卵形鲳鲹基因组调研及其SSR分子标记的开发应用
2020-07-07张永德文露婷罗洪林林勇杜雪松余艳玲韦孜娜黄姻
张永德 文露婷 罗洪林 林勇 杜雪松 余艳玲 韦孜娜 黄姻


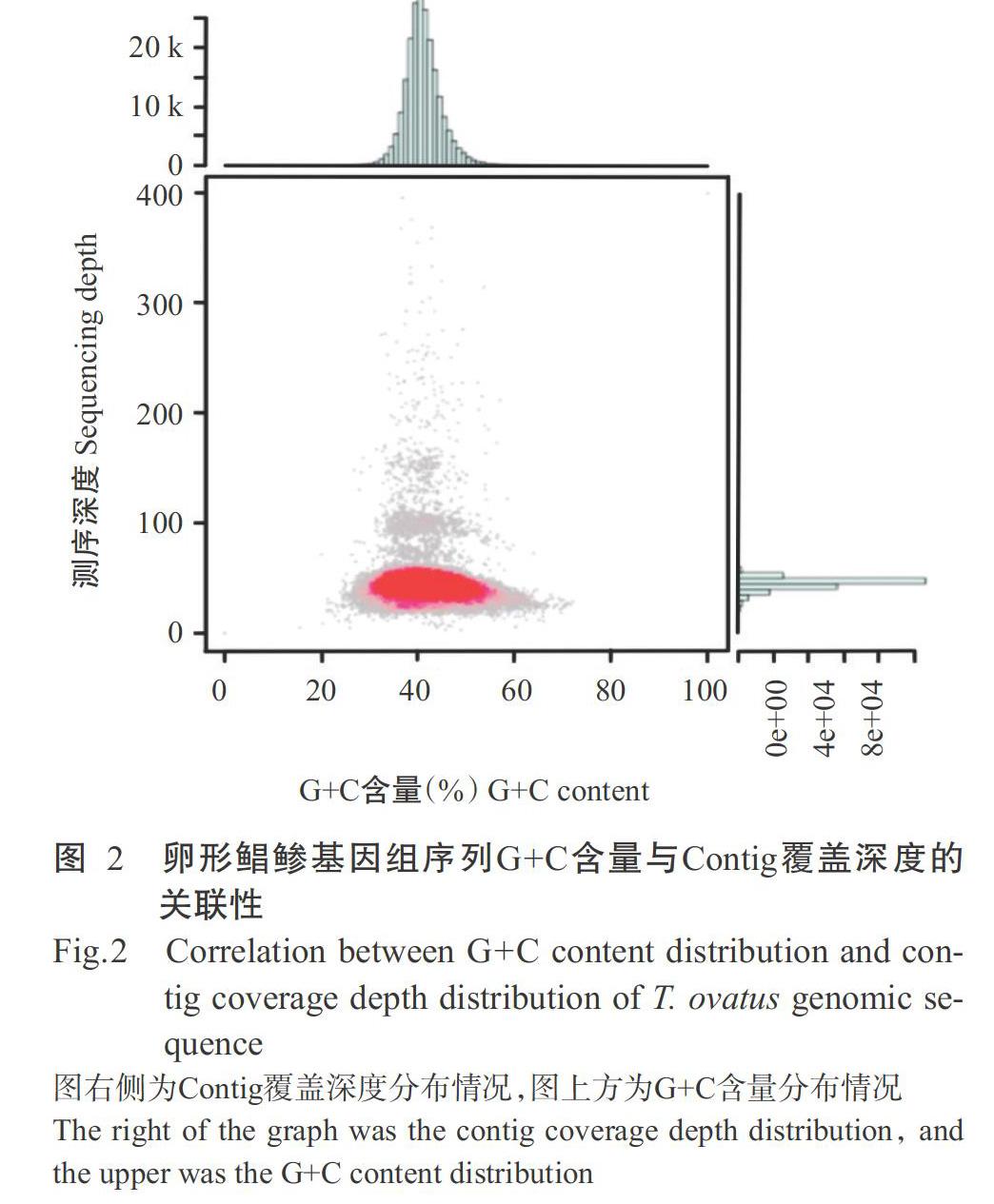
摘要:【目的】通過高通量测序技术调研卵形鲳鲹基因组数据并开发SSR分子标记,为卵形鲳鲹全基因组测序与组装、种质资源保护利用及良种选育提供技术支撑。【方法】通过Illumina Hiseq 2500测序平台对卵形鲳鲹基因组进行调研,采用K-mer方法对基因组大小、杂合率、G+C含量及序列重复性等进行分析,从调研数据中分析SSR的分布特征,筛选出多态性SSR位点,并对卵形鲳鲹养殖群体进行遗传多样性分析。【结果】卵形鲳鲹基因组大小为642.68 Mb,杂合率为0.31%,重复序列比例为30.19%,G+C含量为41.45%,提示卵形鲳鲹基因组为简单基因组。基因组初步组装结果显示,Contig总长度为627.23 Mb,N50、N90分别为8.21和1.71 Mb;Scalffold总长度为628.19 Mb,N50、N90分别为10.19和2.04 Mb。从卵形鲳鲹基因组调研数据中共检测出190121条SSR序列,SSR序列分布密度为295.8条/Mb。在所有SSR序列中,以二核苷酸重复基元最多(115557条),占60.78%;其次是三核苷酸重复基元(54839条),占28.84%;六核苷酸重复基元最少(1172条),占0.62%。在二核苷酸重复基元中以TG和AC的重复数较多,分别占二核苷酸重复基元总数的22.99%和21.76%。从合成的50对SSR引物中筛选获得29对多态性SSR引物,采用这29对SSR引物对卵形鲳鲹群体进行遗传多样性分析,结果发现29个SSR位点共检测到98个等位基因,其有效等位基因数(Ne)为1.3998~3.9123(平均为2.6690),期望杂合度(He)为0.2856~0.7444(平均为0.5965),多态信息含量(PIC)为0.2647~0.6968(平均为0.5195);在29个SSR位点中,高度多态性位点(PIC>0.50)有15个,其余14个为中度多态性位点(0.25 关键词: 卵形鲳鲹;基因组;核苷酸重复基元;SSR分子标记;遗传多样性 中图分类号: S965.331 文献标志码: A 文章编号:2095-1191(2020)05-0983-12 Abstract:【Objective】High throughput sequencing was used to survey the genome data of Trachinotus ovatus and develop SSR molecular markers to provide an effective basis for whole genome sequencing and assembling,protection and utilization of germplasm resources and selective breeding of T. ovatus. 【Method】Illumina Hiseq 2500 sequencing platform was used to survey the T. ovatus genome,and K-mer was used to analyze the genome size, heterozygosity rate, G+C content and sequence repeatability.The distribution characteristics of SSR were analyzed from the survey data. The polymorphic SSR loci were screened and the genetic polymorphism of T. ovatus population was analyzed. 【Result】The genome size of T. ovatus was 642.68 Mb, with a 0.31% heterozygosity rate and 30.19% repeated sequence proportion, and the G+C content was 41.45%, indicating that the T. ovatus genome was a simple genome. Preliminary genome assembly results showed that the total length of contig was 627.23 Mb, N50 and N90 were 8.21 Mb and 1.71 Mb, respectively, while the total length of Scalffold was 628.19 Mb, N50 and N90 were 10.19 Mb and 2.04 Mb, respectively. A total of 190121 SSR sequences were detected from the genomic survey data, with a SSR sequence distribution density of 295.8 SSRs/Mb. Among all SSR sequences, dinucleotide repeat motifs were the most common SSRs, accounting for 60.78%(115557) of the total SSR sequences; followed by trinucleotide repeat motifs, accounting for 28.84%(54839); hexanucleotide repeat motifs were the least, accounting for 1.96%(1172). Among the dinucleotide repeating motifs, TG and AC had the highest frequency in dinucleotide motifs, accounting for 22.99% and 21.76% of the total number of dinucleotiderepeating motifs, respectively. A total of 29 polymorphic SSR primers were screened from 50 SSR primers, and the genetic polymorphism analysis was conducted on 64 T. ovatus with these 29 SSR primers.The results showed that a total of 98 alleles were detec-ted in 29 SSR loci. The effective allele number (Ne) was 1.3998-3.9123 (average 2.6690), the expected heterozygosity (He) was 0.2856-0.7444 (average 0.5965), and the polymorphic information content(PIC) was 0.2647-0.6968(average 0.5195). Among the 29 SSR loci, there were 15 highly polymorphic loci (PIC>0.50), and the remaining 14 were modera-tely polymorphic loci (0.25 Key words: Trachinotus ovatus; genome; nucleotide repeat motif; SSR molecular marker; genetic diversity Foundation item: Guangxi Innovation Driven Development Project(Guike AA17204080-3,Guike AA18242031-2); Basic Research Project of Guangxi Public Welfare Research Institute(CXIF-2016-03) 0 引言 【研究意義】卵形鲳鲹(Trachinotus ovatus)又称金鲳,为暖水性中上层鱼类,主要分布于印度洋、太平洋、大西洋及非洲沿岸的热带和亚热带水域,在我国南海、东海和黄海均有分布(陈伟洲等,2007;Xie et al.,2014;黄小林等,2018)。卵形鲳鲹属于高蛋白低脂肪鱼类,富含多种蛋白及其他营养成分,历来被视为名贵食用鱼类(区又君和李加儿,2005),且具有生长速度快、食性简单及饲料转化率高等特点,是我国海水集约化养殖的主要品种之一。目前,国内外学者针对卵形鲳鲹的研究主要集中在饲料与营养(Wang et al.,2014;胡海滨等,2019)、生长发育(黄小林等,2018;Liu et al.,2019)、免疫与病害(熊向英等,2018;Sun et al.,2019)及其分子生物学(侯树鉴等,2018;Wu et al.,2019)等方面,而有关其遗传育种的研究鲜见报道。遗传育种工作的滞后,导致近年来卵形鲳鲹出现明显的种质退化现象,具体表现为遗传多样性、生长性能、抗逆及抗病性降低,而鱼苗和鱼种死亡率逐年升高等(彭敏等,2011)。因此,急需开展卵形鲳鲹种质资源多样性及遗传育种等相关研究,为加快良种选育及促进其养殖业健康发展提供理论依据。【前人研究进展】分子生物技术的快速发展为开展卵形鲳鲹分子标记辅助选择(MAS)及更高层次的分子设计育种提供了可能,而缺乏基因组信息及分子标记是限制开展卵形鲳鲹MAS的主要因素。在现有的分子标记中,微卫星(SSR)分子标记因具有重复性高、等位基因丰富、共显性及基因组覆盖度高等特点,已发展成为物种遗传多样性分析和连锁作图最常用的方法(Jiao et al.,2012;凌士鹏等,2018;陈海玲等,2019),但SSR分子标记具有种质特异性,需提前进行筛选开发(赵彦花等,2019)。随着转录组测序技术的快速发展及测序成本的进一步降低,大量鱼类相继完成全基因组测序工作,如虹鳟(Oncorhynchus mykiss)(Berthelot et al.,2014)、尼罗罗非鱼(Oreochromis niloticus)(Brawand et al.,2014)、半滑舌鳎(Cynoglossus semilaevis)(Chen et al.,2014)、菊黄东方鲀(Takifugu flavidus)(Gao et al.,2014)、大黄鱼(Larimichthys crocea)(Wu et al.,2014)、鲤(Cyprinus carpio)(Xu et al.,2014)、草鱼(Ctenopharyngodon idellus)(Wang et al.,2015)、亚洲龙鱼(Scleropages formosus)(Bian et al.,2016)、斑点雀鳝(Lepisosteus oculatus)(Braasch et al.,2016)、大菱鲆(Scophthalmus maximus)(Figueras et al.,2016)、大西洋鲑(Salmo salar)(Lien et al.,2016)、海马(Hippocampus erectus)(Lin et al.,2016)、翻车鱼(Mola mola)(Pan et al.,2016)、暹罗斗鱼(Betta splendens)(Fan e al.,2018)、条石鲷(Oplegnathus fasciatus)(Xiao et al.,2019)、西藏高原鳅(Triplophysa tibetana)(Yang et al.,2019)和多鱗白甲鱼(Onychostoma macrolepis)(Sun et al.,2020)等,为这些水生生物的养殖适应性进化研究提供了大数据支撑,其生长、生殖、性别决定、抗病及抗逆等经济性状相关基因挖掘研究也取得重要进展,有效推动了全基因组选择育种、分子模块设计育种、转基因和基因编辑等新型分子育种技术的快速发展。【本研究切入点】对于无参考基因组的物种而言,通过基因组调研数据开发SSR分子标记是一种相对高效的方法(Zhou et al.,2013)。此外,基因组调研可提供有关基因组结构的信息,包括基因组大小、杂合率、G+C含量和重复序列含量等,为物种全基因组测序及序列组装提供参考依据。但至今鲜见有关卵形鲳鲹基因组调研及其SSR分子标记开发的研究报道。【拟解决的关键问题】通过Illumina Hiseq 2500测序平台对卵形鲳鲹基因组进行调研,采用K-mer方法对基因组的大小、杂合率、G+C含量及序列重复性等信息进行分析,从调研数据中挖掘SSR的分布特征,筛选出多态性SSR位点,并对卵形鲳鲹养殖群体进行遗传多样性分析,以期为卵形鲳鲹全基因组测序与组装、种质资源保护利用及良种选育提供技术支撑。 1 材料与方法 1. 1 样品采集与基因组DNA提取 从深圳海域人工养殖的卵形鲳鲹群体中随机选取65尾,在无菌条件下采集其肌肉组织。其中,1份肌肉样品用于基因组测序,另外64份肌肉样品用于群体遗传多样性分析。按照天根海洋动物组织基因组DNA提取试剂盒说明进行卵形鲳鲹基因组DNA提取,并采用NanoDrop 2000超微量分光光度计(ThermalFisher,美国)检测DNA浓度,以1.5%琼脂糖凝胶电泳检测DNA质量。检测合格的DNA置于 -20 ℃冰箱中保存备用。 1. 2 基因组测序及特征分析 随机取1尾卵形鲳鲹的DNA样品,通过Covaris超声波破碎仪随机打断成长度为230 bp的片段,经末端修复、加poly(A)、加测序接头、纯化及PCR扩增等构建小片段文库。采用Illumina Hiseq 2500测序平台进行PE双末端测序,测得的原始数据经数据质控和过滤后,采用K-mer(K=17)对获得的有效数据(Clean data)进行统计分析,估计基因组大小、杂合率及重复率等基因组特征。采用SOAPdenovo 2.01对卵形鲳鲹测序基因组进行初步组装,统计G+C含量和覆盖深度(Luo et al.,2012)。 1. 3 SSR序列查找及引物设计 采用MISA程序搜索检测样品DNA序列中的SSR序列,包括二核苷酸、三核苷酸、四核苷酸、五核苷酸和六核苷酸重复基元(Beier et al.,2017),共重复4次。对获得的SSR序列进行过滤,去除距离过近的SSR序列,最后运用Primer 3.0设计SSR引物(K?ressaar et al.,2018),并挑选其中50对SSR引物委托生工生物工程(上海)股份有限公司合成。 1. 4 SSR位点筛选 采用天根生化科技(北京)有限公司的Golden Easy PCR System-KT221试剂,以合成的SSR引物对卵形鲳鲹进行PCR扩增,反应体系20.0 μL:2×Reaction Mix 10.0 μL,H2O 7.0 μL,上、下游引物(10 ?mol/L)各1.0 μL,DNA模板1.0 μL。扩增程序:95 ℃预变性4 min;95 ℃ 30 s,56~65 ℃ 30 s,72 ℃ 1 min,进行30个循环;72 ℃延伸2 min。PCR扩增产物经1.5%琼脂糖凝胶电泳检测后,挑选特异性强、重复性好、条带清晰的SSR引物再进行梯度PCR扩增,确定其退火温度。以pBR322/MSP I為Marker,随机选取20尾卵形鲳鲹DNA进行SSR引物多态性筛选,经PCR扩增、8% SDS-PAGE电泳、0.1%硝酸银染色及2% NaOH显色后,采用伯乐凝胶成像系统进行拍照,以Bio-Rad Quantity One读取片段大小,分析各SSR引物的多态性。 1. 5 卵形鲳鲹群体遗传多样性分析 以筛选出具有多态性的29对SSR引物对64尾卵形鲳鲹群体进行PCR扩增及PAGE检测分析,读取片段大小。利用PopGen32计算卵形鲳鲹群体的遗传多样性参数,包括等位基因数(Na)、有效等位基因数(Ne)、期望杂合度(He)、Hardy-Weinberg平衡遗传偏离概率(PHWE);采用PICcalc程序(Nagy et al.,2012)计算各SSR位点的多态信息含量(PIC)。 2 结果与分析 2. 1 卵形鲳鲹基因组测序及基因组大小估计结果 采用卵形鲳鲹基因组DNA构建一个230 bp小片段文库,通过Illumina Hiseq 2500测序平台进行PE双末端测序,共获得49.12 G的原始测序数据,其中Q20、Q30分别为95.53%和91.46%,测序错误率为0.04%,说明建库测序成功。对原始测序数据进行数据质控和过滤,获得48.63 G的有效数据。随机抽取过滤后的高质量数据,采用BLAST比对NCBI核苷酸数据库(NT库),结果发现获得的有效数据不存在明显外源污染。对测序数据进行K-mer分析,结果(图1)发现在深度为65时出现主峰值,总K-mer为42406776346,计算得到卵形鲳鲹基因组大小为652.41 Mb,修正后的基因组大小为642.68 Mb。基因组杂合率为0.31%,重复序列比例为30.19%。 2. 2 卵形鲳鲹基因组初步组装情况 采用SOAPdenovo 2.01对卵形鲳鲹测序基因组进行初步组装(K-mer=41),统计结果见表1。其中,Contig总长度为627.23 Mb,N50、N90分别为8.21和1.71 Mb,序列最大长度为95.96 Mb;Scalffold总长度为628.19 Mb,N50、N90分别为10.19和2.04 Mb,序列最大长度为126.91 Mb,基因组G+C含量为41.45%。选取500 bp以上的Contigs,根据其G+C分布及覆盖深度信息绘制散点图(图2),其中红色部分为散点图中点密度较大的部分。从图2右侧的Contig覆盖深度分布情况可看出,在Contig覆盖深度为47处为纯合峰,图中红色散点聚集区域是G+C的主要分布区域;图2上方的G+C含量主峰约出现在41%处,与计算得到的基因组G+C含量一致,且红色散点也分布在G+C含量为41%的附近。在Contig覆盖深度为70~110、G+C含量为30%~50%的区域和Contig覆盖深度为130~170、G+C含量为30%~50%的区域,出现小部分的G+C集中区域,推测这些区域为卵形鲳鲹基因组中的重复区域。 2. 3 卵形鲳鲹基因组SSR特征分析结果 在卵形鲳鲹基因组数据中,过滤掉位于Contig序列两端的SSR序列(距离Contig序列两端小于100 bp)后,共检测出190121条SSR序列,分布密度为295.8条/Mb。对SSR序列长度分布进行统计,结果(图3)发现SSR重复序列长度主要集中在11~24 bp。在所有SSR序列中,以二核苷酸重复基元最多,为115557条,占60.78%;其次是三核苷酸重复基元,为54839条,占28.84%;六核苷酸重复基元最少,仅1172条,占0.62%(图4)。在5种核苷酸重复基元分布(图5)方面,以出现4次重复的核苷酸重复基元最常见(44778条,占23.55%),其次是6次重复(32615条,占17.15%)和7次重复(19014条,占10.00%),而出现20次以上重复的核苷酸重复基元仅有6652条(占3.50%)。此外,在二核苷酸重复基元中以TG和AC的重复数较多,分别占二核苷酸重复基元总数的22.99%和21.76%;三核苷酸重复基元以GAG和AAT的重复数较多,分别占三核苷酸重复基元总数的4.84%和4.81%;四核苷酸重复基元以AAAT的重复数最多,占四核苷酸重复基元总数的7.36%;五核苷酸重复基元以AATTG的重复数最多,占五核苷酸重复基元总数的2.93%;六核苷酸重复基元以CTGATT的重复数最多,占六核苷酸重复基元总数的5.72%。 2. 4 卵形鲳鲹SSR位点筛选及评估结果 以卵形鲳鲹基因组DNA为模板,采用合成的50对SSR引物进行PCR扩增与退火温度筛选,结果表明,有31对SSR引物能扩增出清晰的目的条带,且重复性较好,对应的退火温度介于56.1~64.1 ℃(表2)。SSR引物多态性筛选结果显示,只有TOSR037和TOSR043这2对SSR引物的扩增产物为单态性,其余29对SSR引物的扩增条带均呈多态性。 2. 5 卵形鲳鲹群体的遗传多样性 采用筛选出的29对SSR引物分别对64尾卵形鲳鲹进行PCR扩增(图6)及遗传多样性分析,结果(表3)显示,29个SSR位点共检测到98个等位基因,平均每个SSR位点的Na为3.3793,Ne为2.6690,He为0.5965,PIC为0.5195。在29个SSR位点中,高度多态性位点(PIC>0.50)有15个,其余14个为中度多态性位点(0.25 3 讨论 3. 1 卵形鲳鲹基因组的基本特征 Illumina、Pacific Biosciences和Ion Torrent等高通量测序技术的出现与改进,以及序列组装算法的进步,使得以低成本高效获得动植物全基因组序列成为可能(Quail et al.,2012)。近十年来,各种动植物基因組序列的数量呈指数增长,极大促进了生命科学的快速发展。但不同物种的基因组大小及复杂程度差异明显,对基因组序列组装、测序价格及测序周期均会产生直接影响。大西洋鲑基因组是四倍体,其基因组大小高达2.97 Gb(Lien et al.,2016);太平洋牡蛎基因组大小虽然只有559 Mb,但其SNP序列分布密度较高,约1.22条/100 bp(Zhang et al.,2012);东方牡蛎(Crassostrea virginica)种群的SNP序列分布密度更高,每100 bp就有4.20条(Zhang and Guo,2010)或1.85条(Eierman and Hare,2014)。虾蟹类的核苷酸重复基元数较多,其中凡纳滨对虾(Litopenaeus vannamei)的核苷酸重复基元占其基因组的80%以上(Yu et al.,2015)。当遇到复杂的核苷酸重复序列或扩增不良的区域时,高通量测序技术的短读长和扩增偏好性均可能导致装配碎片化。如GC富集或GC贫乏区通常扩增效果较差,而对基因组序列质量产生明显影响,且对准确性的影响大于完整性(Aird et al.,2011)。这也是造成目前虾类和贝类参考基因组较少,且其基因组序列组装质量通常低于鱼类的重要原因。基因组的大小、杂合率、G+C含量及重复序列比例等信息均可通过K-mer分析进行估计(Shi et al.,2018;Song et al.,2018)。本研究采用K-mer对卵形鲳鲹基因组进行分析,得知卵形鲳鲹基因组大小为642.68 Mb,杂合率为0.31%,重复序列比例为30.19%,G+C含量为41.45%。从K-mer分析指标来看,卵形鲳鲹基因组不算大,其杂合率和重复序列处于中低水平,G+C含量合适,总体上属于简单基因组,后续可进行全基因组测序。卵形鲳鲹基因组序列的初步组装结果显示,Contig总长度为627.23 Mb,N50、N90分别为8.21和1.71 Mb;Scalffold总长度为628.19 Mb,N50、N90分别为10.19和2.04 Mb,即序列组装效果良好。 3. 2 卵形鲳鲹的SSR分布特征 SSR广泛分布于真核生物基因组中,在个体和种群水平上均会表现出多态性(Gadgil et al.,2017)。在大多数物种基因组中,具有短核苷酸重复基元(单核苷酸~三核苷酸)的序列较长核苷酸重复基元(四核苷酸~六核苷酸)的序列更丰富(Jessy et al.,2011)。本研究在卵形鲳鲹基因组调研数据中共检测出190121条SSR序列,SSR序列分布密度为295.8条/Mb。在所有SSR序列中,随着核苷酸重复基元的增加,其数量迅速减少,其中以二核苷酸重复基元最多(115557条),占60.78%,而六核苷酸重复基元最少(1172条),仅占0.62%,与胡子鲶(Clarias batrachus)(Srivastava et al.,2016)和长体圆鲹(Decapterus macrosoma)(孔啸兰等,2019)等鱼类的研究结果相似,但与大西洋鲑鱼、大西洋鳕鱼(Gadus morhua)及红鳍东方鲀(Takifugu rubripes)等鱼类存在差异(Jiang et al.,2014)。说明不同物种的SSR序列存在偏好性。在脊椎动物中,二核苷酸重复基元GT和AC被认为是最常见的SSR重复基元(Zardoya et al.,1996)。孔啸兰等(2019)对长体圆鲹的研究结果显示,二核苷酸重复基元占总SSR序列的53.39%,其中AC/GT类型占二核苷酸重复基元的68.40%。在本研究中,卵形鲳鲹基因组二核苷酸重复基元同样以TG和AC的重复数最多,合计占二核苷酸重复基元总数的44.75%。但相瑜等(2013)研究发现,三疣梭子蟹基因组二核苷酸重复基元以CT和AG的重复数较多,合计占50.00%。这可能与物种间的差异及其选择进化机制不同有关,且不同基因组区域中的SSR可能具有不同特征,从而执行不同的功能(Sonah et al.,2011)。 3. 3 卵形鲳鲹SSR分子标记的开发 目前,SSR引物的获得主要有以下途径:近缘物种引物借鉴法、直接分离法及数据库搜索法。对于亲缘关系非常近的属内种间生物而言,其SSR引物可共用,可根据已发表文献或已发布序列信息寻找所需的目的引物(Das et al.,2018)。相对于大多数研究基础较薄弱的物种,则必须从目标物种的基因组DNA中直接分离出具有多态性的SSR位点。常见的SSR分子标记分离方法主要有经典法、富集法、省略筛库法、ISSR片段扩增法和数据库检索法5种(孙立元,2014)。陈秀荔等(2010)利用生物素—磁珠吸附微卫星富集法筛选获得35个卵形鲳鲹SSR分子标记;孙立元(2014)在采用FIASCO法构建卵形鲳鲹微卫星富集文库的基础上,测序筛选出21个多态性SSR位点。对于一些已公布全基因组数据的物种,则可直接检索其基因组数据而获得SSR位点,极大提升了SSR分子标记开发的效率。 近年來,高通量测序技术的发展有效提升了快速且低成本获得基因组重要测序深度和覆盖范围的能力(Zhou et al.,2014),从而更全面准确地发现物种SSR位点信息。与传统的SSR分子标记开发方法相比,高通量测序更具成本效益,省时且功能强大(Jiang et al.,2015),通过高通量测序获得的基因组或转录组数据是SSR分子标记开发的重要资源(Song et al.,2018;Park et al.,2019;Wang et al.,2019)。与转录组SSR相比,基因组SSR的多态性更高,且在基因组中分布广泛,从而获得更好的图谱覆盖率(Wang et al.,2011)。本研究对卵形鲳鲹基因组调研数据进行分析,共检测出190121个SSR序列,并从测试的50个SSR位点中成功鉴定出29个多态性SSR位点(58.00%)。可见,利用高通量测序技术可获得数量庞大且类型丰富的卵形鲳鲹SSR序列,有助于开展其种群遗传学、遗传作图及数量性状基因座位(QTL)等相关研究,进而为实现卵形鲳鲹分子辅助育种提供技术支撑。 3. 4 卵形鲳鲹群体的遗传多样性 开展SSR分子标记研究可为揭示鱼类的遗传变异和种群结构提供重要信息,但群体遗传变异和种群结构同时受迁移、选择、遗传漂移及地理隔绝等因素的影响。种群遗传多样性是生物生存和发展的一个重要因素(Diz and Presa,2009),遗传多样性丧失会降低种群应对环境变化的能力。在种群遗传多样性研究中,Ne、He和PIC是3个常用的遗传多样性评价参数。在本研究中,卵形鲳鲹群体的Ne为1.3998~3.9123(平均为2.6690),He为0.2856~0.7444(平均为0.5965),PIC为0.2647~0.6968(平均为0.5195),与赵永贞等(2014)对南海区4个卵形鲳鲹群体的研究结果相似,说明卵形鲳鲹具有较丰富的遗传多样性,与该物种目前所处的现状基本吻合。由于卵形鲳鲹属于群游动物,雌雄鱼单独交配成功率极低,且性别难以通过常规方法准确判断,因此难以开展大规模的家系选育,致使一些养殖场直接将野生群体驯化后进行繁育,从而促使卵形鲳鲹种群保持了较高的遗传多样性。但本研究的Hardy-Weinberg平衡性检测结果显示,29个SSR位点中仅TOSR008位点处于Hardy-Weinberg平衡状态,TOSR049位点显著偏离Hardy-Weinberg平衡状态,其余27个SSR位点则极显著偏离Hardy-Weinberg平衡状态。当群体规模较大时,基因频率主要受迁移、选择及同型交配等因素的影响。本研究选取的卵形鲳鲹群体近年来未进行迁移或混杂,造成SSR位点Hardy-Weinberg平衡性丢失的主要原因可能是同型交配或群体内选择,提示稀有等位基因或将面临较高的丢失风险,进而导致种群遗传多样性降低和物种衰退。因此,要保持卵形鲳鲹种群遗传多样性,防止等位基因进一步丢失,必须做好以下措施:(1)不应通过无序养殖和育种计划开展卵形鲳鲹的繁育及育种研究;(2)保持较大的有效种群数量,以提高大量成鱼对繁殖的贡献;(3)采用适当监控手段,如遗传分子标记等监控遗传变异,尤其是稀有等位基因的改变;(4)借鉴现代遗传育种技术开展种群选育工作,保护稀有等位基因,防止种群近交而衰退。 4 结论 卵形鲳鲹基因组为简单基因组,利用基因组调研数据可实现SSR分子标记大规模开发,且新开发的SSR分子标记可用于卵形鲳鲹群体遗传多样性分析。 参考文献: 陈海玲,路雪林,叶泉清,唐绍清. 2019. 基于SSR标记探讨三种金花茶植物的遗传多样性和遗传结构[J]. 广西植物,39(3):318-327. [Chen H L,Lu X L,Ye Q Q,Tang S Q. 2019. Genetic diversity and structure of three yellow Camellia species based on SSR markers[J]. Guihaia,39(3):318-327.] 陈伟洲,许鼎盛,王德强,邓用谋,佘忠明,丘广艳,李远友. 2007. 卵形鲳鲹人工繁殖及育苗技术研究[J]. 台湾海峡,26(3):435-442. [Chen W Z,Xu D S,Wang D Q,Deng Y M,She Z M,Qiu G Y,Li Y Y. 2007. Study on the spawning and hatching technique for Trachinotus ovatus[J]. Journal of Oceanography in Taiwan Strait,26(3):435-442.] 陈秀荔,肖群平,陈晓汉,彭敏,李咏梅. 2010. 卵形鲳鲹微卫星分子标记的筛选[J]. 武汉大学学报(理学版),56(5):564-569. [Chen X L,Xiao Q P,Chen X H,Peng M,Li Y M. 2010. Screening of microsatellite molecular marker in Trachinotus ovatus[J]. Journal of Wuhan University(Na-tural Science Edition),56(5):564-569.] 侯树鉴,黎江,胡舒,何肇强,廖永岩,朱鹏,陆专灵,韦友传. 2018. 卵形鲳鲹组织蛋白酶L基因的克隆及其表达分析[J]. 南方农业学报,49(6):1215-1222. [Hou S J,Li J,Hu S,He Z Q,Liao Y Y,Zhu P,Lu Z L,Wei Y C. 2018. Cloning and expression analysis of cathepsin L gene in Trachinotus ovatus[J]. Journal of Southern Agriculture,49(6):1215-1222.] 胡海滨,解绶启,钱雪桥,贠彪,庄界成. 2019. 饲料中添加玉米蛋白粉或鸡肉粉替代部分鱼粉对卵形鲳鲹生长性能的影响[J]. 动物营养学报,31(6):2752-2764. [Hu H B,Xie S Q,Qian X Q,Yun B,Zhuang J C. 2019. Effects of dietary corn gluten meal or poultry meal partially repla-cing fish meal on growth performance of golden pompano (Trachinotus ovatus)[J]. Chinese Journal of Animal Nutrition,31(6):2752-2764.] 黄小林,张殿昌,林黑着,黄忠,虞为,杨育凯,李涛. 2018. 池塘养殖卵形鲳鲹早期形态性状与体质量的灰色关联分析[J]. 南方农业学报,49(5):1016-1022. [Huang X L,Zhang D C,Lin H Z,Huang Z,Yu W,Yang Y K,Li T. 2018. Grey relational analysis between early morphological traits and body weight of Trachinotus ovatus bred in pond[J]. Journal of Southern Agriculture,49(5):1016-1022.] 孔啸兰,李敏,陈作志,龚玉艳,张俊,张鹏. 2019. 基于RAD-seq技术的长体圆鲹二、三核苷酸重复微卫星标记开发与评价[J]. 南方水产科学,15(3):97-103. [Kong X L,Li M,Chen Z Z,Gong Y Y,Zhang J,Zhang P. 2019. Deve-lopment and evaluation of di-/tri-nucleotide-repeated microsatellites by RAD-seq in Decapterus macrosoma[J]. South China Fisheries Science,15(3):97-103.] 凌士鹏,孙萍,林贤锐,沈建生. 2018. 基于SSR标记的桃种质资源遗传多样性研究[J]. 江西农业学报,30(11):14-18. [Ling S P,Sun P,Lin X R,Shen J S. 2018. A study on genetic diversity of peach germplasm resources based on SSR markers[J]. Acta Agriculturae Jiangxi,30(11):14-18.] 彭敏,陳晓汉,陈秀荔,蒋伟明,杨春玲,李咏梅. 2011. 卵形鲳鲹养殖群体与野生群体遗传多样性的AFLP分析[J]. 西南农业学报,24(5):1987-1991. [Peng M,Chen X H,Chen X L,Jiang W M,Yang C L,Li Y M. 2011. Genetic diversity of wild and cultured Trachinotus ovatus populations by AFLP markers[J]. Southwest China Journal of Agricultural Sciences,24(5):1987-1991.] 区又君,李加儿. 2005. 卵形鲳鲹的早期胚胎发育[J]. 中国水产科学,12(6):786-788. [Ou Y J,Li J E. 2005. Early embryonic development in Trachinotus ovatus[J]. Journal of Fishery Sciences of China,12(6):786-788.] 孙立元. 2014. 卵形鲳鲹分子标记的筛选与应用[D]. 上海:上海海洋大学. [Sun L Y. 2014. The screening and application of molecular markers in Trachinotus ovatus[D]. Shanghai:Shanghai Ocena University.] 相瑜,任丽平,王日昕. 2013. 三疣梭子蟹微卫星标记的筛选及特征分析[J]. 浙江海洋学院学报(自然科学版),32(5):421-426. [Xiang Y,Ren L P,Wang R X. 2013. Isolation and characterization microsatellite markers in Portunus trituberculatus[J]. Journal of Zhejiang Ocean University(Natural Science),32(5):421-426.] 熊向英,黄国强,王志成,文雪. 2018. 广西卵形鲳鲹海豚链球菌基因分型、耐药谱型以及毒力基因检测[J]. 水产学报,42(4):586-595. [Xiong X Y,Huang G Q,Wang Z C,Wen X. 2018. Molecular typing,antibiogram type and detection of virulence genes of Stereptococcus iniae strains isolated from golden pompano(Tranchinotus ovatus) in Guangxi Province[J]. Journal of Fisheries of China,42(4):586-595.] 趙彦花,区又君,温久福,李加儿,周慧. 2019. 基于转录组测序技术的黄唇鱼SSR分子标记筛选[J]. 南方农业学报,50(9):2078-2087. [Zhao Y H,Ou Y J,Wen J F,Li J E,Zhou H. 2019. Development of SSR markers in Bahaba flavolabiata by transcriptome sequencing[J]. Journal of Southern Agriculture,50(9):2078-2087.] 赵永贞,陈秀荔,李咏梅,彭敏,杨春玲,韦嫔媛,彭金霞,陈晓汉. 2014. 南海区卵形鲳鲹遗传多样性的研究[J]. 西南农业学报,27(4):1786-1790. [Zhang Y Z,Chen X L,Li Y M,Peng M,Yang C L,Wei P Y,Peng J X,Chen X H. 2014. Genetic polymorphism of Trachinotus ovatus in Nanhai district[J]. Southwest China Journal of Agricultural Sciences,27(4):1786-1790.] Aird D,Ross M G,Chen W S,Danielsson M,Fennell T,Russ C,Jaffe D B,Nusbaum C,Gnirke A. 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries[J]. Genome Biology,12(2):R18. doi:10.1186/gb-2011-12-2-r18. Beier S,Thiel T,Münch T,Scholz U,Mascher M. 2017. MISA-web:A web server for microsatellite prediction[J]. Bioinformatics,33(16):2583-2585. Berthelot C,Brunet F,Chalopin D,Juanchich A,Bernard M,No?l B,Bento P,Silva C D,Labadie K,Alberti A,Aury J M,Louis A,Dehais P,Bardou P,Montfort J,Klopp C,Cabau C,Gaspin C,Thorgaard G H,Boussaha M,Quillet E,Guyomard R,Galiana D,Bobe J,Volff J N,Genêt C,Wincker P,Jaillon O,Crollius H R,Guiguen Y. 2014. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates[J]. Nature Communications,5:3657. doi:10.1038/ncomms4657. Bian C,Hu Y C,Ravi V,Kuznetsova I S,Shen X Y,Mu X D,Sun Y,You X X,Li J,Li X F,Qiu Y,Tay B H,Thevasagayam N M,Komissarov A S,Trifonov V,Kabilov M,Tupikin A,Luo J R,Liu Y,Song H M,Liu C,Wang X J,Gu D G,Yang Y X,Li W J,Polgar G,Fan G Y,Zeng P,Zhang H,Xiong Z J,Tang Z J,Peng C,Ruan Z Q,Yu H,Chen J M,Fan M J,Huang Y,Wang M,Zhao X M,Hu G J,Yang H M,Wang J,Wang J,Xu X,Song L S,Xu G C,Xu P,Xu J M,O'Brien S J,Orbán L,Venkatesh B,Shi Q. 2016. The Asian arowana(Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts[J]. Scientific Reports,6:24501. doi:10.1038/srep24501. Braasch I,Gehrke A R,Smith J J,Kawasaki K,Manousaki T,Pasquier J,Amores A,Desvignes T,Batzel P,Catchen J,Berlin A M,Campbell M S,Barrell D,Martin K J,Mulley J F,Ravi V,Lee A P,Nakamura T,Chalopin D,Fan S H,Wcisel D,Ca?estro C,Sydes J,Beaudry F E G,Sun Y,Hertel J,Beam M J,Fasold M,Ishiyama M,Johnson J,Kehr S,Lara M,Letaw J H,Litman G W,Litman R T,Mikami M,Ota T,Saha N R,Williams L,Stadler P F,Wang H,Taylor J S,Fontenot Q,Ferrara A,Searle S M J,Aken B,Yandell M,Schneider I,Yoder J A,Volff J N,Meyer A,Amemiya C T,Venkatesh B,Holland P W H,Guiguen Y,Bobe J,Shubin N H, Palma F D,Alf?ldi J,Lindblad-Toh K,Postlethwait J H. 2016. The spotted gargenome illuminates vertebrate evolution and facilitates human-teleost comparisons[J]. Nature Genetics,48(4):427-437. Brawand D,Wagner C E,Li Y I,Malinsky M,Keller I,Fan S H,Simakov O,Ng A Y,Lim Z W,Bezault E,Turner-Maier J,Johnson J,Alcazar R,Noh H J,Russell P,Aken B,Alf?ldi J,Amemiya C,Azzouzi N,Baroiller J F,Barloy-Hubler F,Berlin A,Bloomquist R,Carleton K L,Conte M A,D'Cotta H,Eshel O,Gaffney L,Galibert F,Gante H F,Gnerre S,Greuter L,Guyon R,Haddad N S,Haerty W,Harris R M,Hofmann H A,Hourlier T,Hulata G,Jaffe D B,Lara M,Lee A P,MacCallum I,Mwaiko S,Nikaido M,Nishihara H,Ozouf-Costaz C,Penman D J,Przybylski D,Rakotomanga M,Renn S C P,Ribeiro F J,Ron M,Salzburger W,Sanchez-Pulido L,Santos M E,Searle S,Sharpe T,Swofford R,Tan F J,Williams L,Young S,Yin S Y,Okada N,Kocher T D,Miska E A,Lander E S,Venkatesh B,Fernald R D,Meyer A,Ponting C P,Streelman J T,Lindblad-Toh K,Seehausen O,Di Palma F. 2014. The genomic substrate for adaptive radiation in African cichlid fish[J]. Nature,513(7518):375-381. Chen S L,Zhang G J,Shao C W,Huang Q F,Liu G,Zhang P,Song W T,An N,Chalopin D,Volff J N,Hong Y H,Li Q Y,Sha Z X,Zhou H L,Xie M S,Yu Q L,Liu Y,Xiang H,Wang N,Wu K,Yang C G,Zhou Q,Liao X L,Yang L F,Hu Q M,Zhang J L,Meng L,Jin L J,Tian Y S,Lian J M,Yang J F,Miao G D,Liu S S,Liang Z,Yan F,Li Y Z,Sun B,Zhang H,Zhang J,Zhu Y,Du M,Zhao Y W,Schartl M,Tang Q S,Wang J. 2014. Whole-genomesequence of a flatfish provides insights into ZW sexchromosome evolution and adaptation to a benthiclifestyle[J]. Nature Genetics,46(3):253-260. Das R,Arora V,Jaiswal S,Iquebal M A,Angadi U B,Fatma S,Singh R,Shil S,Rai A,Kumar D. 2018. PolyMorph-Predict:A universal web-tool for rapid polymorphic mi-crosatellite marker discovery for whole genome and transcriptome data[J]. Frontiers in Plant Science,9:1966. doi:10.3389/fpls.2018.01966. Diz A P,Presa P. 2009. The genetic diversity pattern of Mytilus galloprovincialis in Galician Rías(NW Iberian estua-ries)[J]. Aquaculture,287(3-4):278-285. Eierman L E,Hare M P. 2014. Transcriptomic analysis of candidate osmoregulatory genes in the eastern oyster Crassos-trea virginica[J]. BMC Genomics,15(1):503. doi:10. 1186/1471-2164-15-503. Fan G Y,Judy C,Ma K L,Yang B R,Zhang H,Yang X W,Shi C C,Law H C H,Ren Z T,Xu Q W,Liu Q,Wang J H, Chen W B,Shao L B,Gon?alves D,Ramos A,Cardoso S D,Guo M,Cai J,Xu X,Wang J,Yang H M,Liu X,Wang Y T. 2018. Chromosome-level reference genome of the Siamese fighting fish Betta splendens,a model species for the study of aggression[J]. Gigascience,7(11):giy087. doi:10.1093/gigascience/giy087. Figueras A,Robledo D,Corvelo A,Hermida M,Pereiro P,Rubiolo J A,Gómez-Garrido J,Carreté L,Bello X,Gut M,Gut I G,Marcet-Houben M,Forn-Cuní G,Galán B,García J L,Abal-Fabeiro J L,Pardo B G,Taboada X,Fernández C,Vlasova A,Hermoso-Pulido A,Guigó R,?lvarez-Dios J A,Gómez-Tato A,Vi?as A,Maside X,Gabaldón T,Novoa B,Bouza C,Alioto T,Martínez P. 2016. Whole genome sequencing of turbot(Scophthalmus maximus;Pleuronectiformes):A fish adapted to demersal life[J]. DNA Research,23(3):181-192. Gadgil R,Barthelemy J,Lewis T,Leffak M. 2017. Replication stalling and DNA microsatellite instability[J]. Biophysical Chemistry,225:38-48. doi:10.1016/j.bpc.2016. 11.007. Gao Y,Gao Q,Zhang H,Wang L L,Zhang F C,Yang C Y,Song L S. 2014. Draft sequencing and analysis of the genome of pufferfish Takifugu flavidus[J]. DNA Research,21(6):627-637. Jessy L,Murat C,Morin E,Tacon L F,Martin F. 2011. Survey and analysis of simple sequence repeats in the Laccaria bicolor genome,with development of microsatellite markers[J]. Current Genetics,57(2):75-88. Jiang Q,Li Q,Yu H,Kong L F. 2014. Genome-wide analysis of simple sequence repeats in marine animals-A comparative approach[J]. Marine Biotechnology,16(5):604-619. Jiang Q,Wang F,Tan H W,Li M Y,Xu Z S,Tan G F,Xiong A S. 2015. De novo transcriptome assembly,gene annotation,marker development,and miRNA potential target genes validation under abiotic stresses in Oenanthe java-nica[J]. Molecular Genetics and Genomics,290(2):671-683. Jiao Y,Jia H M,Li X W,Chai M L,Jia H J,Chen Z,Wang G Y,Chai C Y,van de Weg E,Gao Z S. 2012. Development of simple sequence repeat(SSR) markers from a genome survey of Chinese bayberry(Myrica rubra)[J]. BMC Genomics,13(1):201. doi:10.1186/1471-2164-13-201. K?ressaar T,Lepamets M,Kaplinski L,Raime K,Andreson R,Remm M. 2018. Primer3_masker:Integrating masking of template sequence with primer design software[J]. Bioinformatics,34(11):1937-1938. Lien S,Koop B F,Sandve S R,Miller J R,Kent M P,Nome T,Hvidsten T R,Leong J S,Minkley D R,Zimin A,Grammes F,Grove H,Gjuvsland A,Walenz B,Hermansen R A,von Schalburg K,Rondeau E B,Genova A D,Samy J K,Olav Vik J,Vigeland M D,Caler L,Grimholt U,Jentoft S,V?ge D I,de Jong P,Moen T,Baranski M,Palti Y,Smith D R,Yorke J A,Nederbragt A J,Tooming-Klunderud A,Jakobsen K S,Jiang X T,Fan D D,Hu Y,Liberles D A,Vidal R,Iturra P,Jones S J M,Jonassen I,Maass A,Omholt S W,Davidson W S. 2016. The Atlanticsalmon genome provides insights into rediploidization[J]. Nature,533(7602):200-205. Lin Q,Fan S H,Zhang Y H,Xu M,Zhang H X,Yang Y L,Lee A P,Woltering J M,Ravi V,Gunter H M,Luo W,Gao Z X,Lim Z W,Qin G,Schneider R F,Wang X,Xiong P W,Li G,Wang K,Min J M,Zhang C,Qiu Y,Bai J,He W M,Bian C,Zhang X H,Shan D,Qu H Y,Sun Y,Gao Q,Huang L M,Shi Q,Meyer A,Venkatesh B. 2016. The seahorse genome andthe evolution of its specialized morphology[J]. Nature,540(7633):395-399. Liu B,Guo H Y,Zhu K C,Guo L,Liu B S,Zhang N,Yang J W,Jiang S G,Zhang D C. 2019. Growth,physiological,and molecular responses of golden pompano Trachinotus ovatus(Linnaeus,1758) reared at different salinities[J]. Fish Physiology and Biochemistry,45(6):1879-1893. Luo R B,Liu B H,Xie Y L,Li Z Y,Huang W H,Yuan J Y,He G Z,Chen Y X,Pan Q,Liu Y J,Tang J B,Wu G X,Zhang H,Shi Y J,Liu Y,Yu C,Wang B,Lu Y,Han C L,Cheung D W,Yiu S M,Peng S L,Zhu X Q,Liu G M,Liao X K,Li Y R,Yang H M,Wang J,Lam T W,Wang J. 2012. SOAPdenovo2:An empirically improved memory-efficient short-read de novo assembler[J]. GigaScience,1(1):18. doi:10.1186/2047-217X-1-18. Nagy S,Poczai P,Cernák I,Gorji A M,Heged?s G,Taller J. 2012. PICcalc:An online program to calculate polymorphic information content for molecular genetic studies[J]. Biochemical Genetics,50(9-10):670-672. Pan H L,Yu H,Ravi V,Li C,Lee A P,Lian M M,Tay B H,Brenner S,Wang J,Yang H M,Zhang G J,Venkatesh B. 2016. The genome of the largestbony fish,ocean sunfish (Mola mola),provides insightsinto its fast growth rate[J]. GigaScience,5(1):36. doi:10.1186/s13742-016-0144-3. Park S,Son S,Shin M,Fujii N,Hoshino T,Park S. 2019. Transcriptome-wide mining,characterization,and development of microsatellite markers in Lychnis kiusiana (Caryophyllaceae)[J]. BMC Plant Biology,19(1):14. doi:10. 1186/s12870-018-1621-x. Quail M A,Smith M,Coupland P,Otto T D,Harris S R,Connor T R,Bertoni A,Swerdlow H P,Gu Y. 2012. A tale of three next generation sequencing platforms:Comparison of ion torrent,pacific biosciences and illumina MiSeq sequencers[J]. BMC Genomics,13:341. doi:10.1186/1471-2164-13-341. Shi L L,Yi S K,Li Y H. 2018. Genome survey sequencing of red swamp crayfish Procambarus clarkii[J]. Molecular Biology Reports,45(5):799-806. Sonah H,Deshmukh R K,Sharma A,Singh V P,Gupta D K,Gacche R N,Rana J C,Singh N K,Sharma T R. 2011. Genome-wide distribution and organization of microsatellites in plants:An insight into marker development in Brachypodium[J]. PLoS One, 6(6):e21298. Song H,Zhang Y X,Yang M J,Sun J C,Zhang T,Wang H Y. 2018. Genome survey on invasive veined rapa whelk (Rapana venosa) and development of microsatellite loci on large scale[J]. Journal of Genetics,97(4):e79-e86. Srivastava S,Kushwaha B,Prakash J,Kumar R,Nagpure N S,Agarwal S,Pandey M,Das P,Joshi C G,Jena J K. 2016. Development and characterization of genic SSR markers from low depth genome sequence of Clarias batrachus (magur)[J]. Journal of Genetics,95(3):603-609. Sun B M,Lei Y,Cao Z J,Zhou Y C,Sun Y,Wu Y,Wang S F,Guo W L,Liu C S. 2019. TroCCL4,a CC chemokine of Trachinotus ovatus,is involved in the antimicrobial immune response[J]. Fish & Shellfish Immunology,86:525-535. Sun L N,Gao T,Wang F L,Qin Z L,Yan L X,Tao W J,Li M H,Jin C B,Kocher D T,Wang D S. 2020. Chromosome-level genome assembly of a cyprinid fish Onychostoma macrolepis by integration of Nanopore sequencing,Bionano and Hi-C technology[J]. Molecular Ecology Resources. doi:10.1111/1755-0998.13190. Wang J,Ai Q H,Mai K S,Xu H G,Zuo R T. 2014. Dietary chromium polynicotinate enhanced growth performance,feed utilization,and resistance to Cryptocaryon irritans in juvenile large yellow croaker(Larmichthys crocea)[J]. Aquaculture,432:321-326. Wang L,Yu H,Li Q. 2019. Development of microsatellite markers and analysis of genetic diversity of Barbatia virescens in the southern coasts of China[J]. Genes Geno-mics,41(4):407-416. Wang Y P,Lu Y,Zhang Y,Ning Z M,Li Y,Zhao Q,Lu H Y,Huang R,Xia X Q,Feng Q,Liang X F,Liu K Y,Zhang L,Lu T T,Huang T,Fan D L,Weng Q J,Zhu C R,Lu Y Q,Li W J,Wen Z R,Zhou C C,Tian Q L,Kang X J,Shi M J,Zhang W T,Jang S H,Du F K,He S,Liao L J,Li Y M,Gui B,He H H,Ning Z,Yang C,He L B,Luo L F,Yang R,Luo Q,Liu X C,Li S S,Huang W,Xiao L,Lin H R,Han B,Zhu Z Y. 2015. The draft genome of the grass carp(Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation[J]. Nature Genetics,47(6):625-631. Wang Y W,Samuels T D,Wu Y Q. 2011. Development of 1030 genomic SSR markers in switchgrass[J]. Theoretical and Applied Genetics,122(4):677-686. Wu C W,Zhang D,Kan M Y,Lv Z M,Zhu A Y,Su Y Q,Zhou D Z,Zhang J S,Zhang Z,Xu M Y,Jiang L H,Guo B Y,Wang T,Chi C F,Mao Y,Zhou J J,Yu X X,Wang H L,Weng X L,Jin J G,Ye J Y,He L,Liu Y. 2014. The draft genome of the large yellow croaker reveals well-developed innate immunity[J]. Nature Communication,5:5227. doi:10.1038/ncomms6227. Wu M,Guo L,Zhu K C,Guo H Y,Liu B S,Zhang N,Jiang S G,Zhang D C. 2019. Molecular characterization of toll-like receptor 14 from golden pompano Trachinotus ovatus (Linnaeus,1758) and its expression response to three types of pathogen-associated molecular patterns[J]. Comparative Biochemistry and Physiology. Part B:Biochemistry and Molecular Biology,232:1-10. doi:10.1016/j.cbpb.2019.02.010. Xiao Y S,Xiao Z Z,Ma D Y,Liu J,Li J. 2019. Genome sequence of the barred knifejaw Oplegnathus fasciatus (Temminck & Schlegel,1844):The first chromosome-level draft genome in the family Oplegnathidae[J]. Giga-Science,8(3):giz013. doi:10.1093/gigascience/giz013. Xie Z Z,Xiao L,Wang D D,Fang C,Liu Q Y,Li Z H,Liu X C,Zhang Y,Li S S,Lin H R. 2014. Transcriptome analysis of the Trachinotus ovatus:Identification of reproduction,growth and immune-related genes and microsatellite markers[J]. PLoS One,9(10):e109419. Xu P,Zhang X F,Wang X M,Li J T,Liu G M,Kuang Y Y,Xu J,Zheng X H,Ren L F,Wang G L,Zhang Y,Huo L H,Zhao Z X,Cao D C,Lu C Y,Li C,Zhou Y,Liu Z J,Fan Z H,Shan G L,Li X G,Wu S X,Song L P,Hou G Y,Jiang Y L,Jeney Z,Yu D,Wang L,Shao C J,Song L,Sun J,Ji P F,Wang J,Li Q,Xu L M,Sun F Y,Feng J X,Wang C H,Wang S L,Wang B S,Li Y,Zhu Y P,Xue W,Zhao L,Wang J T,Gu Y,Lv W H,Wu K J,Xiao J F,Wu J Y,Zhang Z,Yu J,Sun X W. 2014. Genome sequence and genetic diversity of the common carp,Cyprinus carpio[J]. Nature Genetics,46(11):1212-1219. Yang X F,Liu H P,Ma Z H,Zou Y,Zou M,Mao Y Z,Li X M,Wang H,Chen T S,Wang W M,Yang R B. 2019. Chromosome-level genome assembly of Triplophysa tibe-tana,a fish adapted to the harsh high-altitude environment of the Tibetan Plateau[J]. Molecular Ecology Resources,19(4):1027-1036. Yu Y,Zhang X J,Yuan J B,Li F H,Chen X H,Zhao Y Z,Huang L,Zheng H K,Xiang J H. 2015. Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific white shrimp Litopenaeus vannamei[J]. Scientific Reports,5:15612. doi:10.1038/srep15612. Zardoya R,Vollmer D M,Craddock C,Streelman J T,Karl S,Meyer A. 1996. Evolutionary conservation of microsatellite flanking regions and their use in resolving the phylogeny of Cichlid fishes(Pisces:Perciformes)[J]. Procee-dings. Biological Sciences,263(1376):1589-1598. Zhang G F,Fang X D,Guo X M,Li L,Luo R B,Xu F,Yang P C,Zhang L L,Wang X T,Qi H G,Xiong Z Q,Que H Y,Xie Y L,Holland P W H,Paps J,Zhu Y B,Wu F C,Chen Y X,Wang J F,Peng C F,Meng J,Yang L,Liu J,Wen B,Zhang N,Huang Z Y,Zhu Q H,Feng Y,Mount A,Hedgecock D,Xu Z,Liu Y J,Domazet-Lo?o T,Du Y S,Sun X Q,Zhang S D,Liu B H,Cheng P Z,Jiang X T,Li J,Fan D D,Wang W,Fu W J,Wang T,Wang B,Zhang J B,Peng Z Y,Li Y X,Li N,Wang J P,Chen M S,He Y,Tan F J,Song X R,Zheng Q M,Huang R L,Yang H L,Du X D,Chen L,Yang M,Gaffney P M,Wang S,Luo L H,She Z C,Ming Y,Huang W,Zhang S,Huang B Y,Zhang Y,Qu T,Ni P X,Miao G Y,Wang J Y,Wang Q,Steinberg C E W,Wang H Y,Li N,Qian L M,Zhang G J,Li Y R,Yang H M,Liu X,Wang J,Yin Y,Wang J. 2012. The oyster genome reveals stress adaptation and complexity of shell formation[J]. Nature,490(7418):49-54. Zhang L S,Guo X M. 2010. Development and validation of single nucleotide polymorphism markers in the eastern oyster Crassostrea virginica Gmelin by mining ESTs and resequencing[J]. Aquaculture,302(1-2):124-129. Zhou W,Hu Y Y,Sui Z H,Fu F,Wang J G,Chang L P,Guo W H,Li B B. 2013. Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis(Rhodophyta) based on next-generation sequen-cing[J]. PLoS One,8(7):e69909 Zhou Z C,Dong Y,Sun H J,Yang A F,Chen Z,Gao S,Jiang J W,Guan X Y,Jiang B,Wang B. 2014. Transcriptome sequencing of sea cucumber(Apostichopus japonicus) and the identification of gene-associated markers[J]. Molecular Ecology Resources,14(1):127-138. (責任编辑 兰宗宝)
