番木瓜CpTIFY10A-like基因克隆及表达分析
2020-08-11吕金慧王府润陈萍
吕金慧 王府润 陈萍

摘要:【目的】克隆番木瓜CpTIFY10A-like基因序列,并分析其表達特性,为探索该基因功能和抗番木瓜环斑病毒(PRSV)分子机制提供理论依据。【方法】结合前期研究的抗PRSV转录组数据,利用RACE对上调表达基因CpTIFY10A-like进行克隆,获得该基因cDNA全长序列,分析其编码区序列(CDS)及进行生物信息学分析。利用已鉴定的PRSV对3月龄番木瓜植株进行不同处理,以感染PRSV且出现病症的植株(CK+)、1.0 mL/L禾甲安(CTS-N)灌施感染PRSV且出现病症的植株(B)为处理组,以清水灌施(不添加禾甲安)不感染PRSV的植株(CK-)为对照组,实时荧光定量PCR(qRT-PCR)检测不同处理下PRSV基因和CpTIFY10A-like基因在番木瓜叶片中的表达情况。最后将CpTIFY10A-like基因CDS序列连接至pET28a-sumo载体上构建原核表达载体,转化Rosetta(DE3)感受态细胞,通过不同浓度IPTG诱导蛋白表达,利用SDS-PAGE电泳检测原核表达情况。【结果】CpTIFY10A-like基因全长为1352 bp,CDS序列为822 bp,编码273个氨基酸残基;其编码蛋白理论等电点(pI)9.23,不稳定系数62.97,亲水性平均系数-0.458,属于不稳定的亲水性碱性蛋白,定位在细胞核上,其二级结构以无规则卷曲和α-螺旋为主,同时含有少量的β-折叠和延伸链。CpTIFY10A-like蛋白与酸枣和白梨TIFY蛋白的氨基酸序列相似性较高,均含有特定的TIFY结构域,属于TIFY蛋白家族,三者在系统发育进化树上聚在同一分支,表明亲缘关系较近。不同处理的番木瓜叶片中,PRSV基因的相对表达量排序为CK+(1.02)>B(0.39)>CK-(0),CpTIFY10A-like基因的相对表达量排序为B(53.12)>CK+(1.15)>CK-(1.02),且CpTIFY10A-like基因在经禾甲安处理的感染PRSV番木瓜叶片中相对表达量显著升高(P<0.05,下同),而PRSV基因的相对表达量显著降低。CpTIFY10A-like基因在原核细胞中表达蛋白的分子量与理论分子量(29.36 kD)相符,且不同浓度IPTG诱导下,IPTG浓度越高,蛋白表达量越多,表明该基因在原核细胞中成功表达。【结论】禾甲安可诱导CpTIFY10A-like基因高效表达,进而通过茉莉酸通路起调节作用以抵抗PRSV感染,即CpTIFY10A-like基因在番木瓜抗PRSV过程中发挥重要调控作用。
关键词: 番木瓜;CpTIFY10A-like基因;生物信息学;原核表达;禾甲安(CTS-N);番木瓜环斑病毒(PRSV)
Abstract:【Objective】This study cloned the papaya CpTIFY10A-like gene sequence and analyzed its expression chara-cteristics,in order to provide a theoretical reference for exploring the function of this gene and the molecular mechanism of resistance to papaya ring spot virus(PRSV). 【Method】Combined with the anti-PRSV transcriptome data of the pre-vious research of the research group,the up-regulated gene CpTIFY10A-like was cloned using RACE technology. The full-length cDNA was obtained and the coding sequence(CDS) was analyzed, then bioinformatic analysis of this gene was conducted. Three-month-old papaya plants were treated differently with the identified PRSV. With plants infected with PRSV and showing symptoms(CK+), plants infected with 1.0 mL/L CTS-N infected with PRSV and showing symptoms(B) were the treatment groups. The plants irrigated with clear water(without the addition of CTS-N) and not infected with PRSV(CK-) were as the control group. qRT-PCR was used to detect the expression of PRSV gene and CpTIFY10A-like gene in papaya leaves under different treatments. At last,connected the CDS sequence of CpTIFY10A-like gene to pET28a-sumo vector to construct a prokaryotic expression vector. The reconstruction vector was transformed into competent cell Rosetta(DE3) and induced protein expression by different concentrations of IPTG. The prokaryotic expression of this gene was detected by SDS-PAGE electrophoresis. 【Result】The results demonstrated that the gene was 1352 bp in full-length,of which the CDS sequence was 822 bp,and it encoded 273 amino acid residues. The theoretical isoelectric point (pI) of the encoded protein was 9.23,the instability coefficient was 62.97,and the average hydrophilicity coefficient was 0.458. It belonged to unstable hydrophilic basic protein. Subcellular localization predicted that the gene was located on the nucleus. The secondary structure of the protein expressed by this gene was mainly random curl and α-helix,and also contained a small amount of β-turn fold and extended chain. Amino acid sequence of CpTIFY10A-like protein had high similarity with that of TIFY protein in Z. jujuba and Pyrus x bretschneideri, they both had specific TIFY domain and belonged to TIFY family. Phylogenetic analysis revealed that the CpTIFY10A-like gene was clustered into the same branch as of the homologous genes of Ziziphus jujuba. This showed that their kinship was close. Using qRT-PCR to detect papaya leaves in different treatments,it was found that the relative expression of PRSV gene was ranked as CK+(1.02)>B(0.39)>CK-(0),and the relative expression of CpTIFY10A-like gene was ranked as B(53.12)>CK+(1.15)>CK-(1.02),and the expre-ssion of CpTIFY10A-like gene in the leaves of PRSV infected papaya treated with CTS-N was significantly increased(P<0.05, the same below),while the expression of PRSV gene was significantly decreased. The molecular weight of the protein expressed by CpTIFY10A-like gene in prokaryotic cells was consistent with the theoretical molecular weight(29.36 kD). In the induction of different concentrations of IPTG,the higher the IPTG concentration,the greater the protein expression,it indicated that the gene was successfully expressed in prokaryotic cells. 【Conclusion】CTS-N can induce the efficient expression of CpTIFY10A-like gene,which plays a regulatory role in the jasmonic acid pathway to prevent and control PRSV. It indicates that the CpTIFY10A-like gene plays an important regulatory role in the anti-PRSV process of papaya.
Key words:Carica papaya L.; CpTIFY10A-like gene; bioinformatics; prokaryotic expression; CTS-N; papaya ring spot virus (PRSV)
0 引言
【研究意义】番木瓜(Carica papaya L.)是熱带特有的草本果树,又名万寿果、乳瓜和石瓜,被世界卫生组织认定为最具营养价值的十大水果排行之首(陈豪军等,2013)。近年来,番木瓜生产面临的毁灭性病害是由番木瓜环斑病毒(Papaya ring spot virus,PRSV)引起(Tripathi et al.,2010;张荣萍等,2017)。植株感染此病后,初期在茎支脉上出现水浸斑,嫩叶上出现黄绿相间或深或浅的花叶病症,末期新生叶片黄化、畸形,发病严重的叶片呈鸡爪状或线状,最后枯死(Bau et al.,2008)。禾甲安(CTS-N)是一种绿色防控农药试剂,主要成分是壳聚糖,可有效防治番木瓜环斑花叶病毒病(鄢兴祥等,2019),其防治机理是激活番木瓜内源抗病基因,从而达到抗病效果。前人研究发现,番木瓜CpTIFY10A-like基因(GenBank登录号XM_022035541.1)是在禾甲安防治PRSV过程中表达上调的基因之一(安娜,2018),推测其是番木瓜内源抗病基因。因此,开展番木瓜CpTIFY10A-like基因克隆及表达分析对PRSV防治具有重要意义。【前人研究进展】CpTIFY10A-like属于TIFY蛋白家族,该家族具有特定的TIFY结构域,称为花序分生组织中表达的锌指蛋白(ZIM)结构域,包含约28个氨基酸和1个核心基因基序,其中甘氨酸保守存在,其余氨基酸可变(周明,2013),表达的相关蛋白分别为JAZ、TIFY、zimlike(ZML)和PEAPOD(PPD)蛋白(Ebel et al.,2018)。这些蛋白参与茉莉酸通路的应激反应,同时与其他转录因子相互作用,在植物的生长发育过程中发挥重要调节作用(Vanholme et al.,2007;Bai et al.,2011)。Vanholme等(2007)在拟南芥中克隆获得18个表达TIFY蛋白的基因;Zhu等(2013)通过转录组分析发现,栽培大豆中的GsTIFY-10b、GsTIFY-10c和GsTIFY-10d等基因在碳酸盐胁迫条件下上调表达,说明TIFY基因家族成员在植物环境胁迫响应中发挥重要作用;Huang等(2016)对毛竹中的24个TIFY基因家族成员进行分类、鉴别和系统发育分析,揭示了部分TIFY基因家族成员参与非生物胁迫;Zhao等(2016)研究发现,干旱条件下棉花TIFY基因家族成员均高效表达;孙程(2013)研究发现,玉米TIFY基因家族成员具有响应激素和胁迫的能力。【本研究切入点】目前,国内外有关番木瓜抗PRSV的功能基因的研究报道较少。【拟解决的关键问题】基于本课题组前期试验所得的抗PRSV转录组数据(安娜,2018),通过RACE克隆CpTIFY10A-like基因全长序列及对其进行生物信息学分析,实时荧光定量PCR(qRT-PCR)检测CpTIFY10A-like基因表达情况,并构建其原核表达载体分析该基因表达蛋白的情况,为探索CpTIFY10A-like基因的抗病分子机制打下基础。
1 材料与方法
1. 1 试验材料
供试材料为番木瓜品种台农二号,种植于海南大学农科试验基地。本生烟草由中国热带农业科学院生物所提供。禾甲安购自山东三碘生化有限公司;植物总RNA提取的试剂盒购自天根生化科技(北京)公司;cDNA反转录试剂盒购自北京全式金生物技术有限公司;SMARTer RACE 5'/3' Kit试剂盒、BamH I酶、Xho I酶和T4连接酶购自TaKaRa公司;IPTG和5×SDS-PAGE Loading Buffer购自广州赛国生物科技有限公司;大肠杆菌DH5α感受态细胞购自诺唯赞生物科技股份有限公司;Rosetta(DE3)购自上海唯地生物技术有限公司;质粒pET28a-sumo购自武汉淼灵生物科技有限公司;qRT-PCR试剂盒购自普洛麦格(北京)生物技术有限公司。主要仪器设备:AGT9601 96孔PCR仪(杭州安杰思医学科技股份有限公司)、Mini-ES2电泳仪(杭州奥盛仪器有限公司)和Stratagene Mx3005P qRT-PCR仪(美国安捷伦公司)。
1. 2 试验方法
1. 2. 1 总RNA提取及反转录cDNA合成 采集番木瓜叶片,参照离心柱型植物总RNA提取试剂盒说明提取其RNA,并利用TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix试剂盒反转录合成cDNA模板,具体反应程序:25 ℃ 10 min,42 ℃ 30 min,85 ℃ 5 s,4 ℃恒温。合成的cDNA置于-20 ℃保存备用。
1. 2. 2 CpTIFY10A-like基因克隆 以番木瓜叶片总RNA反转录合成的cDNA为模板进行PCR扩增,参考本课题组前期试验的抗PRSV转录组数据注释信息,使用Primer 5.0进行引物设计。5'-RACE和3'-RACE克隆参照RACE试剂盒说明进行操作(Yeku and Frohman,2003)。使用引物为表1中的5'RACE、3'RACE、T0和T1,对CpTIFY10A-like基因的5'端和3'端进行克隆。RACE反应体系50.0 μL:15.5 μL PCR-Grade H2O,2.5 μL cDNA模板,1.0 μL特异性引物(GSP),25.0 μL SeqampTM Buffer(2×),1.0 μL SeqAmp DNA聚合酶,5.0 μL 10×UPM(上、下游混合物,T0+T1)。扩增程序:94 ℃预变性2 min;94 ℃ 30 s,68 ℃ 30 s,72 ℃ 3 min,进行25个循环;72 ℃延伸5 min。CpTIFY10A-like基因中间片段克隆引物为表1中的1667-F和1667-R。反应体系50.0 μL:5.0 μL LA Taq Buffer II(10×),2.5 μL cDNA模板,30.0 μL PCR-Grade H2O,10 μmol/L 1667-F和1667-R各2.0 μL,0.5 μL LA Taq DNA聚合酶,8.0 μL dNTP Mixture。扩增程序:96 ℃预变性3 min;96 ℃ 30 s,58 ℃ 20 s,72 ℃ 50 s,进行35个循环;72 ℃延伸3 min,16 ℃,1 min。PCR产物采用1%琼脂糖凝胶电泳进行检测,切胶回收纯化目标条带,利用T/A连接试剂盒连接至pMD19-T载体后转化DH5α感受态细胞,挑取单克隆菌株进行菌液PCR鉴定,将阳性克隆送至天一辉远生物科技有限公司进行测序。使用DNAMAN对CpTIFY10A-like基因片段及其5'-RACE和3'-RACE进行碱基序列拼接。
1. 2. 3 生物信息学分析 使用ExPASy ProtParam在线工具分析CpTIFY10A-like蛋白的理化性质;利用ProtScale预测CpTIFY10A-like蛋白的亲/疏水性(Zhang et al.,2019);使用DNAMAN对CpTIFY10A-like蛋白进行同源性比對;采用WoLF PSORT对CpTIFY10A-like蛋白进行亚细胞定位;运用IBCP在线工具SOPMA预测CpTIFY10A-like蛋白的二级结构(Geourjon and Deléage,1996)。将CpTIFY10A-like蛋白的氨基酸序列提交至NCBI数据库中与酸枣(XM_016011994.1)和白梨(XM-009338910.2)的同源蛋白进行BLAST比对。运用MEGA 6.0的邻接法构建系统发育进化树(Hall,2013),主要参数设置为距离模型:Maximum composite likelihood;稳健性检测:Bootstrap法;重复次数:1800次;空位缺失数据的处理:Complete deletion。
1. 2. 4 qRT-PCR检测 本课题组前期对番木瓜叶片感染的PRSV进行RT-PCR检测,同时在形态学、细胞学和分子生物学方面均对PRSV进行充分鉴定(安娜,2018)。利用鉴定的PRSV对3月龄番木瓜植株进行不同处理,以感染PRSV且出现病症的植株(CK+)、1.0 mL/L禾甲安灌施感染PRSV且出现病症的植株(B)为处理组,以清水灌施(不添加禾甲安)不感染PRSV的植株(CK-)为对照组,处理1次后,给CK-、CK+和B组正常浇水施肥培养,5 d后采集不同处理番木瓜叶片,以其cDNA为模板,利用Eastep? qPCR Master Mix Kit试剂对叶片中的PRSV基因和CpTIFY10A-like基因进行qRT-PCR检测。反应体系20.0 μL:10.0 μL Eastep? qPCR Master Mix(2×),2.0 μL cDNA模板,10 μmol/L上、下游引物(PRSV-F和PRSV-R/TIFY-F和TIFY-R)各0.4 μL,Nuclease-Free Water补足至20.0 μL。以Actin为内参基因,其qRT-PCR引物见表2。qRT-PCR扩增程序:95 ℃预变性10 min;95 ℃ 30 s,58 ℃ 30 s,72 ℃ 30 s,进行45个循环,在55 ℃退火结束时读取荧光信号。然后测定熔解曲线,熔解曲线步骤:95 ℃预变性30 s,55~95 ℃升温全过程连续读取荧光信号,最后进行数据分析。
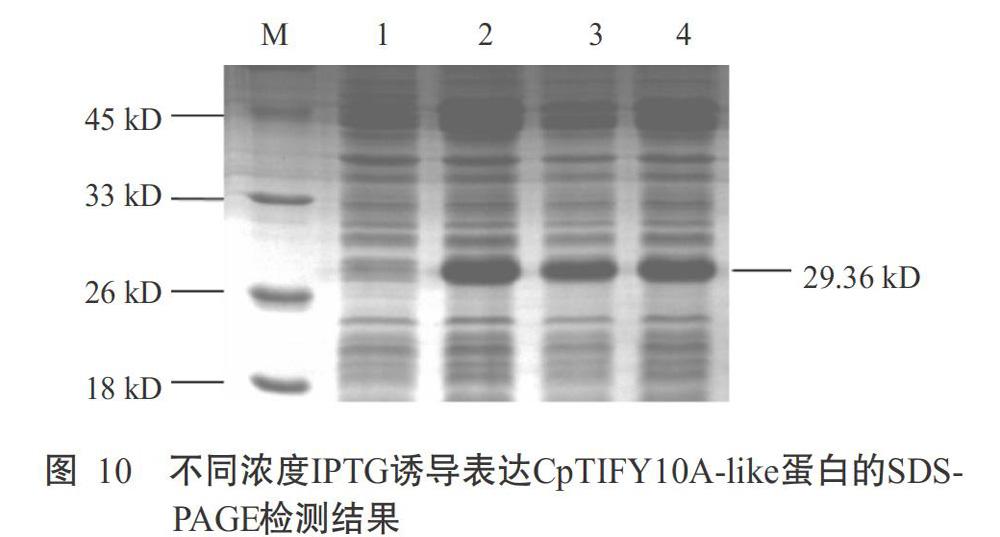
1. 2. 5 原核表达载体构建 使用Primer 5.0设计原核表达的CpTIFY10A-like基因编码区序列(CDS)的上游引物(5'-CGCGGATCCATGTCGAATTCGCCT GAG-3',下划线处为BamH I酶切位点)和下游引物(5'-CCGCTCGAGCTACTGTAACAATGGTGATTGA G-3',下划线处为Xho I酶切位点),用于目的基因CpTIFY10A-like PCR扩增。反应体系50.0 μL:2.5 μL cDNA模板,30.0 μL PCR-Grade H2O,10 μmol/L的上、下游引物各2.0 μL,0.5 μL LA Taq DNA聚合酶,5.0 μL 10×LA Taq Buffer II和8.0 μL dNTP Mixture。扩增程序:96 ℃预变性3 min;96 ℃ 30 s,58 ℃ 20 s,72 ℃ 3 min,进行35个循环;72 ℃延伸5 min。利用BamH I和Xho I双酶切PCR扩增目的基因和表达质粒pET28a-sumo 5 h,T4连接酶16 ℃过夜连接,转化Rosetta(DE3)感受态细胞,菌液PCR验证后挑取阳性单克隆送天一辉远生物科技有限公司测序。测序正确后,将转化菌株接种于LB液体培养基中摇菌OD600为0.6时,加入不同浓度(0.5、0.8和1.0 mmol/L)的IPTG,16 ℃下过夜诱导蛋白表达,以不加IPTG诱导为对照,然后加入5×SDS-PAGE Loading Buffer,100 ℃煮沸10 min,经SDS-PAGE检测蛋白的表达效果。
1. 3 统计分析
采用2-△△Ct法计算目的基因的相对表达量(Livak and Schmittegen,2001),试验所得数据采用Excel 2010进行统计分析及图表绘制,运用SPSS 22.0进行方差分析和显著性分析。
2 结果与分析
2. 1 番木瓜叶片总RNA提取结果
番木瓜叶片总RNA电泳结果如图1所示,28S条带和18S条带明显亮于5S条带,表明提取的RNA效果较好,可用于后续研究。
2. 2 CpTIFY10A-like基因克隆结果
3'-RACE克隆片段长度584 bp(图2-A),5'-RACE克隆片段长度630 bp(图2-B),CpTIFY10A-like基因中间片段长度1027 bp(图2-C),最后拼接获得CpTIFY10A-like基因全长为1352 bp。将其提交至NCBI数据库进行BLAST对比,结果发现与已发表的番木瓜CpTIFY10A-like基因(XM_022035541.1)的核苷酸序列同源性达100%,CDS序列为822 bp,证明克隆获得的CpTIFY10A-like基因属于已知序列,且真实存在,可用于后续基因功能验证。
2. 3 生物信息学分析结果
使用ExPASy ProtParam在线软件分析CpTIFY10A-like蛋白理化性质,结果显示CpTIFY10A-like蛋白由273个氨基酸残基组成,理论分子量29.36 kD,分子式C1283H2054N356O407S12,理论等电点(pI)9.23,为碱性蛋白。CpTIFY10A-like蛋白的不稳定系数62.97,亲水性平均系数-0.458,为不稳定的亲水性蛋白(图3)。使用WoLF PSORT进行CpTIFY10A-like蛋白亚细胞定位,结果显示核定位系数8.0,核质体定位系数6.0,叶绿体定位系数2.0,线粒体定位系数2.0,质体的定位系数2.0,表明CpTIFY10A-like蛋白定位于细胞核。利用SOPMA在线分析CpTIFY10A-like蛋白二级结构,结果如图4所示,α-螺旋占15.02%,延伸链占8.42%,β-折叠占2.20%,无规则卷曲占74.36%,说明该蛋白二级结构以无规则卷曲和α-螺旋为主,同时含有少量的β-折叠和延伸链。
2. 4 CpTIFY10A-like基因同源性分析结果
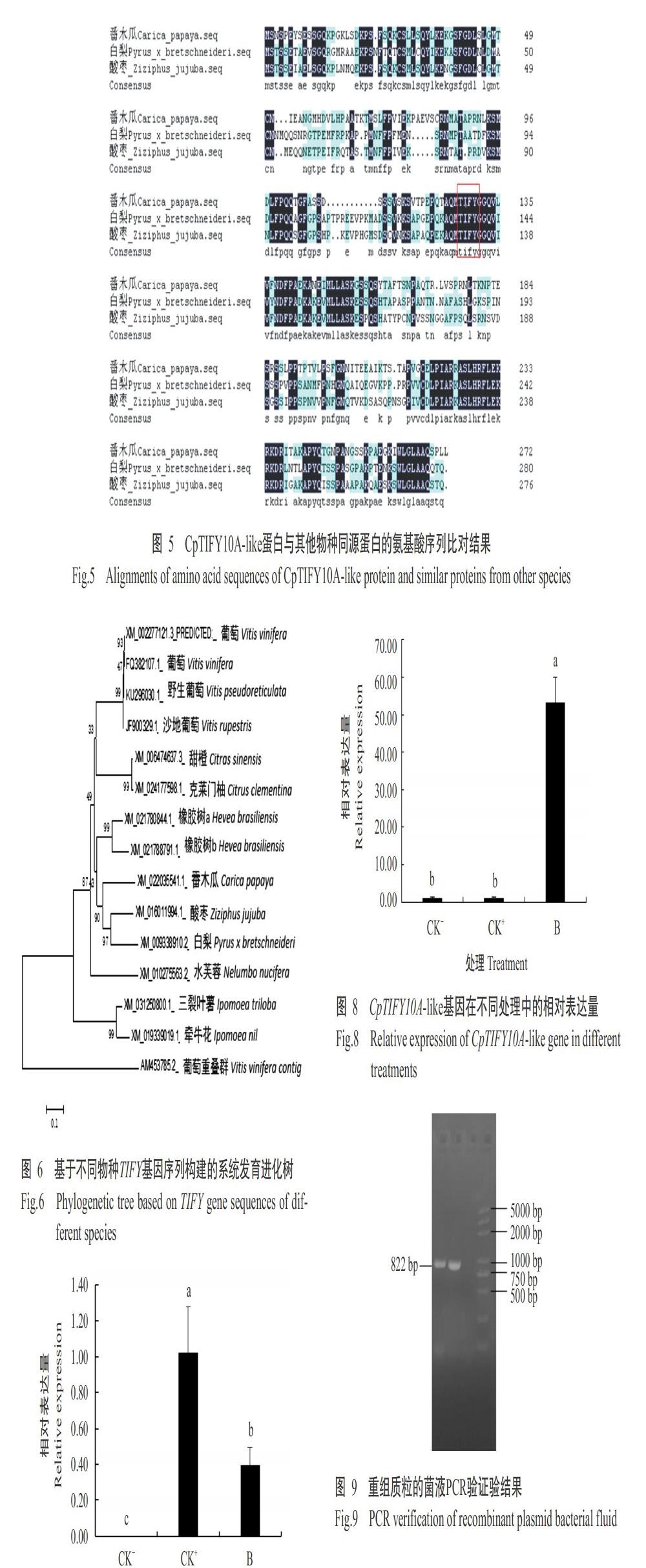
将CpTIFY10A-like基因编码蛋白的氨基酸序列提交至NCBI数据库中进行BLAST比对,结果显示其与酸枣和白梨TIFY蛋白的氨基酸序列相似性达83.21%和81.02%,且多序列比对分析显示三者均含有特定的TIFY结构域(图5)。将CpTIFY10A-like基因与NCBI数据库中同源性较高的物种TIFY基因进行系统发育进化分析,结果(图6)显示CpTIFY10A-like基因与酸枣和白梨的TIFY基因聚为同一分支,即三者亲缘关系较近。
2. 5 qRT-PCR检测结果
qRT-PCR检测结果(图7)显示,不同处理下PRSV基因的相对表达量排序为CK+(1.02)>B(0.39)>CK-(0),三者存在显著差异(P<0.05,下同);CpTIFY10A-like基因的相对表达量排序为B(53.12)>CK+(1.15)>CK-(1.02),其中B处理与CK+处理存在显著差异(图8)。结合图7和图8可知,在经禾甲安处理后的感病植株中CpTIFY10A-like基因表达量显著升高,而PRSV基因表达量显著降低,说明禾甲安可诱导CpTIFY10A-like基因高效表達,且CpTIFY10A-like基因在植株抗PRSV过程中发挥重要调控作用。
2. 6 CpTIFY10A-like基因原核表达分析结果
由菌液PCR验证结果(图9)和测序结果可知,构建的重组质粒中已成功插入目标片段(CpTIFY10A-like基因CDS序列为822 bp),表明成功获得重组质粒pET28a-CpTIFY10A-like-sumo。由图10可知,与对照(泳道1)相比,泳道2、3和4在26~33 kD之间有明显条带,与CpTIFY10A-like蛋白的理论分子量29.36 kD基本相符,且不同浓度IPTG诱导下,IPTG浓度越高,蛋白表达量越多,表明CpTIFY10A-like蛋白在原核细胞中成功表达。
3 讨论
禾甲安可促使番木瓜体内抗病性酶活升高,从而起到防治番木瓜环斑病害的作用(张芮宁等,2020)。在分子层面上发现禾甲安的施加会诱导番木瓜体内抗病相关基因表达,从而达到抗PRSV的效果,而CpTIFY10A-like基因就是其中的一个抗病基因(安娜,2018)。TIFY基因家族广泛存在于陆地植物中(Bai et al.,2011)。本研究的多重序列对比结果显示,CpTIFY10A-like蛋白与其他物种如拟南芥和紫花苜蓿TIFY蛋白均含有特定的TIFY结构域(Ming et al.,2008;Zhu et al.,2014)。前人研究发现,拟南芥、番茄和马铃薯中的TIFY基因家族成员均能影响茉莉酸合成(Grunewald,2009;胡利宗等,2017)。本课题组前期通过转录组数据分析获得的KEGG通路如图11所示,CpTIFY10A-like基因作用于茉莉酸合成通路上,而茉莉酸又与植物的抗病性相关(Vanholme et al.,2007;安娜,2018),故推测CpTIFY10A-like基因参与番木瓜抗逆响应过程。本研究的qRT-PCR检测结果显示,在经禾甲安处理后的感病植株中CpTIFY10A-like基因表达量显著升高,而PRSV基因表达量显著降低,推测禾甲安可诱导CpTIFY10A-like基因高效表达,致使PRSV基因表达量大幅度降低,与霍鹏(2015)研究发现番木瓜通过eIF4E基因沉默以抵抗PRSV的结果相似。此外,本研究通过原核表达分析可知,CpTIFY10A-like基因可在原核细胞中表达,今后可通过相关次生代谢产物和体外蛋白研究进一步探究其抗PRSV的作用机理。
本研究通过分析番木瓜CpTIFY10A-like基因的生物学信息、抗病表达量和原核表达效果,为今后深入探索CpTIFY10A-like基因的功能及表达蛋白互作提供理论参考,同时为研究CpTIFY10A-like基因在番木瓜抗PRSV病毒病过程的分子调控机理打下基础,对番木瓜抗病育种具有重要意义。
4 结论
禾甲安可诱导CpTIFY10A-like基因高效表达,进而通过茉莉酸通路起调节作用以抵抗PRSV感染,即CpTIFY10A-like基因在番木瓜抗PRSV过程中发挥重要调控作用。
参考文献:
安娜. 2018. CTS-N诱导下番木瓜对环斑病毒病抗性研究[D]. 海口:海南大学. [An N. 2018. Study on the resistance of papaya to ringspot virus induced by CTS-N[D]. Haikou:Hainan University.]
陈豪军,潘祖建,周全光,甘卫堂. 2013. 番木瓜优良品种的引进与选育研究[J]. 中国南方果树,42(6):59-63. [Chen H J,Pan Z J,Zhou Q G,Gan W T. 2013. Study on the introduction and breeding of excellent papaya varieties[J]. South China Fruits,42(6):59-63.]
胡利宗,王俊生,赵锦慧,王秋霞,韩娇月,纪秀娥. 2017. 番茄与马铃薯TIFY家族基因的鉴定与比较分析[J]. 分子植物育种,15(4):1192-1203. [Hu L Z,Wang J S,Zhao J H,Wang Q X,Han J Y,Ji X E. 2017. Identification and comparative analysis of the TIFY family genes in tomato and potato[J]. Molecular Plant Breeding,15(4):1192-1203.]
霍鹏. 2015. 利用多重荧光定量PCR鉴定番木瓜病毒及番木瓜eIF4E家族基因干扰效应研究[D]. 海口:海南大学. [Huo P. 2015. Study on the interference effects of papaya virus and papaya eIF4E family genes by multiplex fluorescent quantitative PCR[D]. Haikou:Hainan University.]
孙程. 2013. 玉米TIFY家族ZmJAZ14基因的功能验证[D]. 北京:中国农业科学院. [Sun C. 2013. Functional verification of the ZmJAZ14 gene in the TIFY family of maize [D]. Beijing:Chinese Academy of Agricultural Sciences.]
鄢兴祥,李萍,陈萍,樊俊华,刘蕤丹,张荣萍. 2019. 4种生物农药抗番木瓜花叶病毒病的效果及其生理基础研究[J]. 中国南方果树,48(2):75-81. [Yan X X,Li P,Chen P,Fan J H,Liu Y D,Zhang R P. 2019. Effects of four biological pesticides on papaya mosaic virus disease and its physiological basis[J]. South China Fruits,48(2):75-81.]
張荣萍,樊俊华,陈慧娟,鄢兴祥,陈萍. 2017. 3种生物农药对番木瓜环斑病的防效研究[J]. 中国南方果树,46(3):79-82. [Zhang R P,Fan J H,Chen H J,Yan X X,Chen P. 2017. Study on the control effect of three biological pesticides on papaya ring spot disease[J]. South China Fruits, 46(3):79-82.]
张芮宁,袁舟宇,陈萍. 2020. 禾生素在植物中的应用研究进展[J]. 南方农业,14(5):117-120. [Zhang R N,Yuan Z Y,Chen P. 2020. Research progress of the application of CTS-N in plants[J]. South China Agriculture,14(5):117-120.]
周明. 2013. 玉米TIFY/JAZ蛋白全基因组分析[D]. 雅安:四川农业大学. [Zhou M. 2013. Whole-genome analysis of maize TIFY/JAZ protein[D]. Yaan:Sichuan Agricultu-ral University.]
Bai Y H,Meng Y J,Huang D L,Qi Y H,Chen M. 2011. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family[J]. Genomics,98(2):128-136.
Bau H J,Kung Y J,Raja J A,Chen S J,Chen K C,Chen Y. K,Wu H W,Yeh S D. 2008. Potential threat of a new pathotype of papaya leaf distortion mosaic virus infecting transgenic papaya resistant to papaya ringspot virus[J]. Phytopathology,98(7):848-856.
Ebel C,Benfeki A,Hanin M,Solano R,ChiniA. 2018. Characterization of wheat(Triticum aestivum)TIFY family and role of Triticum durum TdTIFY11a in salt stress tolerance[J]. PLoS One,13(7):e0200566.
Geourjon C,Deléage G. 1996. SOPMA:Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments[J]. Computer Applications in the Biosciences Cabios,11(6):681-684.
Grunewald W,Vanholme B,Pauwels L,Plovie E,Inzé D,Gheysen G,Goossens A. 2009. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin[J]. EMBO Reports,10(8):923-928.
Hall B G. 2013. Building phylogenetic trees from molecular data with MEGA[J]. Molecular Biology and Evolution(5):5.
Huang Z,Jin S H,Guo H D,Zhong X J,He J,Li X,Jiang M Y,Yu X F,Long H,Ma M D,Chen Q B. 2016. Genome-wide identification and characterization of TIFY family genes in Moso Bamboo(Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses[J]. PeerJ,4(10):e2620. doi:10.7717/peerj.2620.
Livak K J,Schmittgen T D. 2000. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method[J]. Methods,25(4):402-408.
Ming R,Hou S B,Feng Y,Yu Q G,Alexandre D L,Jimmy H S,Pavel S,Wang W,Benjamin V L,Kanako L T,Lewis,Steven L S,Lu F,Meghan R J ,Rachel L S,Jan E M,Chen C X,Qian W B,Shen J G,Du P,Moriah E,Eric T,Tang H B,Eric L,Robert E P,Todd P M,Kerr W,Danny W R,Henrik A,Wang M L,Zhu Y J,Michael S,Niranjan N,Ricelle A A,Peizhu G,Andrea B,Wai C M,Christine M A,Ren Y,Liu C,Wang J M,Wang J P,Na J K,Eugene V S,Brian H,Jyothi T,David N,Wang X Y,John E B,Andrea R G,Arthur L D,Ratnesh S,Jon Y S,Savarni T,Kabi N,Wei H R,Beth I,Maya P,Jiang N,Zhang W L,Gernot P,Aaron W,Rafael N P,Manuel J T,Feltus A F,Brad P,Li Y J,Max B,Luo M C,Liu L,David A C,Stephen M M,Paul H M,Tak S,Jiang J M,Mary A S,Vikki F,Thomas M O,Dorothy E S,Claude W D,Jeffrey D P,Michael F,Andrew H P,Dennis G,Wang L,Maqsudul A. 2008. The draft genome of the transgenic tropical fruit tree papaya(Carica papaya Linnaeus)[J]. Nature,452(7190):991-996.
Tripathi S,Suzuki J Y,Ferreira S A,Gonsalves D. 2010. Papaya ringspot virus-P:Characteristics,pathogenicity,sequence variability and control[J]. Molecular Plant Patho-logy,9(3):269-280.
Vanholme B,Grunewald W,Bateman A,Kohchi T,Gheysen G. 2007. The tify family previously known as ZIM[J]. Trends in Plant Science,12(6):239-244.
Yeku O,Frohman M A. 2003. Rapid amplification of cDNA ends(RACE)[J]. Methods in Molecular Biology,226(67):105.
Zhang G X,Wu Y G,Muhammad Z U H,Yang Y Z,Yu J,Zhang J F,Yang D M. 2019. cDNA cloning,prokaryotic expression and functional analysis of 3-hydroxy-3-me-thylglutaryl coenzyme a reductase(HMGCR) in Pogostemon cablin[J]. Protein Expression and Purification,163:105454.
Zhao G,Song Y,Wang C X,Hamama I B,Wang Q H,Zhang C J,Yang Z R,Liu Z,Chen E Y,Zhang X Y,Li F G. 2016. Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton[J]. Molecular Genetics and Genomics,291(6):2173-2187.
Zhu D,Bai X,Luo X,Chen Q,Cai H,Ji W,Zhu Y M. 2013. Identification of wild soybean(Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress[J]. Plant Cell Reports,32(2):263-272.
Zhu D,Li R T,Liu X,Sun M Z,Wu J,Zhang N,Zhu Y M. 2014. The positive regulatory roles of the TIFY10 proteins in plant responses to alkaline stress[J]. PLoS One,9(11):e111984.
(責任编辑 陈 燕)
