. All the variables (including gender,age, Acute Physiology and Chronic Health Evaluation II [APACHE II] score and Sequential Organ Failure Assessment [SOFA] scores, white blood cell [WBC],neutrophil, leukomonocyte, platelet counts, procalcitonin,C-reactive protein, and lactic acid levels) were considered in separate univariate Cox regression models and those with aP-value of 0.05 or less entered into a multivariate Cox regression model. Analyses of correlation between continuous variables were performed using linear regression with coefficient of determination (R2). AP-value of less than 0.05 was considered statistically signif icant.
RESULTS
Patient population
In total, 74 patients diagnosed with sepsis were included(Table 1). Patients were divided into the virus-positive group and the virus-negative group according to the presence or absence of concomitant viral infection confirmed by NGS.There were 27 male and 10 female patients in the virusnegative group, whereas there were 23 male and 14 female patients in the virus-positive group (P>0.05). The mean age of the virus-negative group (53.86±17.03 years) was lower than that of the virus-positive group (62.35±14.43 years) (P<0.05). Patients with concomitant viral infection experienced a longer period of ICU hospitalization than those in the virus-negative group (39.11±28.18 days vs.24.62±28.05 days,P<0.05). Patients in the virus-positive group had higher APACHE II and SOFA scores than those in the virus-negative group (bothP<0.01). However, no statistically significant differences were observed in the laboratory infection parameters.
Top 20 positive pathogenic microorganisms detected by NGS
A total of 118 samples from 74 patients were tested by NGS, which yielded 88.98% positive results over the whole study period.Pseudomonas chlororaphis,Pseudomonas aeruginosa, andStenotrophomonas maltophiliawere the most frequently detected pathogens after the exclusion of typical skin commensals such as coagulase-negative (CoN)Staphylococci. However,despite the current gold standard, blood cultures were unable to identify concomitant viral infection, whereas NGS could give a broad viral prof ile. Human herpesvirus 5 was the most frequently detected pathogen, present in approximately 29.73% (22 of 74) of our patients.
Correlation of viral load with sepsis severity identified by Cox regression analysis and survival time
Owing to the limited number of virus varieties detected in the patients, we did not perform a similar analysis. Instead, we employed Cox regression analysis.Seven variables (sex, WBC count, procalcitonin,lactic acid level, APACHE II score, SOFA score, and concomitant viral infection) were observed to correlate with the severity of sepsis based on univariate Cox regression analysis (Table 2). Further, to correct for possible confounding factors, data for these seven variables were adjusted based on the Cox multivariate regression model. In all patient groups, the concomitant virus infection was closely associated with an increased hazard ratio for sepsis severity (Figure 1). The impact of concomitant viral infection on the survival rate was significantly obvious (P<0.05; Figure 2). In addition,at 25 days after admission, the survival rate decreased continuously for those in the virus-positive group, whereas it did not change significantly in the virus-negative group.These results indicated that concomitant viral infection correlated closely with sepsis severity and had negative effects on the survival of patients with sepsis.

Table 1. Demographic data and clinical characteristics of patients in this study

Table 2. Univariate Cox proportional hazards analysis for the 90-day mortality
No signif icant correlation between bacteria and sepsis severity
As the primary cause of sepsis, pathogenic bacteria play a crucial role. Therefore, we also investigated the correlation between the bacteria varieties and sepsis severity. No significant correlation was observed between the bacteria varieties and sepsis severity in either laboratory results or ICU scores for patients with less than ten types of bacteria (R2=0.15,P=0.20), and for patients with more than ten types of bacteria (R2=0.02,P=0.88). These results indicated that bacteria varieties did not correlate with the severity of sepsis.
DISCUSSION
Sepsis has a significa nt impact on the critically ill population. Rapid diagnosis is thus important not only to enhance patients’ survival rate but also to promote reasonable antibiotic use. Nevertheless, an accurate and comprehensive identification of pathogenic microorganisms is still challenging. In addition, patients’clinical symptoms and laboratory results are sometimes nonspecific.[17]In recent years, whether the concomitant viral infection is a potential causative agent in failed sepsis treatment has emerged as a subject of active debate.[18]Increasing evidence has suggested a close correlation between viral load and poor prognosis.[19-21]A prospective study with 329 patients enrolled for septic shock reported that 112 (34.04%) patients who had multiple concurrent viremia events suffered higher risk of mortality.[22]These results suggested that concurrent viral load could not be ignored. Therefore, a timely and accurate identification of viral load could provide valuable information for later reasonable therapy in critically ill patients.
NGS-based approaches could provide a rapid and comprehensive detection of bacterial, fungal, and viral infections in a single assay.[12]To the best of our knowledge, few published studies have investigated the effect of concomitant viral infection on the survival of patients with sepsis.[23,24]Therefore, we investigated the performance of NGS-based diagnosis technique for the effect of concomitant viral infection on the survival rate in ICU septic patients.

Figure 1. Forest plots of multivariable Cox model adjusted hazard ratios with 95% conf idence interval for the 90-day mortality. The multivariable Cox model was adjusted for sex, WBC count, SOFA, lactic acid, PCT, and concomitant virus infection at admission. WBC: white blood cell;SOFA: Sequential Organ Failure Assessment; PCT: procalcitonin.
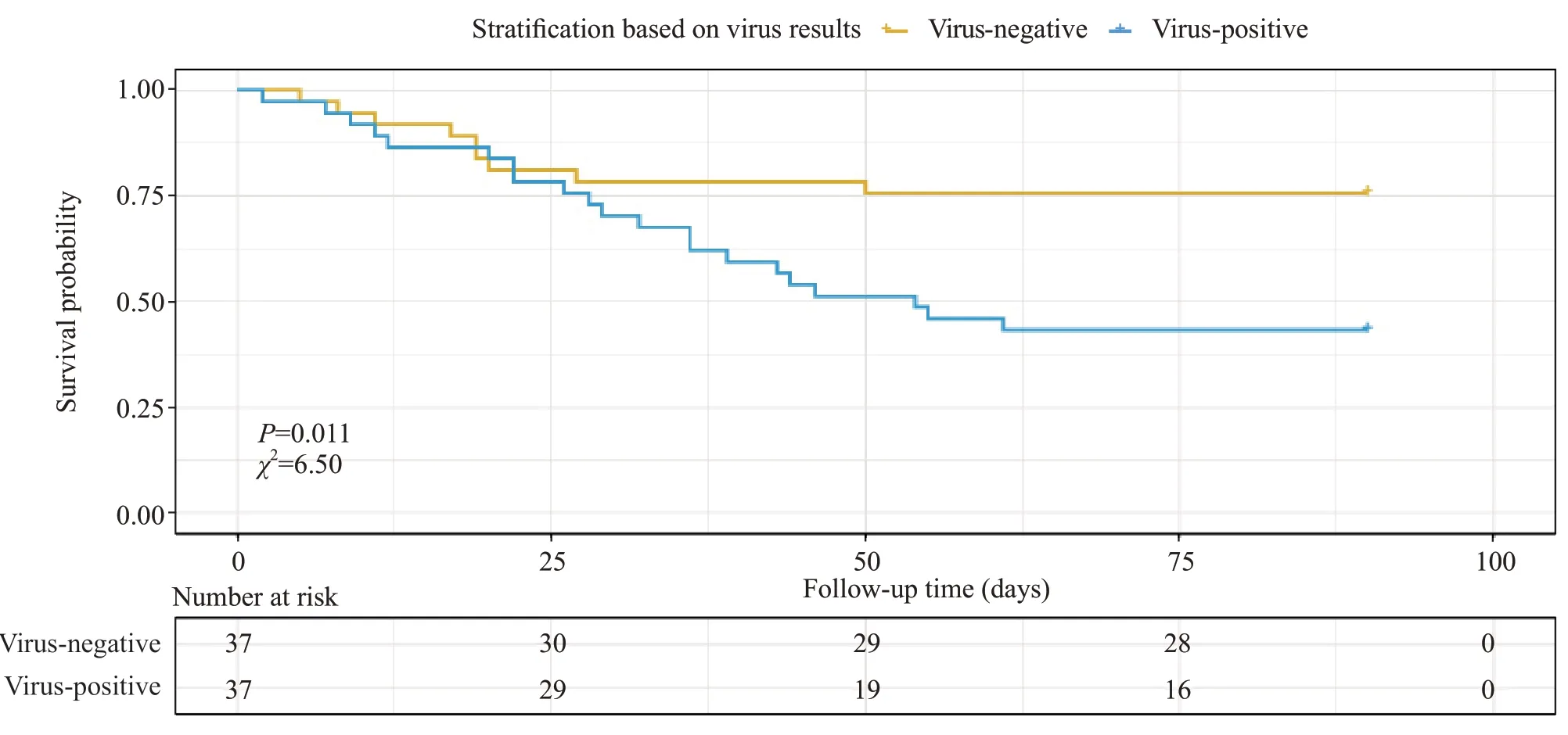
Figure 2. Kaplan-Meier plots of survival curves for ICU sepsis patients with and without virus. ICU: intensive care unit.
Pathogenic bacteria are crucial for the initiation and development of sepsis. We found that NGS exhibited the satisfactory performance in detecting potential bacterial infections, including certain bacteria that were particularly difficult to be cultured, which was consistent with the results of a previous study.[13]Furthermore, NGS-based diagnosis had a higher sensitivity than blood culture, independent of antimicrobial treatment (data not shown). In this present study, we analyzed the effect of bacteria varieties on sepsis severity. However, the results revealed no significant correlation between the bacteria varieties and sepsis severity.Therefore, the concurrent viral load became our focus.
We divided patients into a virus-positive group and a virus-negative group according to the presence or absence of the viral load. Patients with viral load were older and experienced a longer period of ICU hospitalization, with higher ICU scores (including APACHE II score and SOFA score) compared to those with bacterial infection only. These results suggested that patients with concurrent viral load may suffer more severe sepsis. Human herpesvirus 5 was the most frequently detected pathogen, occurring in approximately 29.73% (22 of 74) of the patients included in our study.This was in line with previous studies, which found that treating documented HSV patients with acyclovir could improve outcomes.[10]Cox regression analysis results showed an obvious impact of concurrent viral load on the survival rate. Interestingly, at 25 days after admission,the survival rate in the virus-negative group did not change signif icantly, whereas the survival rate decreased continuously in the virus-positive group. These results indicated that concomitant viral infection may have a negative impact on the survival of patients with sepsis.
Previous studies have shown a close correlation between infection with the virus HSV-1 and the therapeutic effect of ICU sepsis treatment. Traen et al[10]reported that acyclovir treatment was positively linked to shorter in-hospital stay and ICU stay, and lower mortality. Schuierer et al[11]divided patients into three groups: untreated group, acyclovir-treated patients with high (>105HSV copies/mL) and low (103-105HSV copies/mL) viral load groups, and found that prophylactic usage of acyclovir improved median ICU survival time(8 days vs. 22 days,P<0.05) with signif icantly improved circulatory and pulmonary function. Notably, because the prolonged ICU stay would accompany increasing complications, the long-term therapeutic benef it resulting from acyclovir treatment still required further study.Nevertheless, these potential complications could be prevented by proper patient management. Overall, we believe that prophylactic administration of antiviral drugs combined with antibiotics may improve the efficacy of treatment of sepsis patients compared with antibiotics alone. However, the exact mechanism of the effect of viral load on the survival of sepsis patients remains elusive.
The present study has several limitations. Firstly, we only investigated the relationship between concomitant viral infection and the sepsis severity and survival rate of ICU sepsis patients, and did not study the effect of combined prophylactic antiviral and antibiotic treatments on the prognosis of ICU sepsis patients. Given the current lack of direct evidence supporting the use of prophylactic antiviral drugs to increase survival rate, no attending physicians would make this treatment decision aggressively. However,considering the current literature and our study results,we believe that sepsis patients in the ICU would benefit greatly from combined prophylactic antiviral and antibiotic treatments. Secondly, owing to the study period, our sample size was relatively small, so the generalization of the results from this study may not be appropriate. Therefore, further studies with a larger sample size and longer duration are required. Thirdly, it is not possible to conclude from the results of this study whether a causative relationship exists between concurrent viral infection and sepsis severity;however, this possibility seems plausible. As indicated in this study, all patients with viral load had higher ICU scores and lower survival rate compared with patients without concurrent viral infection. Fourthly, during the process of this study, the blood sample was tested in batches. In addition,the majority of the patients enrolled were discharged after their metagenomic next-generation sequencing (mNGS)results were obtained. As a result, we cannot obtain further blood samples from these patients, and a conf irmatory PCR cannot be performed to confirm the viral DNA. However,previous studies have indicated relatively high consistency between mNGS and PCR.[25,26]
CONCLUSIONS
The present study reveals that the concurrent viral load correlates closely with sepsis severity and the survival rate of sepsis patients in the ICU. Therefore,prophylactic administration of antiviral drugs combined with antibiotics may provide several benefits to ICU sepsis patients, including fewer complications, better prognosis, and higher survival rate.
ACKNOWLEDGMENTS
We are grateful to Doctor Kai-qiang Sun for his help in embellishing the manuscript regarding the spell and grammar errors.
Funding: The study was supported by grants from Science and Technology Committee of Shanghai (18411951400); Key Clinical Medical Specialties Project in Shanghai Pudong New Area (PWZzk2017-22); Science and Technology Action Plan(19495810200); Leading Talent Project in Shanghai Pudong New Area Health System (PWRl2018-08).
Ethical approval:This present study was approved by the Ethics Committee Board of our institution.
Conflicts of interest:The authors confirm that no conflict of interest or any f inancial relationship that relates to the content of the manuscript has been associated with this publication.
Contributors:LWD, JLQ, and JW contributed equally to this study. All authors read and approved the f inal version.
杂志排行
World journal of emergency medicine的其它文章
- Trends and challenges of emergency and acute care in Chinese mainland: 2005-2017
- Factors associated with refractory pain in emergency patients admitted to emergency general surgery
- Identifying critically ill patients at risk of death from coronavirus disease
- Clinical correlates of hypotension in patients with acute organophosphorus poisoning
- Effects of metabolic syndrome on onset age and long-term outcomes in patients with acute coronary syndrome
- Predictors of recurrent angina in patients with no need for secondary revascularization
