Cohesin结构及功能研究进展
2020-03-04张雨方玉达
张雨,方玉达
综述
Cohesin结构及功能研究进展
张雨,方玉达
上海交通大学农业与生物学院单细胞生物学联合研究中心,上海 200240
Cohesin是一类在真核生物进化过程中保守的蛋白复合体,由4个重要亚基相互作用形成环状结构,在细胞分裂过程中参与维持染色体的有序排布。在动物中研究发现cohesin还可以作为分子间的连结器介导绝缘子/增强子–启动子间长距离交互,导致基因表达增强或者抑制,但在植物中关于cohesin在调控基因表达和维持染色体构象方面的研究却相对滞后。本文介绍了cohesin的结构特点和主要组成亚基,对调控cohesin在染色质上动态变化的相关因子进行了总结,并结合近年来植物中cohesin的功能研究和动物中cohesin在三维基因组及转录调控中的重要作用,展望了植物中cohesin在转录调控中的潜在功能。
SMC;cohesin;细胞周期;三维基因组;转录调控
细胞核是细胞遗传与代谢的调控中心,遗传物质DNA有序且密集地分布其中。人类细胞核基因组的物理长度约102 cm,即使基因组较小的拟南芥(),其细胞核基因组也有3.8 cm,而这些DNA通过折叠浓缩后储存在仅有几微米的细胞核内。染色体经过折叠形成有序的三维结构,这一过程很大程度上依赖染色体结构维持蛋白(structural maintenance of chromosomes, SMC)的调控[1~3]。SMC复合体从真菌、植物到人类都非常保守,包括cohesin、condensin和SMC5/6三大类。Condensin的功能主要与染色体内部的凝聚相关,当人类细胞敲除condensin后,导致染色体不能凝聚,不能形成正常姐妹染色单体,在分裂后期姐妹染色单体也不能正常分离。SMC5/6功能主要与DNA的损伤修复相关。关于cohesin的功能,早期人们研究发现其在酵母细胞有丝分裂和减数分裂过程中都发挥重要功能。在分裂过程中,cohesin可以维持染色体的正常形态,保证姐妹染色单体及同源染色体在细胞的不同分裂时期正确分布[4~6]。而在间期,cohesin维持染色质形成不同的空间结构,调控基因表达,还与DNA复制、DNA损伤修复相关[5,7~9]。最近的研究还发现cohesin介导的染色质环挤出动态过程对RAG (recombination-activating gene)扫描损伤位点起到促进作用,并在数量众多的V(D)J (variable- diversity-joining)重排和交错转化重组(cross switch recombination, CSR)过程中发挥重要作用[10]。
Cohesin在维持染色质构象及调控转录方面的研究也成为三维基因组学和表观遗传学研究的热点。本文在介绍cohesin结构特点、主要组成亚基及其功能的基础上,对cohesin在染色质上从招募到稳定结合,再到解离过程中调控其动态变化的作用因子进行了总结,并结合近年来在哺乳动物及酵母中的相关研究,讨论了cohesin在植物与动物中功能的保守程度,对植物cohesin在基因表达调控中的潜在功能进行了展望。
1 Cohesin结构
1.1 SMC蛋白复合体的结构特点
真核生物的SMC复合体都是在两个SMC蛋白组成的异源二聚体基础上形成的[11~13]。每个SMC 蛋白由1000~1500个氨基酸组成,中间是球状的铰链(hinge)结构域,铰链结构域两侧延伸形成卷曲螺旋(coiled-coils)结构域[14],卷曲螺旋结构域终端分别为Walker A和Walker B结构域,即SMC蛋白N端的Walker A和C端的Walker B结构域。Walker A含有核苷酸结合结构域(nucleotide-binding domain, NBD),Walker B含有与典型ATP酶同源的ATP结合结构域(ATP-binding cassette, ABC)。单个SMC蛋白以hinge结构为中心,两侧的coiled-coils结构域反向平行相互作用在一起,这使得SMC的N端Walker A和C端Walker B结构域相互靠近在一起,形成有功能ATP酶(ATPase)结构域(图1,A和B)[15~17]。SMC的ATPase位点对于整个SMC蛋白复合体在DNA上的结合和解离至关重要[18]。
1.2 Cohesin主要亚基
Cohesin是SMC复合体中的一类,由SMC1、SMC3和SCC3 (在动物中是Rad21)以及kleisin亚基组成的环状套索结构[14,19]。其中,SMC1与SMC3是典型的SMC 蛋白,SMC1和SMC3的hinge结构域相互作用形成V形的异源二聚体,底部由kleisin亚基将两个SMC蛋白的ATP酶结构域连接形成闭合环状V形复合体(图1B)[20,21]。Kleisin亚基与SCC3亚基相互作用,进而招募SCC3形成完整的cohesin蛋白复合体[22]。酵母中发现SCC3 (SA2)的C端与kleisin相结合。蛋白结构分析发现,SCC3内部凹面可以与kleisin (Rad21/Scc1-M)亚基中间很大一段相互作用[23]。目前在植物中还没有关于cohesin各亚基间相互作用的报道。
拟南芥cohesin的AtSMC1和AtSMC3亚基与酵母和哺乳动物SMC家族相比蛋白同源性很高。拟南芥和单突纯合突变体种子在发育过程中胚和胚乳都存在严重缺陷[24~26],胚胎发育早期就死亡,由此可见cohesin在胚胎发育早期即已经发挥着重要作用。拟南芥中AtSCC3不存在基因冗余现象,与酵母中SCC3蛋白有40%的同源性。动物中SCC3亚基含有HEAT-repeat (Huntingtin, elongation factor 3, protein phosphatase 2A)结构域[27],而拟南芥中AtSCC3却不含有HEAT-repeat结构域。拟南芥纯合突变体在胚胎发育早期缺陷致死,Chelysheva等[28]发现Ws(Wassileskija)拟南芥弱的突变体植株与野生型相比表现出矮小、晚花、育性降低、有丝分裂及减数分裂均发生异常。
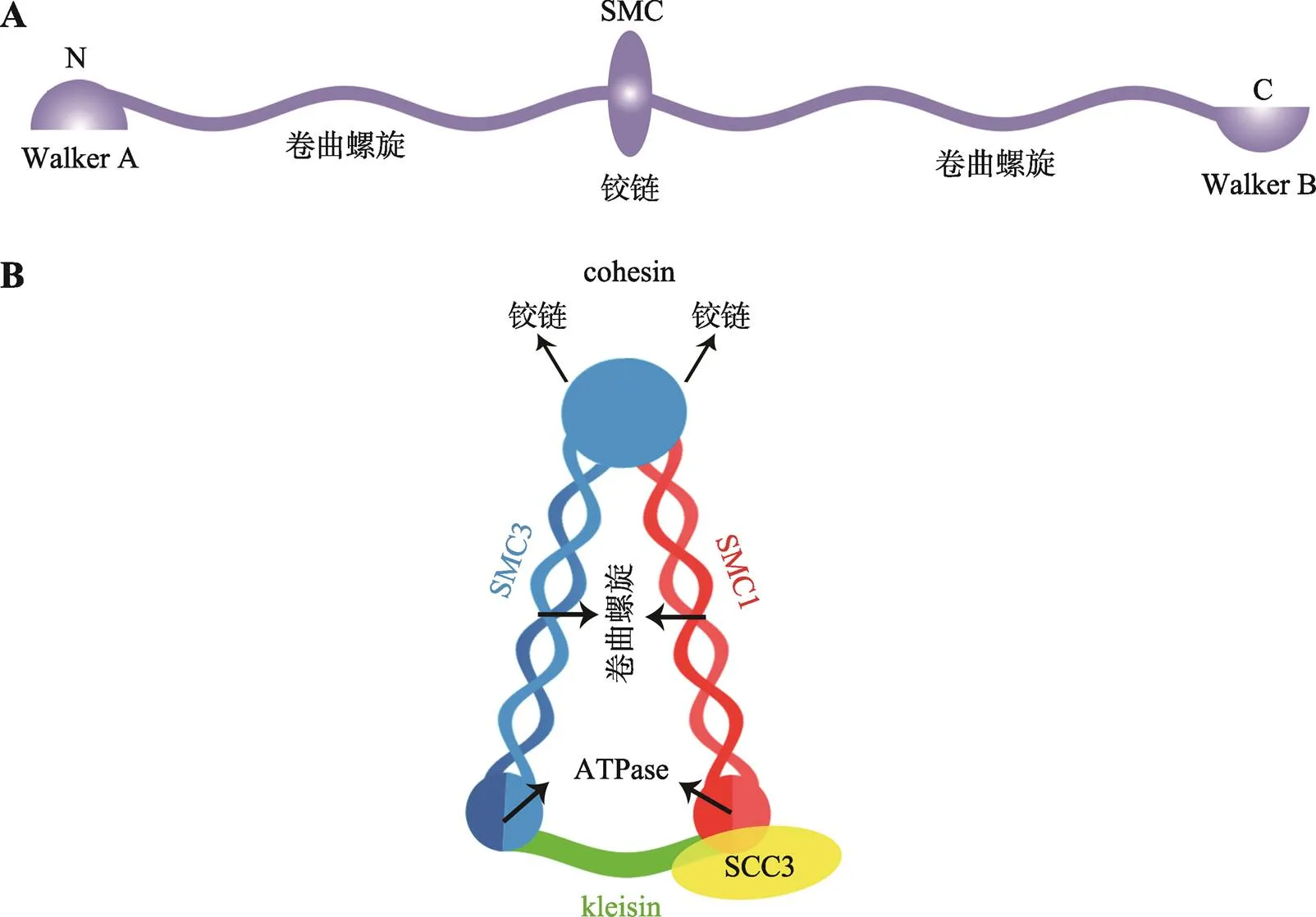
图1 SMC类蛋白及cohesin的结构示意图
A:SMC类蛋白保守结构域示意图;B:Cohesin结构示意图。根据参考文献[18]绘制。
Kleisin亚基在拟南芥、水稻(L.)和玉米(L.)中均有研究。在拟南芥和水稻中kleisin 亚基的4个同源蛋白相对保守,玉米中仅有AFD1一个同源蛋白,酿酒酵母()及脊椎动物中kleisin 亚基有RAD21和REC8两个同源蛋白(表1)[29]。拟南芥中Kleisin亚基的4个同源蛋白分别为:AtSYN1、AtSYN2、AtSYN3和AtSYN4[30~33]。突变体雌雄配子不育,但其营养生长等生长发育过程均正常[34,35],表明AtSYN1蛋白主要在减数分裂形成配子过程中发挥重要功能[34,36]。最近研究发现,拟南芥第一次减数分裂过程中cohesin维持在着丝粒区域依赖两个蛋白磷酸化酶对AtSYN1的去磷酸化作用[37,38]。AtSYN3主要定位在核仁,与rDNA结构维持及rRNA转录和加工成熟有关[33]。AtSYN2和AtSYN4在有丝分裂中发挥重要功能。AtSYN2与种子萌发过程中DNA损伤后修复相关[35],而AtSYN4与苗期体细胞DNA损伤修复相关[39]。酵母双杂交实验证明,AtSYN4 也可以与磷酸化酶PP2A B'α、PP2AB'β和PP2AB'ζ相互作用,磷酸化酶与AtSYN4的相互作用可能与有丝分裂过程中cohesin在着丝粒上的维持相关[37,38]。
水稻中RAD21-4/OsREC8是酵母中REC8的同源蛋白,OsREC8在减数分裂过程中保证同源染色体正确的配对及联会(表1)。突变体及RNAi植株在营养生长阶段与野生型相比无明显差异,但育性均显著降低[40,41]。在水稻各组织中均有表达,但在花和芽中表达明显高于叶和根。在花中高表达。RNAi植株的花粉有丝分裂异常,染色体不能正常分离,且花粉活力严重降低,但花粉的减数分裂并无异常,表明OsRAD21-3在花粉减数分裂后的有丝分裂过程中发挥作用[42]。通过原位杂交发现多在细胞分裂旺盛的组织中高表达,且异位表达后水稻细胞生长迟缓、植株发育异常[43]。
玉米AFD1是REC8的同源蛋白,其功能与同源染色体配对、联会复合体的形成及RAD51在染色体上的分布有关。AFD1会影响染色体在细线期及偶线期的分布。突变体中染色体在偶线期不能呈“花束”形态(bouquet formation)分布,减数分裂发生异常[44]。
2 调节cohesin在染色质上动态变化的因子
Cohesin在细胞分裂过程中重要的功能是维持姐妹染色单体有序地分布。在显微镜下可以观察到,在细胞分裂前期到中期cohesin都结合在染色体臂以及着丝粒区域,维持两条姐妹染色单体粘连在一起,在分裂中后期cohesin从染色体臂上解离下来,末期着丝粒上的cohesin也解离下来,姐妹染色单体得以正常分离。在转录过程中cohesin也随着RNA聚合酶及转录因子从转录起始位点向转录终止位点移动[45,46]。可见cohesin在染色质上的结合是动态变化的。Cohesin在染色质上的动态变化在拟南芥,酵母,线虫()和人类()中均有相关研究[4~6,47,48]。Cohesin的动态变化依赖很多蛋白,如:SCC2负责在DNA上招募cohesin,而cohesin在染色质上的维持依赖CTF7/ECO1 (chromosome transmission fidelity/establishment of cohesion 1)。另外,WAPL (wings apart-like protein)和PDS5 (precocious dissociation of sisters protein 5)因子与cohesin从DNA上解离有关(表1)。
2.1 Cohesin在染色质上的加载
在DNA复制开始前,cohesin的加载因子SCC2和SCC4先在染色质上结合,进而招募cohesin在染色质上结合[6,47,49,50]。在酿酒酵母中,SCC4可以稳定SCC2在染色质上的结合[49,51],两者作用在一起形成cohesin的加载因子,从而招募cohesin[6,52]。Cohesin与SCC2在DNA上的结合位点并不是随机的,两者染色质免疫共沉淀-高通量测序(ChIP-seq)的分析结果发现它们各自结合位点可能没有重叠[53,54],这可能由于cohesin最初是依赖SCC2与DNA的结合,但cohesin在染色质上的结合位置是动态变化的,cohesin会在其他因子的作用下移动,如间期cohesin随着转录过程中RNA聚合酶在染色质上移动。Cohesin多分布在转录相对活跃的地方,且随转录过程在转录终止区域富集[55~57]。SCC2与cohesin结合的程度会影响cohesin在染色质上的移动。当SCC2突变后,cohesin在转录起始位点的结合能力也降低[58]。SCC2和SCC4影响cohesin在染色质上结合的具体机制还不完全清楚。酵母研究发现,SCC2和SCC4突变后,完整的cohesin环可以形成,但不能与DNA结合。和突变体的表型与SMC1和SMC3的ATPase结构域突变后的表型类似,即cohesin可以形成完整的环状复合体,但也不能结合DNA。据此推测,SCC2可以加强SMC3和SMC1的ATPase活性以及催化DNA形成容易被cohesin有效结合的拓扑异构结构,进而影响cohesin在染色质上的结合[14,21,59~61]。

表1 Cohesin亚基及相关调控因子
*根据序列比对得到的同源蛋白。根据参考文献[29]整理。
拟南芥中cohesin加载因子的同源蛋白为AtSCC2和AtSCC4。AtSCC2与动物中同源蛋白有20%的同源性,除了有动物中共有的HEAT-repeat结构域外,AtSCC2还有植物中特有的植物同源结构域(plant homeodomain, PHD)。PHD结构域与组蛋白表观修饰以及基因表达调控相关[62,63]。在植物中,SCC2也是非常重要的蛋白。拟南芥纯合突变体在种子形成过程中胚乳过度增生分裂、发育异常、胚胎早期致死[62]。拟南芥纯合突变体胚胎在心形胚形成阶段不能对称分裂,胚柄处过度增生[64]。拟南芥植株中可观察到减数分裂过程中染色体分离紊乱,同时结合在染色质上的AtSCC3蛋白也减少,并出现姐妹染色单体黏连,染色体桥及分裂后细胞中染色体数目异常的现象[62]。在双突变体背景下,生长素报告基因被限制在胚柄底部细胞中表达,而野生型中报告基因在胚柄顶部细胞中表达。这表明AtSCC2和AtSCC4的缺失会导致胚胎发育过程中胚柄细胞胚胎潜能的改变。植物和酵母中都发现,SCC4可以与SCC2的N端稳定地相互作用在一起,但植物AtSCC4与AtSCC2之间的相互作用不会影响AtSCC4的定位。拟南芥AtSCC2的突变并没有改变植物体细胞核中AtSCC4的定位[54]。此外,有丝分裂间期AtSCC4与kleisin亚基AtSYN4共定位[54],而AtSCC2的主要功能被认为在减数分裂过程中影响cohesin的定位[62],这表明在拟南芥中AtSCC4与AtSCC2功能存在特异性。最近研究发现,玉米中DEK15是SCC4的同源蛋白。在突变体中,姐妹染色单体形态异常,非整倍数细胞增多,且种子胚乳发育异常,胚胎早期死亡率增加。玉米DEK15对于染色体精确的分离非常重要,且可以协同染色质重塑因子促进cohesin在染色质上的结合[65]。
2.2 Cohesin在染色质上的维持
在有丝分裂S期前,cohesin在SCC2和SCC4的招募下与DNA结合。从S期到分裂中期,cohesin一直结合在染色体臂及着丝粒上,维持姐妹染色单体连接在一起,直至后期cohesin从染色体上解离下来。在这个过程中,cohesin复合体在染色质上的维持依赖几个关键蛋白:ECO1 (establishment of cohesion 1)又称为CTF7 (chromosome transmission fidelity 7),以及sororin因子。
酵母中CTF7/ECO1是乙酰转移酶,在S期可以对SMC3的head结构域的两个赖氨酸残基进行乙酰化修饰[66~69]。SMC3的ATPase位点K112和K113位被乙酰化后,ATPase结构域关闭,使kleisin亚基与SMC亚基结合紧密,进而使cohesin环状结构稳定[68~70]。SMC3的这两个赖氨酸残基位点在多种生物中都是非常保守的,在人体细胞中,ESCO1 (establishment of sister chromatid cohesion N-acetyltransferase 1)和ESCO2两个乙酰化酶同样可以乙酰化SMC3[69,70]。酵母CTF7缺失会造成染色质状态混乱,导致cohesin在染色体臂及着丝粒上分布异常,以及细胞周期异常[71,72]。酵母CTF7/ECO1与增值细胞核抗原(proliferating cell nuclear antigen, PCNA)和复制因子C (replication factor C, RFC)复合体直接相互作用,这表明在姐妹染色单体形成过程中,DNA的复制和cohesin作用下的姐妹染色单体粘连是同时进行的[73,74]。
在脊椎动物中,还存在另外一个对cohesin与染色质的稳定结合起到重要作用的sororin因子。由于一些解离因子的存在,仅仅乙酰化的SMC3不足以让cohesin在复制过程中稳定地结合在染色质上,还需要乙酰化结合蛋白sororin来维持整个复合体的稳定。Sororin含有FGF结合序列,可以结合在PDS5 (precocious dissociation of sisters 5)蛋白上,进而起到稳定cohesin-DNA的作用[75~77]。在裂殖酵母()中,PDS5可以加强SMC3的乙酰化[78]。PDS5在间期与sororin相互作用,有协助cohesin结合DNA,并有维持cohesin与DNA稳定结合的功能。在后期,PDS5与解离因子相互作用,促进cohesin从DNA上解离下来,可见PDS5与不同因子相互作用发挥的功能也不同[79~81]。
拟南芥可以互补酵母突变体表型[82~84],这表明cohesin在细胞分裂过程中的功能在拟南芥和酵母中是非常保守的。AtCTF7包含PIP-BOX (PCNA-interacting protein BOX)、一个C2H2锌指蛋白结构域和一个乙酰转移酶结构域[82]。与其他生物相同,AtCTF7功能也有剂量效应,atctf7杂合体雄配子异常,小孢子母细胞发育正常,植物营养生长无明显异常,但育性降低。完全缺失的突变体拟南芥表现出严重的生长缺陷表型:胚胎在发育到球形胚阶段就严重畸形,仅能获得少数纯合植株,表现出极矮小、不育的表型,同时cohesin在染色质上的结合明显减少[82,83]。过表达CTF7也会导致拟南芥胚珠在发育早期死亡[85]。
2.3 Cohesin从染色质上解离
WAPL是调控cohesin从染色质上的解离下来的关键因子。有丝分裂中后期,cohesin开始逐渐从 染色体臂上解离下来,仅保留在着丝粒区域。起始cohesin从染色体臂上解离下来的过程与SCC3亚基的磷酸化相关,这个磷酸化过程依赖于WAPL解离因子[86]。有丝分裂后期,SCC3与sororin被磷酸化,磷酸化后的sororin不再与PDS5相互作用,PDS5与解离因子WAPL相互作用,PDS5-WAPL复合体促进cohesin从染色体壁上解离下来。Cohesin从着丝粒上解离下来的过程依赖蛋白酶对kleisin亚基的水解,整个过程WAPL-PDS5-SCC3协同发挥作用[79,87,88]。
拟南芥中有5个同源基因,在不同器官中检测表达量,发现在种子成熟过程中其表达量明显下降。当植株被γ射线照射后,表达上升。敲除后,减数分裂只轻微受到影响,但是DNA的同源重组修复能力明显减弱[89]。拟南芥中有两个同源基因和[90],而仅有一个拷贝[82],分子及遗传学实验证明AtWAPL和AtCTF7二者功能拮抗[91]。和T-DNA插入突变体在植物生长发育以及育性方面都没有异常[90],纯合双突变体在营养生长阶段与野生型相比没有差异,但雌配子雄配子活性下降,植株育性降低。在减数分裂方面,双突变体的同源染色体配对异常,纺锤体形成异常,且cohesin在染色体臂上滞留,出现黏连在一起的姐妹染色单体,在后期不能正常分离[90]。WAPL在许多生物有丝分裂过程中发挥重要功能,减数分裂中的研究较少。对拟南芥AtWAPL的研究发现,其在植物减数分裂中同样发挥重要功能。拟南芥atctf7杂合子突变体植株育性降低,纯合突变体植株生长发育严重缺陷,并且不育[83]。Kuntal De等[91]在研究AtCTF7和AtWAPL功能时发现,将纯合突变体与atctf7突变体杂交,获得三突纯合突变体,其生长发育与野生型无明显差异,但育性比和atctf7低,可见AtWAPL蛋白缺失可以抵消突变体在有丝分裂过程中cohesin不能结合到染色体上的缺陷。同时表明作为调控cohesin动态变化的因子,AtWAPL和AtCTF7在功能上相互拮抗。
3 Cohesin功能
早期关于cohesin的研究大多集中在细胞分裂过程中,其中在有丝分裂和减数分裂过程中cohesin对于姐妹染色单体间有序的凝聚在一起发挥着重要功能。近期研究表明cohesin还可以在分子间起到连接的作用,在长距离范围内影响DNA的交互,进而调控转录。另外,cohesin在DNA损伤修复方面也发挥重要功能,Scc1亚基就是在酵母中筛选易发生DNA损伤突变体时发现的[92,93]。对非洲爪蟾()和鸡()的细胞进行持续γ射线照射会导致染色体的断裂,此过程伴随着cohesin在DNA上的结合增多,以及cohesin动态变化会更加活跃[94]。
3.1 Cohesin在细胞分裂中的功能
一个细胞在分裂成两个不同细胞的过程中需要很多蛋白协同发挥作用,并要经历几个重要时期以确保正常的细胞能继续完成整个细胞周期,阻止异常的细胞进行分裂。有丝分裂过程中,G1期需要完成细胞健康与否的分拣,正常的细胞进入S期,异常的细胞不再进行分裂。G2期确保细胞完成了正确的DNA复制过程,才能进入分裂期。在S期染色体经历了复制过程,产生两个一样的姐妹染色单体。从S期DNA开始复制起cohesin就将两个姐妹染色单体有序地黏连在一起,直到分裂后期才完全从染色体上解离下来。这个机制在所有真核生物中都是非常保守的[95~99]。
体细胞进行有丝分裂的过程中,G1期SCC2和SCC4招募cohesin与DNA结合,这个过程也依赖SMC蛋白ATP水解酶活性。SMC1和SMC3形成的hinge结构是DNA链进入cohesin环的“入口”[60]。Cohesin与DNA结合后,从间期到中期,在染色体上的维持依赖于ECO1/CTF7这个乙酰转移酶对SMC3亚基的乙酰化作用,以及sororin-PDS5蛋白的结合抑制了WAPL蛋白打开cohesin环的作用[77,100,101]。在S期,cohesin在DNA上的加载与DNA的复制过程协同进行[102]。在前期–中期转换的阶段,染色体臂上的cohesin开始解离下来,这个过程依赖一些有丝分裂激酶的作用。以哺乳动物为例,cohesin的SA (SCC3)亚基被Plk1磷酸化以及sororin蛋白被Cdk1和Aurora B磷酸化都与cohesin从染色体臂上的解离相关,其中WAPL也发挥重要作用[103,104]。但在有丝分裂后期姐妹染色单体分离之前,cohesin会一直结合在着丝粒上,此时SGO1以及PP2A会保护SA及sororin不被磷酸化,从而使cohesin维持在着丝粒上[103,105]。中后期纺锤体上的微管向细胞两极牵引,此时着丝粒上的cohesin产生的内聚力可以抵消掉部分纺锤体的牵引力。在中期赤道板上的姐妹染色单体有了分别向两极移动的重新定向,确保染色体可以正常移动到两极后,才进行后期着丝粒解凝聚。这时cohesin的kleisin亚基在蛋白水解酶作用下水解,致使cohesin从着丝粒上解离下来,姐妹染色单体向两极移动[12,106]。
在减数分裂过程中cohesin同样发挥着重要作用。在哺乳动物生殖细胞中,与体细胞相比cohesin的SMC1α亚基及SA1和SA2亚基绝大多数被SMCβ及STAG3/SA3代替,SMCβ及STAG3/SA3是减数分裂特异的cohesin亚基[107~109]。生殖细胞中的kleisin亚基为REC8和RAD21L,这也是哺乳动物中减数分裂特异的亚基(表2)。减数分裂过程中cohesin在DNA上的结合和维持过程同样是依赖SCC2和SCC4、PDS5以及sororin,且这些调控因子在减数分裂和有丝分裂中的功能保守[110~112]。在减数分裂过程中,kleisin亚基与cohesin在染色质上的时空分布相关。在哺乳动物减数分裂前期,REC8类cohesin在DNA复制前结合到染色质上,大量REC8类cohesin与DNA的结合会贯穿整个减数分裂过程,直到第二次减数分裂中期。而RAD21L类cohesin大多是在DNA复制完成之后与染色体结合,且在第一次减数分裂的粗线期后期就从染色体上解离下来[113~115]。减数分裂过程中cohesin从染色体上的解离过程同样依赖WAPL[116],其机制也与有丝分裂相同。在酿酒酵母中,减数分裂SMC亚基与有丝分裂亚基相同,都为PSM1和PSM2。Klesin亚基与有丝分裂不同,为减数分裂特异的REC8,有丝分裂中的PSC3亚基在减数分裂中为REC11。目前已知拟南芥cohesin亚基中只有SYN1是减数分裂特有的(表2)。
无论是有丝分裂还是减数分裂,cohesin对维持姐妹染色单体凝聚在一起发挥着重要功能,这种凝聚力从间期DNA复制开始一直持续到中后期姐妹染色单体分开。如果缺少了分子间的凝聚力,会导致基因组不稳定、非整倍体细胞增多、DNA修复力下降、染色体异位等异常[117~119]。

表2 Cohesin亚基在有丝分裂及减数分裂中的比较
3.2 Cohesin在维持染色质构像及基因表达调控中的功能
最早是在果蝇()中发现cohesin具有转录调控的功能。Cohesin的加载因子 Nipped-B(SCC2)发生突变后,基因的表达受到抑制,Nipped-B可以介导基因区域增强子–启动子的相互作用。如果cohesin不能结合到基因上,启动子不能与增强子互作,基因转录水平降低[120]。同样,当人缺失了cohesin 加载因子CdLS(SCC2)会造成科妮莉亚·德·兰格发育综合征(Cornelia de Lange syndrome),这是一种引起上肢发育畸形、智力缺陷的疾病,其致病原因是由于CdLS的缺失导致下游基因转录调控异常[121,122]。
CTCF(CCCTC-binding factor)是协同cohesin维持染色质三维结构及调控转录的关键因子。染色质在细胞核内相互作用形成拓扑异构相关结构域(topologically associating domain, TAD),TADs是与染色质三维结构功能相关的重要区域,TADs内部染色质交互密集,TADs之间染色质交互频率低[123]。有研究提出TAD的主要作用是限制启动子和增强子间的相互作用[124,125]。不同TAD之间被边界区域(boundary)隔开,边界区域富集CTCF和cohesin(图2)[126],且多富集转录相对活跃的管家基因[127~130]。边界区域基因表达相对活跃,与染色质结构相对松散,以及富集着一些与活跃染色质相关的组蛋白修饰标记(H3K4me3和H3K36me3)相关。
拟南芥中,染色体组织形态上没有明显的TAD。同时,拟南芥中也缺少动物中经典的CTCF绝缘蛋白,这与拟南芥中缺少典型的TAD存在相关性。仅有很少的可信证据表明在拟南芥中存在类似于绝缘元件的DNA (insulator-like DNA)序列。然而,在对拟南芥进行高分辨率的全基因组染色质构象捕获(Hi-C)后发现超过1000个类似TAD(TAD-like)的区域[131]。拟南芥中这些区域和动物中的TAD有着相似的特性:在TAD内部,染色质交互密集;在TAD之间,染色质的交互受到限制。同样它们在染色体松散的地方以及基因表达活跃的地方富集[131,132]。但植物中还没有关于cohesin与三维基因组的相关报道。
研究发现cohesin加载因子、SMC和kleisin不同亚基在全基因组上的结合位点与CTCF有显著重叠,并且cohesin与CTCF共同对这些基因转录起到抑制的作用。尽管CTCF和cohesin在很多环状DNA结构处共同结合,但是它们在维持染色质构象上的功能不尽相同。CTCF与转录抑制相关,而cohesin除了与CTCF共同作用的位点外,还在很多基因位点与转录激活相关[127]。根据染色质包装紧密程度可以将cohesin的结合位点分类:在包装紧密的DNA结合位点,通常是cohesin与CTCF共同结合的位点;染色质包装松散的DNA结合位点,通常没有CTCF结合,这些区域大多为启动子或增强子区[133~139]。Cohesin还和一些其它的调控因子如调控蛋白复合体(mediator complex)相互作用发挥转录激活作用[133~137]。可见cohesin作为分子间桥梁,通过影响长距离范围内DNA上调控元件如:绝缘子/增强子-启动子(insulator/enhancer-promoter)之间的染色质交互来调控转录。Cohesin将增强子-启动子拉近在一起时,可以起到转录激活作用,此时cohesin多与转录因子或mediator共同起作用;当cohesin将绝缘子–启动子拉近在一起时,可以起到转录抑制功能(图3)[18,140],此时cohesin多与CTCF共同发挥作用。
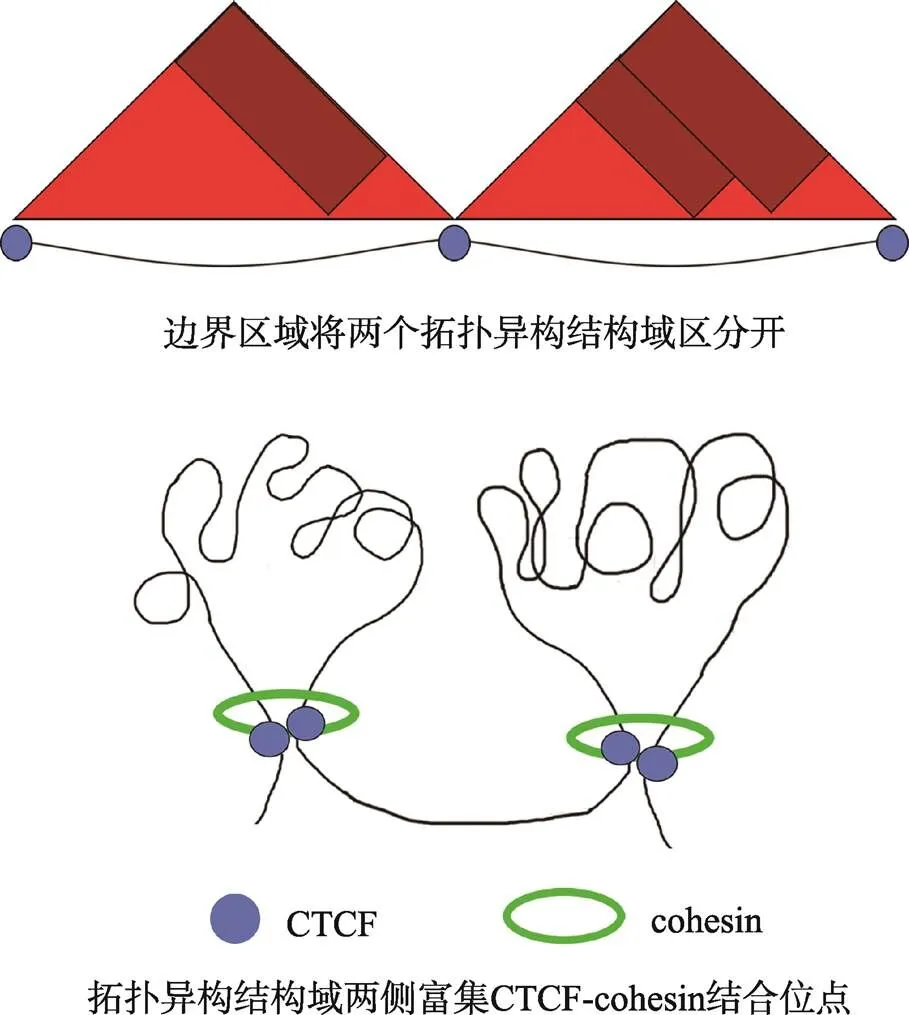
图2 拓扑异构结构域的二维结构示意图
根据参考文献[126]绘制。
在复制过程中关于cohesin在染色质上结合机制的研究相对较多,最近Murayama等[141]人用详尽巧妙的体外实验探索了cohesin在DNA复制过程中动态结合DNA的机制:cohesin在加载因子及ATP的存在下可以结合在双链DNA上,当cohesin先结合一分子DNA双链上后,仅能再结合一分子单链DNA,这个过程也是依赖加载因子和ATP的作用。Cohesin结合了单链DNA后,体外再给予单链DNA、DNA聚合酶和dNTP等条件会稳定整个DNA-cohesin结构。这个过程成功模拟了复制叉形成及推进过程中cohesin动态结合DNA的过程[142,143]。而在转录过程中,是否会形成DNA-cohesin-RNA复合体,目前还不是很清楚,还没有直接的证据表明cohesin能在转录过程中可以沿着DNA移动。最近Peters和他的同事利用遗传学结合基因组学方法研究发现在转录过程中cohesin在转录复合体的作用下可以随转录进程移动。在突变体中,cohesin在转录起始位点结合增加30%。在双突变体中,cohesin在转录终止位点下游滞留。基因的转录程度不同,cohesin的分布也随之不均一,转录活跃区域的基因上的cohesin会被推到转录不活跃区域[144]。这些结果暗示转录过程中cohesin可能在PolII-TFs的推动下从转录起始位点向转录终止位点移动。全基因组范围内PolII-ChIP-seq,RAD21-ChIP-seq也表明cohesin不仅可以通过控制长距离范围内DNA上转录元件的结合调控转录,也可以直接与转录复合体等相关因子在转录起始位点发挥作用[144]。

图3 Cohesin在基因表达调控中的功能
根据参考文献[18]绘制。
4 结语与展望
对cohesin复合体研究至今已有30多年的时间,除了其主要组成亚基SMC1、SMC3、kleisin和SCC3以外,许多与其功能相关的因子也被发现,包括cohesin加载因子SCC2、SCC4和解离因子WAPL等。这些研究使得cohesin在细胞分裂过程中的功能及机制逐渐清晰。在细胞周期中,cohesin对于维持染色体的正常形态和有序排布是至关重要的。在拟南芥和水稻中,cohesin在有丝分裂及减数分裂中的功能与酵母,哺乳动物高度保守。此外,cohesin在植物中对于胚胎发育、育性及DNA损伤修复也发挥重要作用。
在cohesin的作用下,两条姐妹染色单体靠凝聚力联系在一起,这对于染色体在整个细胞周期中正确的动态变化和正确的分布是至关重要的。在真核生物有丝分裂过程中,DNA复制同时cohesin就已经开始发挥作用。在S期DNA成功复制后就形成了联系在一起的姐妹染色单体,直到末期cohesin从染色体上解离下来,新的子细胞形成。在第一次减数分裂中期,cohesin确保联会合复合体形成,保证第一次减数分裂后期同源染色体间可以正常交换和分离。当植物缺失cohesin的SMC亚基后在胚胎发育早期就死亡;一些敲低cohesin表达的植物有丝分裂,减数分裂染色体形态分布严重异常,育性明显降低。这都表明,cohesin对于一个物种的生存和繁衍有着重要的影响。
近些年,ChIP-seq技术及Hi-C技术的应用,为cohesin调控染色质间相互作用、影响基因表达提供了很多证据。在动物中,cohesin是一个研究染色质长距离交互、三维基因组与转录调控关系的重要蛋白复合体,在植物中却缺少相关研究。在哺乳动物中发现cohesin与染色质构像及转录调控相关功能与CTCF这个关键因子紧密联系,但在拟南芥中并不存在CTCF的同源蛋白。另外,拟南芥染色体组织形态上没有明显的TADs,但有超过1000个类似TAD的区域,并且这些TAD-like的区域性质与动物中TAD的特性相类似。拟南芥基因组中没有典型的TAD结构域,这可能与缺少CTCF相关,但TAD-like区域与TAD性质相似,推测拟南芥cohesin在维持三维基因组结构及转录调控中可能同样会发挥功能。动物细胞中发现cohesin与一些中介因子(mediator)、转录因子及转录复合体相互作用,并且它们在基因组上有显著共同结合位点。在植物缺少CTCF的情况下,cohesin是否能与一些其他类型转录因子相互作用来调控基因表达及是否参与植物三维基因组产生和维持是值得进一步研究的。拟南芥中有研究发现在RNAi植株中,与同源染色体联合及染色体同源重组相关基因表达水平发生变化[145],以及在突变体中等基因的转录水平也发生了变化[83,85]。这些基因转录水平的变化是否直接由cohesin引起的并不清楚,其中的机制也没有研究,有待进一步探索。
[1] van Ruiten MS, Rowland BD. SMC complexes: universal DNA looping machines with distinct regulators., 2018, 34(6): 477–487.
[2] Hirano T. Condensin-Based chromosome organization from bacteria to vertebrates., 2016, 164(5): 847– 857.
[3] Hassler M, Shaltiel IA, Haering CH. Towards a unified model of SMC complex function., 2018, 28(21): R1266–R1281.
[4] Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis., 1999, 400(6743): 461–464.
[5] Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis., 1999, 98(1): 91–103.
[6] Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins., 2000, 5(2): 243– 254.
[7] Watrin E, Peters JM. Cohesin and DNA damage repair., 2006, 312(14): 2687–2693.
[8] Patel L, Kang R, Rosenberg SC, Qiu YJ, Raviram R, Chee S, Hu R, Ren B, Cole F, Corbett KD. Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase., 2019, 26(3): 164–174.
[9] Peng L, Zhang FX. The structure and function of SMC proteins., 2001, 23(2): 173–276.彭莉, 张飞雄. SMC蛋白的结构和功能. 遗传, 2001, 23(2):173–276.
[10] Zhang Y, Zhang XF, Ba ZQ, Liang ZY, Dring EW, Hu HL, Lou JM, Kyritsis N, Zurita J, Shamim MS, Aiden AP, Aiden EL, Alt FW. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination., 2019, 573(7775): 600–604.
[11] Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of theprotein., 1997, 89(4): 511–521.
[12] Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1., 1999, 400(6739): 37– 42.
[13] Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews EA, Sweeney S, Shinagawa H, Watts FZ, Lehmann AR. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex., 2005, 25(1): 172–184.
[14] Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex., 2002, 9(4): 773–788.
[15] Cobbe N, Heck MM. The evolution of ATPase activity in SMC proteins., 2006, 63(3): 685–696.
[16] Ames GF, Lecar H. ATP-dependent bacterial transporters and cystic fibrosis: analogy between channels and transporters., 1992, 6(9): 2660–2666.
[17] Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily., 2000, 101(7): 789–800.
[18] Jeppsson K, Kanno T, Shirahige K, Sjögren C. The maintenance of chromosome structure: positioning and functioning of SMC complexes., 2014, 15(9): 601–614.
[19] Volkov A, Mascarenhas J, Andrei-Selmer C, Ulrich HD, Graumann PL. A prokaryotic condensin/cohesin-like complex can actively compact chromosomes from a single position on the nucleoid and binds to DNA as a ring-like structure., 2003, 23(16): 5638– 5650.
[20] Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners., 2003, 11(3): 571–575.
[21] Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring., 2003, 112(6): 765–777.
[22] Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes., 2005, 74:595–648.
[23] Gligoris T, Löwe J. Structural insights into ring formation of cohesin and related SMC complexes., 2016, 26(9): 680–693.
[24] Liu Cm CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D. Condensin and cohesin knockouts inexhibit a titan seed phenotype., 2002, 29(4): 405–415.
[25] Liu CM, Meinke DW. The titan mutants ofare disrupted in mitosis and cell cycle control during seed development., 1998, 16(1): 21–31.
[26] Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD. Mutations incondensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis., 2003, 130(14): 3283–3295.
[27] Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase., 2000, 151(4): 749–762.
[28] Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mézard C, Grelon M. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis., 2005, 118(Pt 20): 4621–4632.
[29] Yuan L, Yang X, Makaroff CA. Plant cohesins, common themes and unique roles., 2011, 12(2): 93–104.
[30] Peirson BN, Bowling SE, Makaroff CA. A defect in synapsis causes male sterility in a T-DNA-taggedthaliana mutant., 1997, 11(4): 659–669.
[31] Cai X, Dong F, Edelmann RE, Makaroff CA. TheSYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing., 2003, 116(Pt 14): 2999–3007.
[32] Dong F, Cai X, Makaroff CA. Cloning and characterization of twogenes that belong to the RAD21/ REC8 family of chromosome cohesin proteins., 2001, 271(1): 99–108.
[33] Jiang L, Xia M, Strittmatter LI, Makaroff CA. Thecohesin protein SYN3 localizes to the nucleolus and is essential for gametogenesis., 2007, 50(6): 1020–1034.
[34] Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene ofis required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family., 1999, 19(4): 463–472.
[35] da Costa-Nunes JA, Bhatt AM, O'Shea S, West CE, Bray CM, Grossniklaus U, Dickinson HG. Characterization of the threeRAD21 cohesins reveals differential responses to ionizing radiation., 2006, 57(4): 971–983.
[36] Bai X, Peirson BN, Dong F, Xue C, Makaroff CA. Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in., 1999, 11(3): 417–430.
[37] Zhang YL, Zhang H, Gao YJ, Yan LL, Yu XY, Yang YH, Xu WY, Pu CX, Sun Y. Protein phosphatase 2A B'α and B'βprotect the centromeric Cohesion during meiosis I., 2019, 179(4): 1556–1568.
[38] Yuan GL, Ahootapeh BH, Komaki S, Schnittger A, Lillo C, De Storme N, Geelen D. Protein phoshatase 2A B'αand βmaintain centromeric sister chromatid cohesion during meiosis in., 2018, 178(1): 317–328.
[39] da Costa-Nunes JA, Capitão C, Kozak J, Costa-Nunes P, Ducasa GM, Pontes O, Angelis KJ. The AtRAD21.1 and AtRAD21.3cohesins play a synergistic role in somatic DNA double strand break damage repair., 2014, 14: 353.
[40] Shao T, Tang D, Wang KJ, Wang M, Che LX, Qin BX, Yu HX, Li M, Gu MH, Cheng ZK. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis., 2011, 156(3): 1386–1396.
[41] Zhang LR, Tao JY, Wang SX, Chong K, Wang T. The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis., 2006, 60(4): 533–554.
[42] Tao J, Zhang L, Chong K, Wang T. OsRAD21-3, an orthologue of yeast RAD21, is required for pollen development in., 2007, 51(5): 919– 930.
[43] Gong C, Li T, Li Q, Yan L, Wang T. Rice OsRAD21-2 is expressed in actively dividing tissues and its ectopic expression in yeast results in aberrant cell division and growth., 2011, 53(1): 14–24.
[44] Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ. Alleles of afd1 dissect REC8 functions during meiotic prophase I., 2006, 119(Pt 16): 3306–3315.
[45] Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork., 2009, 462(7270): 231–234.
[46] Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase., 2011, 21(19): 1624–1634.
[47] Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts., 2004, 14(17): 1598–1603.
[48] Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, Zickler D. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division., 2003, 17(21): 2675–2687.
[49] Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression., 2006, 16(9): 863– 874.
[50] Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality., 2008, 24: 105–129.
[51] Hinshaw SM, Makrantoni V, Kerr A, Marston AL, Harrison SC. Structural evidence for Scc4-dependent localization of cohesin loading., 2015, 4: e06057.
[52] Hinshaw SM, Makrantoni V, Harrison SC, Marston AL. The kinetochore receptor for the cohesin loading complex., 2017, 171(1): 72–84.e13.
[53] D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes., 2008, 22(16): 2215–2227.
[54] Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, Spitz F. Two independent modes of chromatin organization revealed by cohesin removal., 2017, 551(7678): 51–56.
[55] Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription., 2004, 430(6999): 573–578.
[56] Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex., 2011, 21(1): 12–24.
[57] Fernius J, Nerusheva OO, Galander S, Alves Fde L, Rappsilber J, Marston AL. Cohesin-dependent association of scc2/4 with the centromere initiates pericentromeric cohesion establishment., 2013, 23(7): 599– 606.
[58] Rhodes J, Mazza D, Nasmyth K, Uphoff S. Scc2/Nipbl hops between chromosomal cohesin rings after loading., 2017, 6: e30000.
[59] Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes., 2003, 13(22): 1941–1953.
[60] Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge., 2006, 127(3): 523–537.
[61] Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance., 2006, 4(8): e242.
[62] Sebastian J, Ravi M, Andreuzza S, Panoli AP, Marimuthu MP, Siddiqi I. The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in., 2009, 59(1): 1–13.
[63] Shi ZH, Li ZQ, Zhang GF. The mechanism of histone lysine methylation of plant involved in gene expression and regulation., 2014, 36(3): 208– 219.施子晗, 李泽琴, 张根发. 植物组蛋白赖氨酸化修饰参与基因表达调控的机理. 遗传, 2014, 36(3): 208– 219.
[64] Minina EA, Reza SH, Gutierrez-Beltran E, Elander PH, Bozhkov PV, Moschou PN. Thehomolog of Scc4/MAU2 is essential for embryogenesis., 2017, 130(6): 1051–1063.
[65] He YH, Wang JG, Qi WW, Song RT. Maize Dek15 encodes the cohesin-loading complex subunit SCC4 and is essential for chromosome segregation and kernel development., 2019, 31(2): 465–485.
[66] Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion., 2008, 321(5888): 563–566.
[67] Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion., 2008, 321(5888): 566–569.
[68] Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouët F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity., 2009, 33(6): 763– 774.
[69] Zhang JL, Shi XM, Li YH, Kim BJ, Jia JL, Huang ZW, Yang T, Fu XY, Jung SY, Wang Y, Zhang PM, Kim ST, Pan XW, Qin J. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast., 2008, 31(1): 143–151.
[70] Hou FJ, Zou H. Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister-chromatid cohesion., 2005, 16(8): 3908–3918.
[71] Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery., 1999, 13(3): 307–319.
[72] Milutinovich M, Unal E, Ward C, Skibbens RV, Koshland D. A multi-step pathway for the establishment of sister chromatid cohesion., 2007, 3(1): e12.
[73] Kenna MA, Skibbens RV. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes., 2003, 23(8): 2999–3007.
[74] Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F. Establishment of sister chromatid cohesion at thereplication fork., 2006, 23(6): 787–799.
[75] Schmitz J, Watrin E, Lénárt P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase., 2007, 17(7): 630–636.
[76] Feytout A, Vaur S, Genier S, Vazquez S, Javerzat JP. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1., 2011, 31(8): 1771– 1786.
[77] Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM. Sororin mediates sister chromatid cohesion by antagonizing Wapl., 2010, 143(5): 737–749.
[78] Vaur S, Feytout A, Vazquez S, Javerzat JP. Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction., 2012, 13(7): 645–652.
[79] Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin., 2006, 127(5): 955–967.
[80] Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction., 2009, 19(6): 492–497.
[81] Chan KL, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation., 2012, 150(5): 961–974.
[82] Jiang L, Yuan L, Xia M, Makaroff CA. Proper levels of thecohesion establishment factor CTF7 are essential for embryo and megagametophyte, but not endosperm, development., 2010, 154(2): 820–832.
[83] Bolaños-Villegas P, Yang XH, Wang HJ, Juan CT, Chuang MH, Makaroff CA, Jauh GY.CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis., 2013, 75(6): 927–940.
[84] Singh DK, Andreuzza S, Panoli AP, Siddiqi I. AtCTF7 is required for establishment of sister chromatid cohesion and association of cohesin with chromatin during meiosis in., 2013, 13: 117.
[85] Liu DS, Makaroff CA. Overexpression of a truncated CTF7 construct leads to pleiotropic defects in reproduction and vegetative growth in., 2015, 15: 74.
[86] Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2., 2005, 3(3): e69.
[87] Ouyang ZQ, Zheng G, Song JH, Borek DM, Otwinowski Z, Brautigam CA, Tomchick DR, Rankin S, Yu HT. Structure of the human cohesin inhibitor Wapl., 2013, 110(28): 11355–11360.
[88] Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase., 2006, 16(24): 2406–2417.
[89] Pradillo M, Knoll A, Oliver C, Varas J, Corredor E, Puchta H, Santos JL. Involvement of the cohesin cofactor PDS5 (SPO76) during meiosis and DNA repair in., 2015, 6: 1034.
[90] De K, Sterle L, Krueger L, Yang X, Makaroff CA.WAPL is essential for the prophase removal of cohesin during meiosis., 2014, 10(7): e1004497.
[91] De K, Bolaños-Villegas P, Mitra S, Yang X, Homan G, Jauh GY, Makaroff CA. The opposing actions of arabidopsis CHROMOSOME TRANSMISSION FIDELITY7 and WINGS APART-LIKE1 and 2 differ in mitotic and meiotic cells., 2016, 28(2): 521–536.
[92] Birkenbihl RP, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair., 1992, 20(24): 6605–6611.
[93] Heo SJ, Tatebayashi K, Kato J, Ikeda H. The RHC21 gene of budding yeast, a homologue of the fission yeast rad21+ gene, is essential for chromosome segregation., 1998, 257(2): 149–156.
[94] Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, Earnshaw WC, Takeda S. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells., 2001, 1(6): 759– 770.
[95] Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase., 1992, 117(5): 921–934.
[96] Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation., 2008, 117(2): 123–135.
[97] Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei., 2010, 12(2): 185–192.
[98] Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance., 2006, 17(5): 2287–2302.
[99] Zhao JP, Wang B. Genetic and biochemical control mechanism regulating entry into and exit from mitosis in eucaryotes., 1994, 16(4): 40–45.赵吉平, 王斌. 真核生物细胞有丝分裂起始、终止的遗传与生化调控机制. 遗传, 1994, 16(4): 40–45.
[100] Alomer RM, da Silva EML, Chen J, Piekarz KM, McDonald K, Sansam CG, Sansam CL, Rankin S. Esco1 and Esco2 regulate distinct cohesin functions during cell cycle progression., 2017, 114(37): 9906–9911.
[101] Carretero M, Ruiz-Torres M, Rodríguez-Corsino M, Barthelemy I, Losada A. Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres., 2013, 32(22): 2938–2949.
[102] Rhodes JDP, Haarhuis JHI, Grimm JB, Rowland BD, Lavis LD, Nasmyth KA. Cohesin can remain associated with chromosomes during DNA replication., 2017, 20(12): 2749–2755.
[103] Nishiyama T, Sykora MM, Huis in't Veld PJ, Mechtler K, Peters JM. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin., 2013, 110(33): 13404–13409.
[104] Liu H, Rankin S, Yu HT. Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis., 2013, 15(1): 40–49.
[105] Wolf PG, Cuba Ramos AC, Kenzel J, Neumann B, Stemmann O. Studying meiotic cohesin in somatic cells reveals that Rec8-containing cohesin requires Stag3 to function and is regulated by Wapl and sororin., 2018, 131(11): pii:jcs212100.
[106] Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation., 2000, 2(8): 492–499.
[107] Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1., 2001, 21(20): 6984–6998.
[108] Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1βis required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination., 2004, 6(6): 555–562.
[109] Bayés M, Prieto I, Noguchi J, Barbero JL, Pérez Jurado LA. Evaluation of the Stag3 gene and the synaptonemal complex in a rat model (as/as) for male infertility., 2001, 60(3): 414–417.
[110] Fukuda T, Hoog C. The mouse cohesin-associated protein PDS5B is expressed in testicular cells and is associated with the meiotic chromosome axes., 2010, 1(3): 484–494.
[111] Gómez R, Felipe-Medina N, Ruiz-Torres M, Berenguer I, Viera A, Pérez S, Barbero JL, Llano E, Fukuda T, Alsheimer M, Pendás AM, Losada A, Suja JA. Sororin loads to the synaptonemal complex central region independently of meiotic cohesin complexes., 2016, 17(5): 695–707.
[112] Jordan PW, Eyster C, Chen JR, Pezza RJ, Rankin S. Sororin is enriched at the central region of synapsed meiotic chromosomes., 2017, 25(2): 115–128.
[113] Ishiguro K, Kim J, Shibuya H, Hernández-Hernández A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, Höög C, Watanabe Y. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes., 2014, 28(6): 594–607.
[114] Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing., 2011, 12(3): 267–275.
[115] Lee J, Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis., 2011, 192(2): 263– 276.
[116] Brieño-Enríquez MA, Moak SL, Toledo M, Filter JJ, Gray S, Barbero JL, Cohen PE, Holloway JK. Cohesin removal along the chromosome arms during the first meiotic division depends on a NEK1-PP1γ-WAPL axis in the mouse., 2016, 17(4): 977–986.
[117] Llano E, Herrán Y, García-Tuñón I, Gutiérrez-Caballero C, de Álava E, Barbero JL, Schimenti J, de Rooij DG, Sánchez-Martín M, Pendás AM. Meiotic cohesin complexes are essential for the formation of the axial element in mice., 2012, 197(7): 877–885.
[118] Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination., 2011, 146(3): 372– 383.
[119] Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of meiotic cohesin by SCC-2 is required for early processing of DSBs and for the DNA damage checkpoint., 2011, 21(17): 1421–1430.
[120] Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes., 1999, 152(2): 577– 593.
[121] Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B., 2004, 36(6): 631–635.
[122] Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome., 2004, 36(6): 636–641.
[123] Zhu ZH, Wang XD. Roles of cohesin in chromosome architecture and gene expression.,2019, 90(4): 187–193.
[124] Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. Functional and topological characteristics of mammalian regulatory domains., 2014, 24(3): 390–400.
[125] 郭亚, 吴强. 采用DNA片段编辑技术反转CTCF结合位点改变基因组拓扑结构和增强子与启动子功能. 遗传, 2015, 37(10): 1073–1074.
[126] Rowley MJ, Corces VG. Organizational principles of 3D genome architecture., 2018, 19(12): 789– 800.
[127] Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IJcken WF, Grosveld FG, Ren B, Wendt KS. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells., 2014, 111(3): 996–1001.
[128] Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions., 2012, 485(7398): 376–380.
[129] Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping., 2014, 159(7): 1665–1680.
[130] Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation., 2019, 20(9): 535–550.
[131] Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. Genome-wide analysis of local chromatin packing in., 2015, 25(2): 246–256.
[132] Ning CY, He MN, Tang QZ, Zhu Q, Li MZ, Li DY. Advances in mammalian three-dimensional genome by using Hi-C technology approach., 2019, 41(3): 215–233.宁椿游, 何梦楠, 唐茜子, 朱庆, 李明洲, 李地艳. 基于Hi-C技术哺乳动物三维基因组研究进展. 遗传, 2019, 41(3): 215–233.
[133] Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms., 2008, 132(3): 422–433.
[134] Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin., 2008, 105(24): 8309–8314.
[135] Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators., 2008, 27(4): 654–666.
[136] Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC- binding factor., 2008, 451(7180): 796–801.
[137] Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture., 2010, 467(7314): 430–435.
[138] Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B. A map of the cis-regulatory sequences in the mouse genome., 2012, 488(7409): 116–120.
[139] Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription., 2010, 20(5): 578–588.
[140] Uhlmann F. SMC complexes: from DNA to chromosomes., 2016, 17(7): 399–412.
[141] Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. Establishment of DNA-DNA interactions by the cohesin ring., 2018, 172(3): 465–477.e415.
[142] Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, Bartlett DA, Zhao PA, Washburn BK, Nora EP, Kraft K, Mundlos S, Bruneau BG, Ljungman M, Fraser P, Ay F, Gilbert DM. Identifying cis elements for spatiotemporal control of mammalian DNA replication., 2019, 176(4): 816–830.e18.
[143] Villa-Hernández S, Bermejo R. Cohesin dynamic association to chromatin and interfacing with replication forks in genome integrity maintenance., 2018, 64(5): 1005–1013.
[144] Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl., 2017, 544(7651): 503–507.
[145] Yuan L, Yang XH, Ellis JL, Fisher NM, Makaroff CA. TheSYN3 cohesin protein is important for early meiotic events., 2012, 71(1): 147–160.
Progresses on the structure and function of cohesin
Yu Zhang, Yuda Fang
Cohesin is an evolutionarily conserved protein complex in eukaryotes. The four subunits of cohesin form a ring structure that plays an important role in maintaining the orderly arrangement of chromatin during cell division. In addition, metazoan cohesin was found to act as an intermolecular linker, which regulates insulator/enhancer–promoter interactions, leading to either enhancement or inhibition of gene expressions. However, little is known about the role of cohesin in the transcriptional regulation in plants. In the review, we introduce the structure and core subunits of cohesin, and summarize the factors that regulate its dynamic changes on chromatin. Based on the functional study of plant cohesin in recent years and researches in animals about the roles of cohesin in the three-dimensional genome organization and transcriptional regulation, we prospect the potential functions of plant cohesin in regulating transcription.
SMC; cohesin; cell cycle; three-dimensional genome; transcriptional regulation
2019-11-07;
2019-12-10
国家自然科学基金项目(编号:31871230)资助[Supported by the National Natural Science Foundation of China (No. 31871230)]
张雨,博士后,研究方向:染色质结构与功能。E-mail: zhangyu2065@sjtu.edu.cn
方玉达,博士,特聘教授,博士生导师,研究方向:植物细胞核与染色质的结构和功能。E-mail: yuda.fang@sjtu.edu.cn
10.16288/j.yczz.19-288
2019/12/19 14:30:58
URI: http://kns.cnki.net/kcms/detail/11.1913.R.20191218.1615.005.html
(责任编委: 史庆华)
