纳米金和荧光DNA 的腺苷和钾离子一体化检测
2020-12-31贺克伍董华泽余永强胡进明
郑 斌, 贺克伍, 程 盛, 董华泽, 余永强, 胡进明
(1. 合肥师范学院化学与化学工程学院,合肥 230061;2. 安徽医科大学第一附属医院放射科,合肥 230022;3. 合肥工业大学分析测试中心,合肥 230009;4. 中国科学技术大学高分子科学与工程系,合肥 230026)
Sensor is a concept that is inspired by biological system which can respond to stimuli and generate corresponding outputs. It is extremely useful in the fields of research and practical applications, including diagnostics in biomedicine,monitoring in environment and quality control in industry[1,2]. Up to now, a number of studies on single analyte detection have been reported, and significant outcomes have been obtained for the constructions of single analyte sensors[3-5]. However, the simultaneous detection of multiple analytes is still challenging, because multiple detection procedures should be performed under the same conditions at the same time while generate different signals. This is pretty important in biological system where the synergistic or antagonistic effects of different substrates will have enormous effects on human body. Therefore, simultaneous monitor or detection of different substances is of great importance.
Ghosh et al[6]reported the detection of dopamine and ions simultaneously using trypsin mediated gold nanoclusters as signal reporters. The detection limits for carbidopa, dopamine, Cu2+, Co2+and Hg2+were 6.5, 0.14, 5.2,0.007 8 nmol/L and 0.005 nmol/L, respectively. However, this method was based on a fluorescence turning off mechanism, leading to negative effects on the results. A tripodal receptor containing both nitrogen and oxygen binding sites was investigated to bind Cu2+selectively with a detection limit of 4.6 μmol/L. The receptor-Cu2+complex presented good recognition capacity towards Br-which can be used as a potential chemosensor for multiple analytes detection[7]. This sensor only detected Br-in case the receptor-Cu2+complex was formed, therefore, the simultaneous detection is limited by the adding sequence. Wang et al[8]synthesized a terbium metal-organic framework (Ln-MOF)sensor that can not only detect, but also monitor nitromethane as well as trace amounts of nitro-aromatic compounds in aqueous solutions. Cyclodextrin modified supramolecular functional polymers were synthesized using molecular imprinting technique which can simultaneously recognize nitrophenol and bisphenol. The system presented good selectivity in practical water detection. However, the synthetic process prevented the large-scale production of the polymers[9]. A phospholipid removal micro-elution solid phase extraction method for sample preparation was developed for liquid chromatography coupled with mass spectroscopy (LC-MS)/mass spectroscopy(MS) instrument which can detect six antipsychotics simultaneously[10]. Gold nanoparticles (AuNPs) and nanostructures were also investigated for multiple analytes detection[11]. Good effects were arrived, but the detection was relied on expensive instrumentations.
To construct a biocompatible sensor, the selection of sensing element is a critical step. Antibodies, due to their high specificity, have long been utilized for sensing. However, the production and stability have limited their largescale preparations and applications[12]. Nucleic acids, also one kind of biological molecules, are found to specifically bind with targets and can be separated and sequenced using Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technique[13]. Since nucleic acids have higher stability and can be synthesized in vitro, a new name aptamer for this kind of nucleic acids with high affinity for target binding has been established since it was first discovered in 1990s[14,15].
Different strategies were adopted for the construction of multiple detection. Liu et al[16]advanced a new and general method utilizing the disassembly of AuNPs during encountering targets. Luo et al[17]immobilized DNA modified with fluorophores with different emission wavelengths onto polystyrene for multiple detections. To minimize strands for one target detection, Li et al[18]used two-strand linking AuNPs strategy for multiple analyses. However, the simultaneous modification of different strands onto the same nanoparticle led to the problem of uneven arrangement of DNA with different strand lengths. Liu et al[19]explored one single strand DNA with fluorescence label for simultaneous detection of two heavy ions. This method was much simpler but had little generality. Mancuso et al[20]utilized the aggregations of AuNPs and silver nanoparticles as multiple sensing signals which had no overlapping spectra to distinguish the signals. Utilizing the porous characteristics of metal-organic frameworks (MOFs), electroactive dyes were capped using dsDNA as the capper. The surfaces of MOFs were modified with aptamers that could recognize different tumor biomarkers which would trigger the release of signals from MOFs[21]. The resulting sensor showed a detection limit as low as 3.6 fmol/L. Desirable progress has been made, yet multiple strands modified onto different nanoparticles with different fluorescences on each strand has not been explored.
In our previous work, the single detection of biological elements has been achieved, such as the monitor of pH[22],adenosine[23], ATP[24-26], lead[27], potassium ions[28], ochratoxin A[29]and saxitoxin[30,31]. To pave the way for future practical application, simultaneous detection of adenosine and potassium ions in one-pot was investigated in this study.Utilizing the special recognition capability of aptamer, potassium and adenosine aptamers are linked onto AuNPs.Another partly complementary strand with fluorescence label can base pair with aptamer in case the target cannot induce fluorescence quenching, while fluorescence turning on when target is presented.
1 Experiment
1.1 Chemicals
Trisodium citrates、hydrogen tetrachloroaurate (Ⅲ) (HAuCl4) (Sinopharm Chemical Reagent Co., Shanghai,China); 6-mercaptohexal-1-ol (MCH)、 tris (2-carboxyethyl) phosphine (TCEP)、 adenosine、 uridine、 cytidine guanosine (Sigma). All chemicals were in their highest purity unless otherwise stated. All oligonucleotide strands were synthesized by Shanghai Sangon Biotechnology Co.. The strands used in this study are listed as follows:
Adenosine complemental strand (Adocpl): 5′-CCCAGGTTCTCTT-6-FAM (FITC/FAM)-3′;
Adenosine aptamer (Adoapt): 5′-SH-C6-AAGAGAACCTGGGGGAGTATTGCGGAGGAAGGT-3′;
Potassium complementary strand (Kcpl): 5′-CTAACCCTTAGAT-ROX-3′;
Potassium aptamer (Kapt): 5′-SH-C6-GGGATTGGGATTGGGATTGGGA-3′;
Thymine strand (T strand): 5′-SH-C6-TTTTCTTTCTTTC-3′.
Aptamers can bind specifically to targets and form secondary structures which will hinder the hybridization between aptamer and its complementary strands. The oligonucleotides were dissolved in 10 mmol/L Tris-HCl buffer(pH=7.0). Different concentrations of adenosine and potassium ions were prepared in the buffer containing 10 mmol/L tris (hydroxymethyl) aminomethane (Tris) acetate at pH 8.2. Millipore MilliQ water was used throughout the study.
1.2 Synthesis and Functionalization of AuNPs
AuNPs were prepared by citrate reduction of HAuCl4according to the literature[20]. Briefly, 10 mL of 38.8 mmol/L sodium citrate was immediately added into 100 mL of 1.0 mmol/L HAuCl4refluxing solution under stirring, and the resulting mixture was kept boiling for 15 min. The solution color turned to a wine red, indicating the formation of AuNPs. The solution was cooled to room temperature with continuous stirring. The size of the AuNPs were verified by scanning electron micrograph (SEM).
The process of probe DNA labeling was performed as follows. An appropriate amount of thiolated DNA was activated with 10 mmol/L acetate buffer (pH=5.2), and TCEP with a concentration of 100 folds more than thiolated DNA was added and incubated for 1 h, and then freshly prepared AuNPs was added and the mixture was shaken gently overnight. To give upright DNA conformation for further hybridization, a mole ratio of T strand to adoapt or Kapt of 1∶3 was adopted. Over the course of 16 h, the DNA-AuNPs conjugates were aged in salts (0.1 mol/L NaCl, 10 mmol/L acetate buffer) for 24 h. Excess reagents were removed by centrifuging at 15 000 r/min for 30 min. The obtained red precipitate was washed, recentrifuged, and dispersed in 1 mL of hybridization buffer.
1.3 DNA-AuNP Characterization
The concentration of DNA-AuNPs was determined by UV/Vis spectroscopy. To measure the amount of modified DNA onto the surfaces of nanoparticle (NP), the NP degradation method was adopted. Briefly, the particles were oxidatively dissolved using KCN solution (200 mmol/L) to liberate mercapto-DNA, the resulting solutions were analyzed using the commercially available assay (Oligreen, Invitrogen). The average DNA numbers on each NP were calculated by the division between DNA concentration and NP concentration.
1.4 Instrumentation
A pH meter (Mettler Toledo, FE20) was used to test the pH value of all buffer solutions with an accuracy of 0.01 pH unit. Fluorescence spectra were recorded on a Horiba Fluoromax-4 spectrophotometer. Transmission electron microscope (TEM) images were recorded on a JEOL JEM-2100F TEM (Tokyo, Japan).
1.5 Fluorescence Experiments
Targets were first mixed with DNA-AuNPs complex for 30 min, afterward fluorescently labeled DNA was added and balanced for 10 min before fluorescence spectra were detected. For adenosine detection, the excitation wavelength was set at 495 nm and the spectra were measured from 505 nm to 550 nm. The potassium ions detection was performed with excitation wavelength at 588 nm and spectra collected from 598 nm to 650 nm.
2 Results and Discussion
It was reported that the linking of two strands with base numbers below seven would induce the dissociation of double strands[32]. Based on this principle, two strands with twelve base pairs are designed. When targets exist, the longer strand containing sequences specific to targets will combine with analytes, inducing the conformational changes and reducing the base pairs which eventually lead to the dissociation of double strands. In this work, the longer strands are attached onto gold nanoparticles with linkages of mercaptol groups. Another short strand was fluorescently labelled at one end. AuNPs can act as fluorescence quencher. Fig. 1 depicts the sensing principle of the multiple detection. The two fluorescent labeled DNAs have non-interfering emission peaks endowing the simultaneous sensing of two targets without affecting the signals of each other.

Fig. 1 A schematic representation of multiple detection by fluorescent DNA and AuNPs
TEM images indicated that the as-prepared AuNPs resembled uniform spheres, with diameters averagely around 13 nm, and were well dispersed in citrate solution (Fig. 2). It was reported that the average modification numbers of DNA on each NP would be in the range of 55 to 100[32]. Since the spacing strand has been used for sequence standing up, the estimated loading number of mercapto-DNA would be 20-50 per NP. The modification ratio was further confirmed by the assay as stated above to be 35 per NP for adenosine detection and 40 for potassium ions detection.
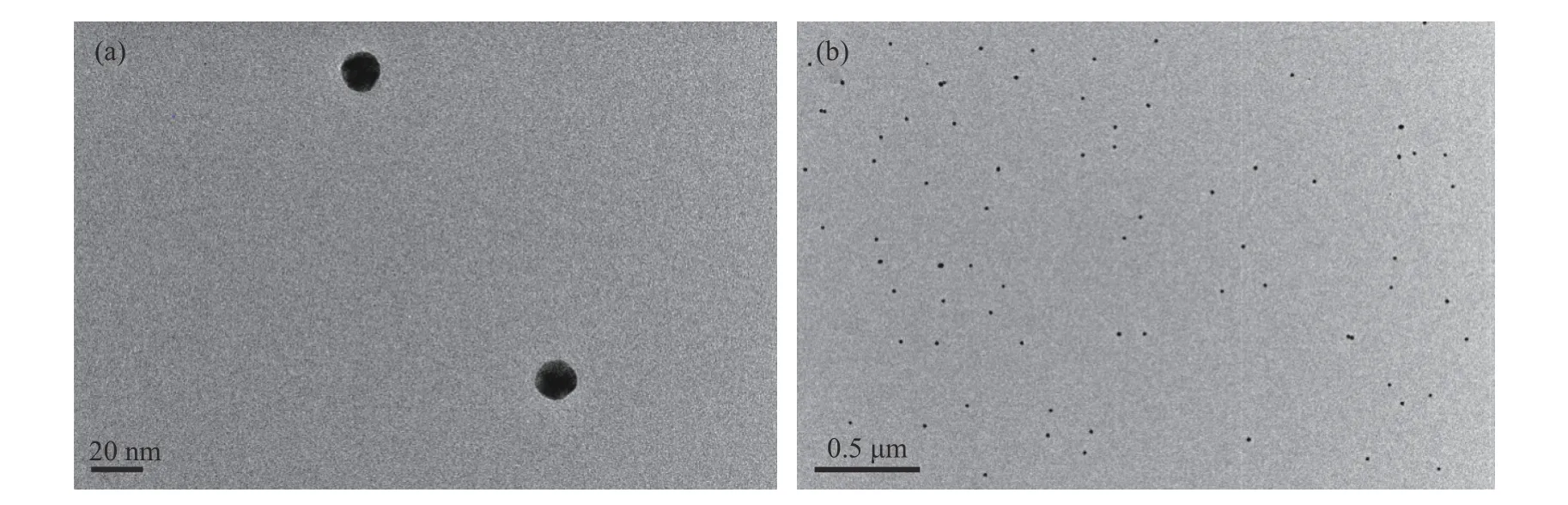
Fig. 2 TEM images of AuNPs at different magnifications
To test the feasibility of this method, the analyte detection was evaluated individually. The experimental conditions were first explored. The influences of complementary strand on sensing were investigated. As shown in Fig. 3(a),the fluorescence intensity was enhanced with the increase of complementary strand bearing fluorophores. The addition of target adenosine liberating the fluorophore from AuNPs gave rise to the increment of fluorescence signal. The fluorescence intensity differences between complementary with and without adenosine became more distinct as the complementary strand concentration increased. The influences of complementary strand concentrations on detection range were explored. At an adocpl concentration as low as 20 nmol/L (Fig. 3(b)), a plot of the intensity versus adenosine concentration gave a linear response with the equation Y = BX + A = 4.529 25X + 965.843 1 (where X is the concentration of adenosine (10-6mol/L) and Y is the intensity) with a coefficient r = 0.994 23 and r2= 0.986 18. The detection limit was 387.9 nmol/L, which was estimated using the signal-to-noise ratio of the blank solution for three times. The detection range had no platform indicating the unsaturation point of detection. To examine the detection range modulation, a higher adocpl concentration and 100 nmol/L adocpl strand were used. The characteristic peak at 520 nm showed a gradual increase with adenosine concentrations (Fig. 4). The higher adocpl concentration showed a sigmoid sensing curve versus adenosine concentrations (Fig. 5). The detection range was between 2 mmol/L and 15 mmol/L. The platform above 15 mmol/L suggests the saturation detection limit. Thus, the modulation of detection range by changing adocpl strand concentration is feasible.

Fig. 3 (a) Fluorescent signals at 520 nm with (red) and without (black) 10 mmol/L adenosine in response to varying aptcpl concentrations;(b) Linear relationship between fluorescence emission intensity at 520 nm and adenosine concentration in the range of 0-15 μmol/L(Error bars represent the standard deviation of three measurements)
As a preliminary exploration, the detection of potassium ions was also studied, as shown in Fig. 5. With the addition of potassium ions, the fluorescence intensity at 605 nm showed a gradual increase, indicating a“fluorescence on” detection method (Fig. 6(a)). The detection was inspected from 0 to 8 μmol/L (Fig. 6(b)). The method showed good repeatability (Fig. 7(a)). A plot of the intensity versus adenosine concentrations gave a linear response with the equation Y = BX + A = 58.532 81X - 79.385 23 (where X is the concentration of adenosine(10-6mol/L) and Y is the intensity) with a coefficient r = 0.991 56 and r2= 0.979 83. The detection limit was 1.6 μmol/L, which was estimated using the signal-to-noise ratio of blank solution for three times (Fig. 7(b)).

Fig. 4 Fluorescence spectra of detection system in response to adenosine with concentrations ranging from 1 mmol/L to 20 mmol/L
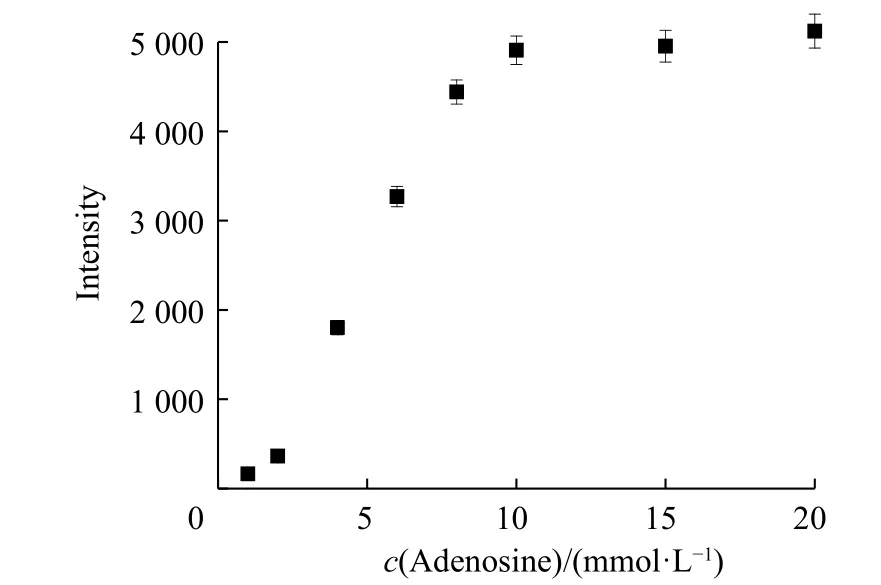
Fig. 5 Fluorescence emission at 520 nm as a function of adenosine concentrations (Error bars represent the standard deviation of three measurements)
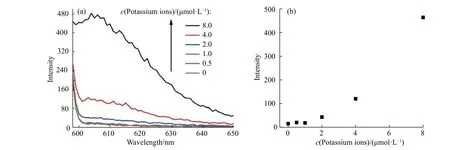
Fig. 6 (a) Fluorescence spectra of detection system in response to potassium ions concentrations from 0 to 8.0 μmol/L; (b) Fluorescence emission at 605 nm as a function of potassium ion concentrations

Fig. 7 (a) Fluorescence spectra of detection system in response to potassium ions concentrations from 0.3 μmol/L to 6.0 μmol/L. (b) Linear relationship between fluorescence emission intensity at 605 nm and potassium ions concentrations in the range of 2-6 μmol/L (Error bars represent the standard deviation of three measurements)
In addition to the multiple detection of analytes, the specificity was also examined. As shown in Fig. 8, the introduction of adenosine induced a high fluorescence signal while other analogues led to only slight emission. The fluorescence of adenosine target was 5 folds more than that of uridine, cytidine and guanosine. Similarly, the fluorescence of potassium ions exceeded at least 2 folds higher than those of other lithium and sodium ions (Fig. 9).These results confirm the good specificity of our multiple detection system.

Fig. 8 Specificity of multiple detection system over analogues:adenosine, uridine, cytidine and guanosine (all 3 mmol/L)

Fig. 9 Specificity of multiple detection system over analogues:potassium ions (3 μmol/L), lithium ions and sodium ions(1 mmol/L for both)
3 Conclusion
In conclusion, a multiple-detection method based on AuNPs and fluorescent labelled DNA strands has been designed. Utilizing the special recognition feature of DNA aptamer, two DNA strands were used to distinguish two targets, adenosine and potassium ions, in one system. In the presence of the targets, DNA strands modified onto AuNPs can bind with targets, by which the conformation of the DNA strands is changed,and leads to combination of partly complementary strand with fluorescence labels, ultimately leading to fluorescence “on” signal. When there are no targets, the complementary strands will base pair with DNA on AuNPs. The fluorescence on complementary strands will be quenched by AuNPs due to the short distance,resulting in turning off fluorescence signal. Since the two fluorescence groups have different emission wavelengths, the simultaneous detection with no signal overlap were realized. This method is simple and shows high selectivity.
Acknowledgements
We are grateful for the financial support provided by the National Natural Science Foundation of China(21504021, 31501576), Foundations of Educational Committee of Anhui Province ( KJ2019A0719), the Excellent Talent Foundation of Education Department of Anhui Province ( gxyq2019066) and the 136 talent plan of Hefei Normal University, Quality Engineering Project of Anhui Province ( 2018sjjd073).
