cDNA-SCoT Analysis of Differential Expressed Genes in Allium tuberosum Induced by Botrytis cinerea
2020-11-30MinghuiCHENZhiluZHANGYanjiaoLIPengqiangYAOShipingCHENGGanqingZHAO
Minghui CHEN, Zhilu ZHANG , Yanjiao LI, Pengqiang YAO, Shiping CHENG, Ganqing ZHAO
Key Laboratory of Ecological Restoration in Hilly Area, Pingdingshan University, Pingdingshang 467000, China
Abstract [Objectives] To investigate the molecular mechanism of gene differential expression in A.tuberosum under Botrytis cinerea stress. [Methods] The leaf of A. tuberosum was inoculated with B. cinerea to induce resistance, then leaf samples were taken at 2, 4, 8, 12 and 24 h, respectively, after inoculation, and used for cDNA-SCoT analysis to detect the gene differential expressions. [Results] More than 800 bands with length of 100-1 800 bp were obtained using 60 SCoT primers. A total of 40 differentially expressed EST sequences were screened out, and 18 non-redundant ESTs with high quality were obtained by cluster analyses of the ESTs sequencing. The results of BlastN showed that the differential genes of A. tuberosum mainly related to disease defense, signal transduction, and protein metabolism. [Conclusions] Further analysis of gene function indicated that brassinosteroid biosynthesis-like proteins, NBS-LRR type disease resistance protein, jasmonic acid induced protein, abscisic stress ripening protein may be involved in the process of the incompatible interaction between the A. tuberosum and B. cinerea.
Key words Allium tuberosum, Grey mould, cDNA-SCoT, Differential genes
1 Introduction
Grey mould is a major disease that is harmful toAlliumtuberosumin the world. It is caused byBotrytiscinereasignificant fungal diseases inAlliumtuberosum. The mycelial growth ofB.cinereahad strong adaptability to environmental conditions[1]. Grey mould often causesA.tuberosumleaves to die and rot, which can not be eaten, directly affecting the yield. Current research onA.tuberosumgrey mould disease focuses on the pathogenic identification[2], communication and control[3], detection technology[4]and other aspects, while there is still no report both at home and abroad aboutB.cinereapathogen infectingA.tuberosum, and the molecular mechanisms.
Start codon targeted polymorphism (SCoT) markers is Collard and Mackill[5]development of a target gene molecular marker technology. It is based on maintaining plants amplified gene ATG translation start site flanking sequences, primers were designed and the genome was amplified. The molecular marker technology has the primer design is simple, low cost, good repeatability, can produce traits associated with marked characteristics. Currently SCoT marking technology has been successfully applied to peanuts[6], longan[7], mango[8]and other plants. But there is still no research report about SCoT marked technology inA.tuberosuminteracted withB.cinereapathogen. Therefore, we usedB.cinereapathogens disseminatedA.tuberosumfor processing, and excludingB.cinereapathogen ofA.tuberosumprocessing plant of its own age as a control. Using RNA mixing pool reverse transcribed into cDNA first chain mixing pool, we performed cDNA-SCoT difference research and obtained the gene information related to expression of resistance toB.cinereainA.tuberosum. In this study, we were intended to provide a reference for exploring the molecular mechanism ofA.tuberosumgene expression underB.cinereapathogen stress andA.tuberosumdisease resistance breeding.
2 Materials
A.tuberosumvarieties healthy seedlings were purchased from Pingdingshan Academy of Agricultural Sciences. HealthyA.tuberosumplants with the same growth were selected, and the leaves were artificially inoculated with fluid containingB.cinereapathogen, and the same-age plants treated with water withoutB.cinereapathogen were taken as control. Plant leaves were taken at 2, 4, 8, 12 and 24 h after inoculation, respectively, and stored at -70 ℃ after quick-freezing by liquid nitrogen.
3 Methods
3.1B.cinereapathogen infection confirmedDNA was extracted using Ying X-M’s[9]methods, and the presence ofB.cinereapathogen was confirmed by PCR amplification. The primer sequences used for amplification were F: 5′-CCCTAGTCCGGAGATTAGCC-3′ and R: 5′-TGGCTCTCATGTTTTCTTCC[10]. PCR amplification was performed on Biometra Tprofessional PCR instrument. 25 μL reaction system was used, including 50 ng template DNA, upstream and downstream primers 25 μL, 12.5 μL and high-fidelity 2×GoldstarTaqMaster Mix, and DEPC treated water was added up to 25 μL. The PCR amplification procedure was pre-denaturation at 95 ℃ for 10 min. Denaturation at 95 ℃ for 30 s, annealing at 50 ℃ for 30 s, elongation at 72 ℃ for 1 min, and cycling for 40 times; Elongation at 72 ℃ for 5 min. At the end of the reaction, 4 μL bromophenol blue was added and mixed. The 6 μL amplified product was electrophoresed in 1.2% agarose gel, stained color photographs.
3.2 RNA extraction and reverse transcriptionTRIzol reagent was used to extract the total RNA fromA.tuberosumleaves, and 1.0% agarosol gel electrophoresis was used to detect the integrity of RNA, an ultraviolet spectrophotometer was used to detect the purity and concentration of RNA. Equal amounts of total RNA of leaf samples at each time period were mixed to construct the corresponding RNA mixing pool. According to the operating instructions of M-MLV reverse transcriptase kit, the first strand of cDNA was synthesized by reverse transcription using mixed pool RNA as template, diluted 10 times with deionized water, and stored at -20 ℃ for later use.
3.3 cDNA-SCoT amplification and PCR detectionPrimers were designed according to Collard and Mackil[10], Luoetal.[11]. The primer was synthesized by Sangon Biological Engineering (Shanghai) Co., Ltd. A SCoT molecular-labeled primer was used as the upstream primer, and a 3’ side anchored primer (GGCCACGCG TCGACTAGTAC) was used as the downstream primer. PCR amplification was performed using the mixed first strand cDNA of the treated and control samples as the template. The cDNA-SCoT PCR amplification system was 25 μL: high-fidelity 2×GoldstarTaqMaster Mix 12.5 μL, 10 μmol/L upstream and downstream primers each 1μl, and the cDNA template was 1 μL, and DEPC treated water was added up to 25 μL. The A amplification procedure of cDNA-SCoT PCR was: pre-denaturation at 95 ℃ for 10 min. Denaturation at 95 ℃ for 30 s, annealing at N ℃ for 1 min (N ℃=Tm value of SCoT primer decreased by 7 ℃), elongation at 72 ℃ for 2 min, 38 cycles in total; it was extended at 72 ℃ for 8 min. PCR amplification was performed using 1.5% agarose gel electrophoresis.
3.4 Recovery, cloning, sequencing and analysis of differential fragmentsAfter PCR products were electrophoreted by agarose gel, the treated and control samples were selected to amplify the bands between 100 and 1 800 bp with different expression, and the bands were recovered and purified by agarose gel recovery kit. The target fragment was connected with TaKaRa’s PMD18-T vector, and the strain of Escherichia coli DH5α was transformed to perform blue-white spot screening. The positive bacteria solution was identified by PCR and sent to Sangon Bioengineering (Shanghai) Co., Ltd. for sequencing. With BioXM2.6 software for sequencing for sequence splicing, after the sequence submitted NCBI Blast (http://www.ncbi.nlm.nih.gov/blast) for homology retrieval, using Amigo website (http://amigo.Geneontology. org/cgi-bin/amigo/go.cgi) to functional annotation analysis.
4 Results and analysis
4.1B.cinereapathogen detectionThe DNA ofA.tuberosumleaves before and after infection withB.cinereawas extracted, and the target fragment was obtained of the plant after infection withB.cinereaby PCR, which was not found the target fragment in the leaves of the plant no infection withB.cinerea(Fig.1). The target fragment was recovered and purified for bidirectional sequencing, and the obtained sequence length was 789 bp, which was completely matched with the corresponding section of ITS ofB.cinereapathogen rDNA published on NCBI, and it was confirmed thatA.tuberosumwere infected withB.cinereapathogen.
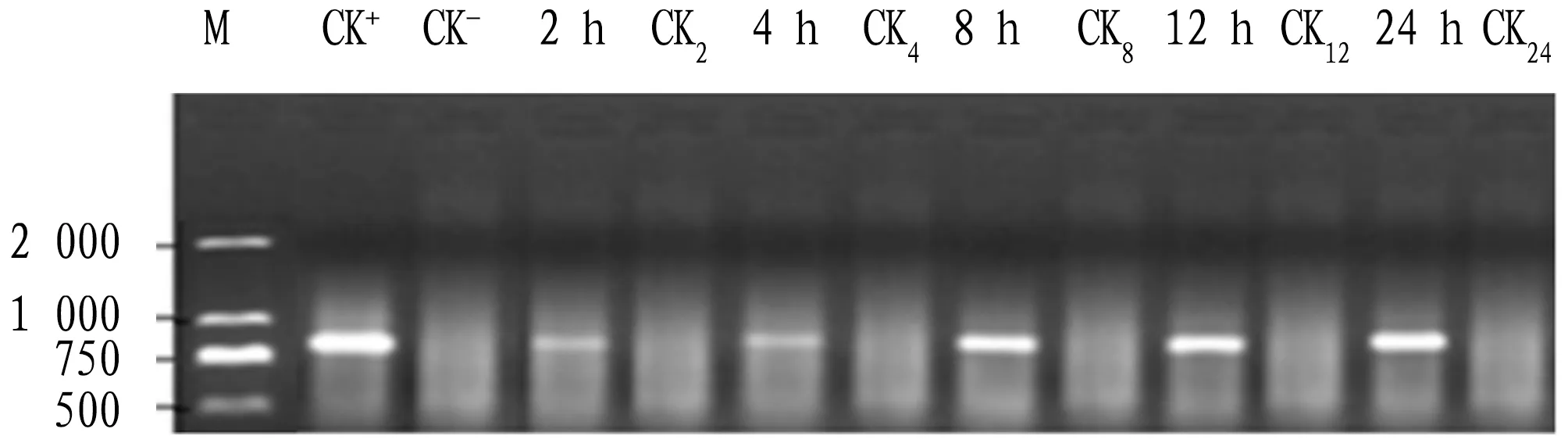
Note: M: DL 2 000 marker; CK+: positive control; CK-: negative control; 2, 4, 8, 12, and 24 h: sampling time after infection; CK2, CK4, CK6, CK8, and CK10: controls in corresponding to sampling time.Fig.1 PCR detection for healthy and infected leaves of Allium tuberosum
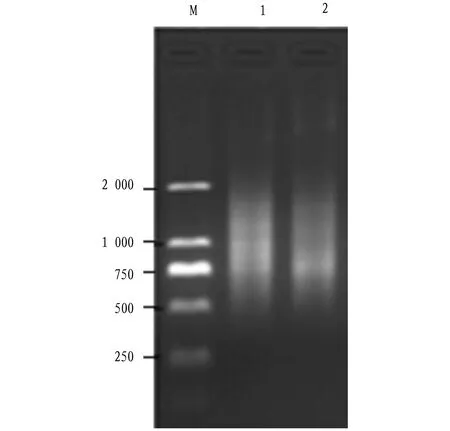
4.2 RNA extraction and reverse transcriptionAs shown in Fig.2, the band type of RNA was complete and its brightness was clear, indicating that the RNA was of good quality. TheOD260/OD280value of the RNA samples was between 1.80 and 2.00 by ultraviolet spectrophotometer, indicating that the RNA extracted was of high purity. As can be seen from Fig.3, the dispersion bands of the products after reverse transcription of total RNA were evenly distributed and of good quality. The results showed that the total RNA and reverse transcriptional products obtained met the experimental requirements.

Note: 1: total RNA in control; 2: total RNA in B. cinerea treatment.Fig.2 Detection of total RNA from Allium tuberosum leaf
4.3 cDNA-SCoT analysis of differentially expressed genes inA.tuberosuminfected withB.cinereaEighteen ESTs with unique functions were obtained after eliminating the duplicates. Among them, there are 14 EST sequences of predictable functions and 4 EST sequences of unknown functions (Table 1). According to the classification criteria of plant gene function proposed by Bevanetal.[12], the genes with known functions were analyzed. The comparison results showed that there were the most genes related to disease resistance, including brassinolide synthesis protein, NBS-LRR resistance protein, jasmonic acid induction protein, cysteine protein, ketoacyl coenzyme A synthetase enzymes. This indicated that afterA.tuberosumwas infected byB.cinereapathogen, the resistance immunity program in plant body started a series of response responses, which were expressed as the diversity of mRNA response in gene, and it also indicated that the resistance mechanism ofA.tuberosumtoB.cinereawas a complex process which was co-regulated by multiple genes.

Table 1 Comparison of Homology between differential expression genes and the documented genes in GenBank
5 Discussions
SCoT technology is a new target molecular marker technology based on translation initiation site. It is a molecular marker technology with PCR reaction as the core, and its core is primer design. In this study, we used SCoT labeling technology to explore the molecular response involved in the interaction betweenB.cinereaandA.tuberosum, obtained multiple cDNA fragments differentially expressed or preferentially expressed inA.tuberosum, and preliminarily analyzed the regulation mechanism ofB.cinereapathogen infection.
Note: M: DL 2 000 marker; 1: cDNA in control; 2: cDNA in B. cinerea treatment.Fig.3 Detection of reverse transcription product cDNA
Brassinolactone (BR) is a phytosterol hormone substance and has important functions in plants. Studies have proved that BR can regulate plant physiological metabolism, improve crop quality and yield, and regulate processes of plant growth and development[13]. In terms of improving plant stress resistance, BR can increase leaf area, chlorophyll content and soluble protein content of wheat growing under drought conditions[14]. SOD and CAT activities of peanut leaves under drought stress were increased[15], proline content of Apricot dalmatians was increased, and its drought resistance was significantly enhanced[16]. It can be considered that whenA.tuberosumleaves were infected byB.cinerea, BR urged the generation of stress signal, induced a series of disease-resistant substances in the plant to enhance the resistance of the plant, so as to inhibit the growth of pathogenic bacteria. Therefore, these genes related to plant defense response are preferentially expressed, which can be used as target genes to improve disease resistance ofA.tuberosum, so as to further explore their functions.
NBS-LRR gene is an intracellular receptor protein gene that contains nucleotide binding site (NBS) and leucine-rich repeats (LRR), and is the most important class of disease-resistant genes[17]. In recent years, there have been many reports on the studies on the homologous sequences of NBS-LRR resistance genes. For example, some homologous DNA fragments of such resistance genes have been obtained from rice[18], soybean[19]and other plants, which have become candidate genes for resistance genes. In this study, the NBS-LRR resistance gene was obtained by using the cDNA-SCoT difference analysis technology, which indicated that after the infection ofB.cinereapathogen,A.tuberosumcould rapidly activate the defense system and improve the expression of some resistance genes through its own regulation ability to resist the invasion of foreign bacteria.
In addition to the above related genes, many genes related to biological and abiotic stress were also found in this study, including α-tubulin, β-tubulin, ABA stress mature protein, 60S ribosomal protein, proline-rich protein, jasmonic acid-induced protein translation initiation factor eif-2b α subfamily, endogenous 1,3-β-glucosidase enzymes. Accordingly, it is speculated that there may be a mechanism of external stress resistance involved in protein metabolism, signal transduction and other joint effects inA.tuberosum, and whether the expression of these genes will affect and to what extent affect the resistance ofA.tuberosum, or whether it is the resistance gene ofA.tuberosum, still need further study.
6 Conclusions
Using the cDNA-SCoT method, we successfully isolated 18 high-quality differential gene fragments, and performed homology comparison and functional classification between the 18 differential gene fragments and non-redundant database BLAST, mainly involving energy metabolism, defense response, signal transduction, protein metabolism and other aspects. In this study, we established the mixed CDNA-SCOT difference library ofA.tuberosumleaves at different time points from 2 to 24 h afterB.cinereainfection.
杂志排行
Asian Agricultural Research的其它文章
- Application of Robust Strategies in Location Selection of Logistics Distribution Center for Fresh Agricultural Products
- Empirical Study on Impact of China’s Economic Growth on Agricultural Trade
- Efficiency Assessment of Land Use Types in Tan Binh Commune, Dak Doa District, Gia Lai Province, Vietnam
- Some Thoughts on China’s Territorial Spatial Planning in the New Period
- Functional Positioning of Pastoral Complex in the Context of Rural Revitalization Strategy: A Case Study of Pavlo Eco Valley in Gaoan City of Jiangxi Province
- Identification and Detection of Dyeing Components in Counterfeit Cinnabar (Elutriation)
