MYB转录因子调控蔬菜花青素生物合成的研究进展
2020-02-04彭亚丽高倩董文熊安平秦玉芝林原熊兴耀胡新喜
彭亚丽 高倩 董文 熊安平 秦玉芝 林原 熊兴耀 胡新喜


摘 要:MYB是调节花青素生物合成的重要转录因子,通过激活和抑制结构基因的表达,维持植物器官内花青素积累量的平衡。R2R3MYB转录激活因子单独或与bHLH、WD40形成复合体调控植物花青素的生物合成。转录抑制因子有R2R3MYB和R3MYB 2种类型。R2R3MYB类抑制因子有2种作用方式,其中一种可以直接作用于结构基因的启动子,使结构基因表达量下调,花青素的生物合成减少;另一种需要借助辅助因子bHLH抑制MBW复合物在花青素生物合成中的表达,减少花青素的积累。R3MYB类转录抑制因子都需要与bHLH蛋白形成复合物,主动或被动地抑制结构基因的表达,阻止花青素的合成积累。笔者对调控蔬菜花青素生物合成的MYB转录因子及其调控作用机制进行了综述,并对今后的研究重点进行了展望。
关键词:花青素;MBW复合物;激活因子;抑制因子
中图分类号:S63+S64+S65 文献标志码:A 文章编号:1673-2871(2020)12-001-07
Abstract:MYB is an important transcription factor that regulates anthocyanins biosynthesis and maintains the balance of anthocyanin accumulation in plant organs by activating and inhibiting the expression of structural genes. Transcription activators R2R3MYB regulate the biosynthesis of anthocyanins alone or by forming a regulatory complex with bHLH and WD40 in plant. Transcription repressors include both R2R3MYB type and R3MYB type. There are two modes of action of R2R3MYB inhibitors, one of which can directly act on the promoters of structural genes to down-regulate the expression of structural gene and decreased anthocyanin biosynthesis. The other inhibits the expression of MBW complex in anthocyanin biosynthesis by binding the cofactor bHLH, and reduces the accumulation of anthocyanin. R3MYB transcription inhibitors need to form a complex with bHLH protein, then inhibit the expression of structural genes actively or passively, and prevent the synthesis and accumulation of anthocyanins. We review the progress of the MYB transcription factors that regulate vegetable anthocyanins biosynthesis and the mechanism of their regulation and prospect the future researches.
Key words:Anthocyanins; MBW complex; Activators; Repressors
花青素是一类广泛存在于高等植物组织中的类黄酮次生代谢产物,主要分布于植物的叶、花和果实中,主要为蓝、紫和红3种颜色[1]。研究表明,紫甘薯、血橙和红肉苹果等都有大量的花青素积累[2-4],关于玉米、拟南芥等的研究已经证明花青素的生物合成是通过苯丙烷途径完成的[5-6],PAL(苯丙氨酸解氨酶)、CHS(查尔酮合成酶)、CHI(查尔酮异构酶)、F3H(黄烷酮羟化酶)、DFR(二氢黄酮醇4-还原酶)、ANS(花青素合酶)等直接编码相关酶的合成[7]。同时,花青素的生物合成也受到MYB(v-myb avian myeloblastosis viral oncogene homolog)、bHLH(basic helix-loop-helix)、WD40等轉录因子的调控[8],共同完成花青素的生物合成过程。如茄子里的Sm TTG1、Sm GL3和Sm TT8都与Sm MYB相互作用,共同完成茄子花青素的合成[9]。笔者对调控花青素生物合成的转录因子特别是MYB转录因子及其调控作用机制进行综述,并对今后的研究重点进行展望。
1 调控蔬菜花青素生物合成的转录因子
转录因子主要通过调控结构基因的转录和表达,间接调控花青素的生物合成过程[8]。调控花青素生物合成途径的转录因子家族主要有MYB、bHLH和WD40三大类[10]。MYB类转录因子作为植物代谢调控中最大的转录因子,也是目前真核生物中研究数目最多和作用最广的转录因子家族之一[11],主要参与植物的次生代谢调控、细胞学形态建成、环境胁迫和急速应答等过程[12]。其中,与花青素生物合成相关的是MYB家族最大的亚类R2R3MYB[13]。C1基因调控玉米花青素的生物合成,首次证明了MYB转录因子在植物中的调节作用[14]。何琼[15]发现,BrMYB2是紫心大白菜花青素合成的关键基因,可以在苗期准确区别紫色和白色叶球。
近些年来,与R2R3MYB蛋白功能完全相反的一类单一MYB蛋白R3MYB被报道,其在花青素生物合成过程中起抑制作用[16]。CPC作为一个R3MYB类抑制子,首先在拟南芥中被发现,主要抑制与毛状体和根毛形成相关的MBW复合物[17-18]。bHLH类转录因子是调控植物次生代谢过程的第二大类转录因子,其N端包含约18个亲水性的碱性氨基酸,紧跟2个由环隔开的两亲性α螺旋,有利于HLH区与bHLH蛋白的二聚化[19]。同时,bHLH蛋白也可以通过识别E-box和DNA序列来调节转录水平[16]。Ludwig等[20]在玉米中发现,R基因编码一个bHLH蛋白,bHLH首次被报道。Nesi等[21]在拟南芥中发现了原花色素调节因子TT8和腺毛调节因子GL3,首次证明了基因bHLH在MBW复合物中起作用。席浩淳等[22]发现,SmbHLH13基因可以正调控茄子内基因F3H和CHS的表达,完成茄子花青素的合成。WD40重复蛋白存在一个包含40个残基的核心区域,该区域由甘氨酸—组氨酸二肽(GH)和色氨酸—天冬氨酸(WD)二肽组成,进行β螺旋折叠而成[23]。Miller等[24]发现,仅有4个或5个WDR家族的TTG1基因控制拟南芥的色素合成,其同源序列在其他物种中也有相似功能。综上所述,在蔬菜花青素合成的过程中,MYB转录因子起着关键性作用。
2 转录因子MYB在蔬菜花青素合成中的调节作用
2.1 參与蔬菜花青素合成的MYB蛋白结构
MYB蛋白的N端具有一段高度保守的DNA结合域,即MYB结构域。根据其结构域的数目,可分为4个亚类(图1),即RMYB、R2R3MYB、3RMYB 和 4RMYB[25]。每一个结构域中含有约52个氨基酸残基,形成3个α螺旋,其中第2、第3个螺旋构成1个HLH结构[26]。R2R3MYB亚类主要参与植物花青素合成,作为植物MYB蛋白家族中最大的亚类,包含着2个与c-MYB的R2和R3最相似的MYB重复序列[16]。保守的N末端MYB结构域和可变的C末端是R2R3MYB的一个固定结构,通常C末端会有一个激活域或抑制域[27]。此外,R3MYB有1个高度保守的[D/E]LX2[R/K]X3LX6LX3R和WXM序列,能与bHLH蛋白相结合[28]。
2.2 转录因子MYB在蔬菜花青素合成中的激活作用
MYB转录因子对花青素生物合成的激活作用贯穿EBGs和LBGs两个阶段。在EBGs阶段,MYB转录因子主要调节花青素合成过程中关键酶基因的转录和表达。在苹果的研究中发现,MdMYBA基因能在低温下与ANS的启动子进行特异性结合,促进花青素的积累[30]。ZHANG等[31]将拟南芥中的PAP1基因在丹参中过表达,检测出与花青素合成相关的PAL、CHS等关键酶基因的表达明显上调,花青素的积累量也明显增加。番茄中的SlAN2基因能在胁迫下诱导果实花青素不均匀积累[32]。功能性的SlAN2-like基因在果皮花青素合成中起主要调控作用,能激活相关酶基因及其调控因子的表达[33]。过表达SlMYB75基因,能诱导紫番茄营养和生殖器官内花青素的大量积累[34]。BoPr基因可以使花青素在观赏性羽衣甘蓝中富集,叶子呈紫色[35]。Wang等[36]分离出BrPur基因,发现其可以使中国白菜的叶片内花青素大量积累,呈现紫色。
在花青素合成的LBGs阶段,MYB转录因子的作用机制主要是与其他蛋白相互作用,其中主要有bHLH蛋白和WD40蛋白[8]。在葡萄中的研究表明,MYB转录因子可以与bHLH类基因VvMYC1相互作用,诱导花青素合成[24]。通过对白色、红色和紫色马铃薯表皮的StAN1、StMYBA1和StMYB113基因功能分析,发现马铃薯的MYBs能在烟草中与bHLH相互作用,增加烟草花青素的积累[37]。WD40家族基因StAN11通过上调StDFR基因的表达,增加马铃薯块茎花青素的积累[38]。在马铃薯中超表达StANS基因,在块茎中发现大量花青素积累[39]。烟草瞬时转化试验发现,RsMYB1和RsTT8在萝卜的花青素生物合成中相互作用,Lim和Lai等[40-41]发现,RsMYB1a基因和RsbHLH4相互作用,可以在主根中大量积累花青素。Xu等[42]在紫色胡萝卜中发现了一个基因DcMYB6,在拟南芥中过表达,能够诱导花青素的合成。紫花菜中一个半显性基因Pr能调节BobHLH1的表达,同时使花青素晚期合成的BoF3H、BoDFR等基因表达量明显上调[43]。紫叶芥菜中的BjDFR、BjTT19、BjTT8基因表达量显著高于绿叶芥菜,这些基因与花青素的后期合成与转运相关[44]。
2.3 转录因子MYB在蔬菜花青素合成中的抑制作用
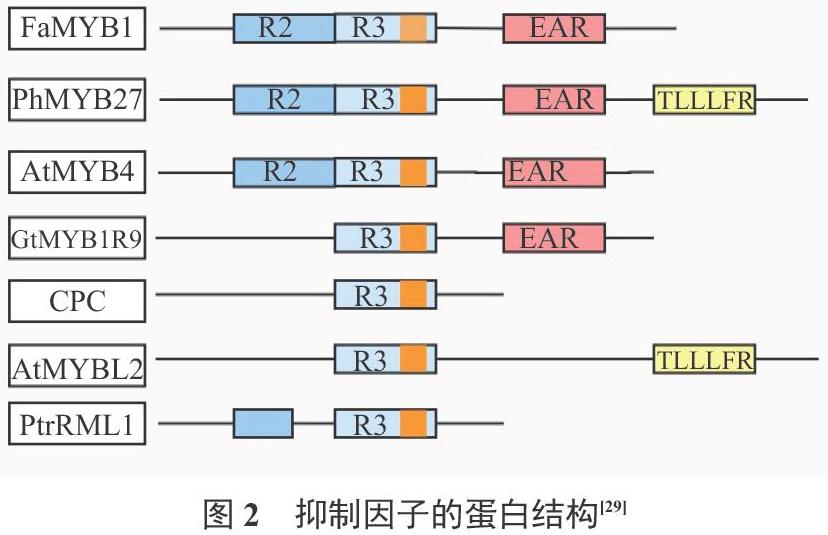
2.3.1 MYB抑制因子的蛋白结构 MYB蛋白的抑制类型取决于C末端的不同结构域,这些保守的结构域和MYB蛋白的抑制活性有关。MYB类的转录抑制因子主要有R2R3MYB和R3MYB两大类[45]。R2R3MYB类抑制子可以分为一般苯丙烷、木质素和类黄酮三大类[29]。它的C端包含一个DNA结构结合域,含有GIDP和EAR两个基团,EAR基团是抑制子的重要结构[46]。AtMYB4基因能够抑制拟南芥的次生代谢过程,从而确定了EAR基团的抑制功能[47-48]。R3MYB类抑制子仅包含一个单MYB重复的DNA结构域, C端保守结构域不同,则作用机制存在差异[49]。其N端与R2R3MYB类抑制子一样,都含有DNA结构结合域,但只有与花青素和原花青素合成相关的抑制子能够与bHLH转录因子结合,共同作用[50-51]。AtCPC基因的C端不包含任何抑制基团,仅保留一个与bHLH蛋白相结合的结构域,与R2R3MYB类转录激活因子竞争bHLH蛋白完成抑制作用[52-53]。AtMYBL2中含有一个R3结构域和部分不完整的R2结构域,可能是在染色体的串联复制过程中,R2结构域发生了大缺失[54]。Matsui等[53]在2008年的研究中发现,AtMYBL2基因中有一个新的基团TLLLFR(图2),在拟南芥次生代谢中表现为抑制作用。
2.3.2 R2R3MYB类抑制因子 目前已经报道的R2R3MYB类抑制因子中,有22种与木质素、一般苯丙烷的生物合成相关[29]。拟南芥中的AtMYB3基因抑制苹果酸介子碱和花青素的合成[55],AtMYB32可以影响木质素在花粉中的合成,并参与其他花粉发育过程[56]。苯丙烷代谢途径中,许多次生代谢产物之间相互影响。大叶杨中的PtoMYB156基因过表达,能减少苯基丙烷基因的表达量,进而抑制木质素生物合成,同时,大叶杨的次生细胞壁黏性、酚类物质和类黄酮含量都有不同程度的降低[57]。
对类黄酮的相关研究主要集中在花青素和原花青素富集方面。Espley等[58]研究发现,R2R3MYB类抑制因子在抑制花青素的合成过程中,需要辅助因子bHLH。FaMYB1、PtrMYB182等抑制因子不能直接与靶向基因的启动子结合,但能与bHLH结合,抑制MBW复合物在花青素合成中的表达,进而减少花青素的积累[54]。Pattanaik等[59]在矮牵牛试验中发现,PhMYB27与MBW复合物结合,抑制了复合物的表达活性;过表达PhMYB27基因,矮牵牛花和叶子中的花青素则明显减少,同时种子中的原花青素积累减少;干扰表达PhMYB27,花青素积累量增加。酵母双杂交试验发现,PhMYB27通过bHLH蛋白与MBW复合物相互作用而下调花青素代谢的表达,同样适用于原花青素[45]。
R2R3MYB转录抑制因子中,有一类抑制子如MdMYB16、AtMYB4、AtMYB60和NtMYB2等,能直接與花青素合成相关结构基因的启动子发生作用,使结构基因表达下调。苹果中的MdMYB16基因能够直接抑制MdUFGT和MdANS关键酶基因在花青素合成中的表达;但是,在得到的过表达MdMYB16愈伤组织中再进行MdbHLH过表达,发现MdbHLH对花青素合成的抑制作用减弱,说明MdbHLH能影响MdMYB16基因的表达,但具体机制尚不清楚[60]。过表达AtMYB60可以使花青素合成中的DFR基因表达量减少[61]。在转基因烟草中过表达NtMYB2基因,UFGT的表达量显著下降[62]。Fornale等[52]研究发现,拟南芥中另一个AtMYB4的同源基因AtMYB7可以抑制DFR和糖基转移酶基因UGT的表达,从而下调黄酮醇的生物合成。在AtMYB7的突变植株内,黄酮生物合成代谢显著增强,含量显著增加。
2.3.3 R3MYB类抑制因子 基因CPC作为R3MYB类抑制因子,在拟南芥的表皮细胞中被发现。研究表明,CPC可以抑制毛状体的分裂和无毛细胞的分化[63]。从英国梧桐中分离出PaTRY和PaCPC-lke基因,将其在拟南芥中进行过表达,可以抑制拟南芥毛状体的形成[64]。CPC型抑制因子也能抑制蔬菜花青素的形成,SlMYBATV基因能负调控番茄果实中花青素的合成[65-66]。StMYBATV可以和bHLHs蛋白结合,抑制马铃薯花青素的形成[67]。CPC型抑制子的表达量与非生物胁迫相关,低氮环境下,CPC能与激活因子PAP1竞争bHLH(GL3),阻止PAP1对关键酶基因DFR的激活作用,导致花青素的合成受阻[68]。Nemie-Feyissa等[69]对拟南芥进行氮胁迫处理,发现CPC基因在缺氮时的表达量显著增加,莲座叶中缺氮可以引起花青素积累的负反馈调节。
AtMYBL2作为另一种R3MYB类抑制因子,在拟南芥中过表达,相关酶基因和花青素的积累均受到抑制[52,26]。AtMYBL2型的转录抑制子,与R2R3MYB类转录抑制子FaMYB1等作用机制相似[70]。AtMYBL2在强光下表达量减小[71]。AtMYBL2型抑制子与花青素的表达量呈负相关[52],如紫甘蓝中检测不到BoMYBL2-1基因的表达[72]。在PtrRML1基因的N末端发现一个EAR基团,拟南芥中进行过表达时,它不仅可以抑制花青素的积累,同时还能影响拟南芥毛状体的生长发育(图2)[73]。
3 MYB转录激活因子和抑制子的互作调控
在花青素合成过程中,MYB转录激活因子和抑制子之间存在竞争关系,可以争夺辅助因子bHLH蛋白和关键酶基因的启动子。类黄酮类MYB抑制子能干扰MBW复合物中激活因子和bHLH蛋白的结合。酵母三杂交试验分析矮牵牛中的PhMYB27基因,发现MYB转录激活因子和抑制子可以同时与bHLH蛋白结合[45]。酵母双杂交进行激活因子、抑制子与bHLH蛋白的结合强度比较,发现启动子上的功能可能与该细胞中MYB激活因子和抑制子间的相对丰度有关[74]。茄子中的SmMYBL1基因通过与MYB激活因子竞争bHLH结合位点,抑制MYB复合物的表达,进而抑制茄子花青素的生物合成[75]。同一个基因可以在不同的合成途径中有不同的功能。MYB75又名PAP1,在拟南芥中过表达,会有大量花青素积累,是一个控制花青素合成的激活因子。Teng等[76]发现了一个MYB75突变体植株,在高糖浓度下,花青素含量积累极少。同时,MYB75也与拟南芥中木质素的沉淀相关,试验中发现,MYB75突变体植株中次生细胞壁的总形成量增加,编码木质素和次生细胞壁多糖生物合成中关键酶基因的表达量升高[77],此时的MYB75是一个抑制因子。CsMYB3是柑橘中重要的抑制因子,在拟南芥中过表达CsMYB3基因,转基因植株的花青素积累量显著低于野生株,但种子中的原花青素合成并没有受到影响。徐强对其调控机制进行研究发现,CsMYB3基因可以平衡CsRuby1基因的激活作用,防止柑橘中花青素的积累过量,形成一个激活—抑制反馈调节环(图3)[78]。
4 展 望
花青素是很好的医用保健物质,也是蔬菜自身应对复杂环境的一种调控反应物质。转录因子通过与启动子或其他转录因子结合介导结构基因的表达,已经成为植物界最常见的调控机制。MYB家族作为调控蔬菜花青素合成的最大类转录因子,既能激活花青素的合成,又能抑制花青素的积累,在花青素的生物合成中占有很重要的作用。最近一些研究报道,MYB类转录激活因子和抑制因子之间存在着复杂的分级反馈调节机制,进而使得我们对MBW复合物有了新的认知。但是关于MYB转录抑制因子的上游研究和具体调节机制依旧存在很多未知之处。MYB类转录抑制因子与MBW复合物的特异结合可能与其转录激活因子相关,抑制因子与MBW复合物间的作用强度或许与抑制强弱相关。相关胁迫对抑制因子表达的潜在影响,是否能在蔬菜作物育种等其他方面得到进一步应用有待后续深入研究。
参考文献
[1] HARBORNE J B.The flavonoids advances in research since 1986[J].Chapman and Hall London,1993:619-652.
[2] MANO H,OGASAWARA F,SATO K,et al.Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato[J].Plant Physiology,2007,143(3):1252-1268.
[3] ZHANG L Y,HU J,HAN X L,et al.A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour[J].Nature Communications,2019,10(1):1494.
[4] BUTELLI E,LICCIARDELLO C,ZHANG Y,et al.Retrotransposons control fruit-specific,cold-dependent accumulation of anthocyanins in blood oranges[J].Plant Cell,2012,24(3):1242-1255.
[5] KOES R,VERWEIJ W,QUATTROCCHIO F.et al.Flavonoids:a colorful model for the regulation and evolution of biochemical pathways[J].Trends in Plant Science,2005,10(5):236-242.
[6] NAING A H and KIM C K.Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants[J].Plant Molecular Biology,2018,98:1-18.
[7] SAITO K,YONEKURA-SAKAKIBARA K,NAKABAYASHI R,et al.The flavonoid biosynthetic pathway in Arabidopsis:structural and genetic diversity[J].Plant Physiology and Biochemistry,2013,72:21-34.
[8] XU W J,DUBOS C,LEPINIEC L.Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J].Trends in Plant Science,2015,20(3):176-185.
[9] 劉新宇,韩洪强,葛海燕,等.茄子花青素合成中SmTTG1、SmGL3和SmTT8的表达及其蛋白质间的相互作用[J].园艺学报,2014,41(11):2241-2249.
[10] 许志茹,李春雷,崔国新,等.植物花青素合成中的蛋白[J].植物生理学通讯,2008,44(3):597-604.
[11] 王萃铂,张瓒,张晓雪,等.菊花转录因子CmMYB59 的克隆与表达特性分析[J].南京农业大学学报,2016,39(1):63-69.
[12] 官丽莉,张雪,韩怡来,等.红花转录因子 Ct MYB1 基因的克隆及原核表达[J].中草药,2015,46(17):2603-2609.
[13] AZUMA A,KOBAYASHI S,MITANI N,et al.Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin[J].Theoretical and Applied Genetics,2008,117(6):1009-1019.
[14] PAZ-ARES J,GHOSAL D,WIENAND U,et al.The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators[J].The EMBO Journal,1987,6(12):3553-3558.
[15] 何琼.紫心大白菜花青素合成和积累的分子机理研究[D].陕西杨凌:西北农林科技大学,2018.
[16] FELLER A,MACHEMER K,BRAUN E L,et al.Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J].The Plant Journal,2011,66(1):94-116.
[17] WANG S C,HUBBARD L,CHANG Y,et al.Comprehensive analysis of single-repeat R3MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis[J].BMC Plant Biology,2008,8(1):81.
[18] WESTER K,DIGIUNI S,GEIER F,et al.Functional diversity of R3 single-repeat genes in trichome development[J].Development,2009,136(9):1487-1496.
[19] MASSARI M E, MURRE C.Helix-loop-helix proteins:regulators of transcription in eukaryotic organisms[J].Molecular and Cellular Biology,2000,20(2):429-440.
[20] LUDWIG S R,HABERA L F,DELLAPORTA S L,et al.Lc,a member of the maize R gene family responsible for tissue-specific anthocyanin production,encodes a protein similar to transcriptional activators and contains the myc homology region[J].Proceedings of the National Academy of Sciences of the USA,1989,86(18):7092-7096.
[21] NESI N,DEBEAUJON I,JOND C,et al.The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques[J].The Plant Cell,2000,12(10):1863-1878.
[22] XI H C,HE Y J,CHEN H Y.Functional characterization of SmbHLH13 in anthocyanin biosynthesis and flowering in eggplant [J/OL].Horticultural Plant Journal [2020-08-28].https://doi.org/10.1016/j.hpj.2020.08.006.
[23] MISHRA A K ,PURANIK S,PRASAD M.Structure and regulatory networks of WD40 protein in plants[J].Journal of Plant Biochemistry and Biotechnology,2012,21(1):32-39.
[24] MILLER J C,CHEZEM W R, CLAY N K.Ternary WD40 repeat-containing protein complexes:evolution,composition and roles in plant immunity[J].Frontiers in Plant Science,2015,6:1108.
[25] OGATA K,MORIKAWA S,NAKAMURA H,et al.Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices[J].Cell,1994,79(4):639-648.
[26] DUBOS C,STRACKE R,GROTEWOLD E,et al.MYB transcription factors in Arabidopsis[J].Trends in Plant Science,2010,15(10):573-581.
[27] STRACKE R,WERBER M, WEISSHAAR B.The R2R3-MYB gene family in Arabidopsis thaliana[J].Current Opinion in Plant Biology,2001,4(5):447-456.
[28] ZIMMERMANN I M,HEIM M A,WEISSHAAR B,et al.Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins[J].The Plant Journal,2004,40(1):22-34.
[29] MA D W and CONSTABLE C P.MYB repressors as regulators of phenylpropanoid metabolism in plants[J].Trends in Plant Science,2019,24(3):275-289.
[30] BAN Y,HONDA C,HATSUYAMA Y,et al.Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin[J].Plant and Cell Physiology,2007,48(7):958-970.
[31] ZHANG Y,YAN Y P,WANG Z Z,et al.The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza[J].Journal of Agricultural and Food Chemistry,2010,58(23):12168-12175.
[32] KIFERLE C,FANTINI E,BASSOLINO L,et al.Tomato R2R3-MYB proteins SlANT1 and SlAN2:same protein activity,different roles[J].PLOS ONE,2015,10(8):e0136365.
[33] SUN C L,DENG L,DU M M,et al.A transcriptional network promotes anthocyanin biosynthesis in tomato flesh[J].Molecular Plant,2020,13(1):42-58.
[34] JIAN W,CAO H H,YUAN S,et al.SlMYB75,an MYB-type transcription factor,promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits[J].Horticulture Research,2019,6:22-36.
[35] LIU X P,GAO B Z,HAN F Q,et al.Genetics and fine mapping of a purple leaf gene,BoPr,in ornamental kale(Brassica oleracea L.var.acephala).[J] BMC Genomics,2017,18(1):230-238.
[36] WANG W H,ZHANG D S,YU S C,et al.Mapping the BrPur gene for purple leaf color on linkage group A03 of Brassica rapa[J].Euphytica,2014,199(3):293-302.
[37] LIU Y H,LIN-WANG K,ESPLEY R V,et al.Functional diversification of the potato R2R3 MYB anthocyanin activators AN1,MYBA1,and MYB113 and their interaction with basic helix-loop-helix cofactors[J].Journal of Experimental Botany,2016,67(8):2159-2176.
[38] LI W,WANG B,WANG M,et al.Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation[J].Journal of Integrative Plant Biology,2014,56(4):364-372.
[39] ZHANG H L,ZHAO X J,ZHANG J P,et al.Functional analysis of an anthocyanin synthase gene StANS in potato[J].Scientia Horticulturae,2020,15:272-279.
[40] LIM S H,KIM D H,KIM J K,et al.A radish basic helix-loop-helix transcription factor,RsTT8 acts a positive regulator for anthocyanin biosynthesis[J].Frontiers in Plant Science,2017,8:1917.
[41] LAI B,CHENG Y Y,LIU H,et al.Differential anthocyanin accumulation in radish taproot:importance of RsMYB1 gene structure[J].Plant Cell Reports,2020,39(2):217-226.
[42] XU H F,WANG N,LIU J X,et al.The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdBHLH33 genes[J].Plant Molecular Biology,2017,94(1/2):149-165.
[43] CHIU L W,ZHOU X J,BURKE S,et al.The purple cauliflower arises from activation of a MYB transcription factor[J].Plant Physiology,2010,154(3):1470-1480.
[44] ZHANG D W,LIU L L,ZHOU D G,et al.Genome-wide identification and expression analysis of anthocyanin biosynthetic genes in Brassica juncea[J].Journal of Integritive Agricuture,2020,19(5):1250-1260.
[45] ALBERT N W,DAVIES K M,LEWIS D H,et al.A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots[J].The Plant Cell,2014,26(3):962-980.
[46] KRANZ H D,DENEKAMP M,GRECO R,et al.Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana[J].The Plant Journal,1998,16(2):263-276.
[47] OHTA M,MATSUI K,HIRATSU K,et al.Repression domains of class II ERF transcriptional repressors share an essential motif for active repression[J].The Plant Cell,2001,13(8):1959-1968.
[48] KAZAN K.Negative regulation of defence and stress genes by EAR-motif-containing repressors[J].Trends in Plant Science,2006,11(3):109-112.
[49] DU H,WANG Y B,XIE Y,et al.Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants[J].DNA Research,2013,20(5):437-448.
[50] YOSHIDA K,MA D W,CONSTABEL C P.The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes[J].Plant Physiology,2015,167(3):693-710.
[51] WANG S C, CHEN J G.Regulation of cell fate determination by single-repeat R3 MYB transcription factors in Arabidopsis[J].Frontiers in Plant Science,2014,5:133.
[52] FORNALE S,LOPEZ E,SALAZAR-HENAO J E,et al.AtMYB7,a new player in the regulation of UV sunscreens in Arabidopsis thaliana[J].Plant Cell Physiology,2014,55(3):507-516.
[53] MATSUI K,UMEMURA Y,OHME-TAKAGI M.AtMYBL2,a protein with a single MYB domain,acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis[J].The Plant Journal,2008,55(6):954-967.
[54] CHEN L H,HU B,QIN Y H,et al.Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors[J].Plant Physiology and Biochemistry,2019,136: 178-187.
[55] ZHOU M L,ZHANG K X,SUN Z M,et al.LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis[J].Plant Physiology,2017,174(3):1348-1358.
[56] PRESTON J,WHEELER J,HEAZLEWOOD J,et al.AtMYB32 is required for normal pollen development in Arabidopsis thaliana[J].The Plant Journal,2004,40(6):979-995.
[57] YANG L,ZHAO X,RAN L Y,et al.PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar[J].Scientific Reports,2017,7:41209.
[58] ESPLEY R V,HELLENS R P,PUTTERILL J,et al.Red colouration in apple fruit is due to the activity of the MYB transcription factor MdMYB10[J].The Plant Journal,2007,49(3):414-427.
[59] PATTANAIK S,PATRA B,SINGH S K,et al.An overview of the gene regulatory network controlling trichome development in the model plant,Arabidopsis[J].Frontiers in Plant Science,2014,5:259.
[60] XU Z S,FENG K,QUE F,et al.A MYB transcription factor,DcMYB6,is involved in regulating anthocyanin biosynthesis in purple carrot taproots[J].Scientific Reports,2017(7):45324.
[61] PARK J S,Kim J B,CHO K J,et al.Arabidopsis R2R3- MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce(Lactuca sativa)[J].Plant Cell Reports,2008,27(6):985-994.
[62] ANWAR M,WANG G Q,WU J C,et al.Ectopic overexpression of a novel R2R3-MYB,NtMYB2 from Chinese narcissus represses anthocyanin biosynthesis in tobacco[J].Molecules,2018,23(4):781-799.
[63] CAVALLINI E,MATUS J T,FINEZZO L,et al.The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine[J].Plant Physiology,2015,167(4):1448-1470.
[64] ZHANG Y P,ZHANG J Q,SHAO C S,et al.Single-repeat R3 MYB transcription factors from Platanus acerifolia negatively regulate trichome formation in Arabidopsis[J].Planta,2019,249:861-877.
[65] CAO X,QIU Z K,WANG X T,et al.A putative R3 MYB repressor is the candidate gene underlying atroviolacium,a locus for anthocyanin pigmentation in tomato fruit[J].Journal of Experimental Botany,2017,68(21/22):5745-5758.
[66] COLANERO S,PERATA P,GONZALI S,et al.The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants[J].Frontiers in Plant Science,2018,9:830.
[67] VINCENZO D,CLIZIA V,GIORGIA B,et al.Genetic and epigenetic dynamics affecting anthocyanin biosynthesis in potato cell culture[J].Plant Science,2020,298:110597.
[68] ZHU H F,FITZAIMMONS K,KHANDELWAL A,et al.CPC,a single-repeat R3 MYB,is a negative regulator of anthocyanin biosynthesis in Arabidopsis[J].Molecular Plant,2009,2(4):790-802.
[69] NEMIE-FEYISSA D,OLAFSDOTTIR S M,HEIDARI B,et al.Nitrogen depletion and small R3-MYB transcription factors affecting anthocyanin accumulation in Arabidopsis leaves[J].Phyto Chemistry,2014,98:34-40.
[70] GATES D J,OLSON B J S C,CLEMENTE T E,et al.A novel R3 MYB transcriptional repressor associated with the loss of floral pigmentation in Iochroma[J].New Phytologist,2018,217(3):1346-1356.
[71] NAKATSUKA T,YAMADA E,SAITO M,et al.Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers[J].Plant Cell Reports,2013,32:1925-1937.
[72] SONG H,YI H,LEE M,et al.Purple Brassica oleracea var.capitata F. rubra is due to the loss of BoMYBL2-1 expression[J].BMC Plant Biology,2018,18(1):18.
[73] HU Q N,YANG L,LIU S D,et al.A repressor motif-containing poplar R3 MYB like transcription factor regulates epidermal cell fate determination and anthocyanin biosynthesis in Arabidopsis[J].Journal of Plant Biology,2016,59:525-535.
[74] ANDREA M,FRANCESCO E F,SERGIO I,et al.Identification of a new R3 MYB type repressor and functional characterization of the members of the MBW transcriptional complex involved in anthocyanin biosynthesis in eggplant(S.melongena L.)[J].PLOS ONE,2020,15(6):e0235081.
[75] MA D,REICHLET M,YOSHIDA K.et al.Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar[J].The Plant Journal,2018,96(5):949-965.
[76] JULIE M,KASHCHANDRA G R,AJAY J,et al.A genome-wide transcriptional analysis using Arabidopsis thaliana affymetrix gene chips determined plant responses to phosphate deprivation[J].Proceedings of the National Academy of Science of the USA,2005,102(33):11934-11939.
[77] BREVITZ J O,XIA Y,BLOUNT J,et al.Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis[J].Plant Cell,2000,12(12):2383-2394.
[78 ] HUANG D,TANG Z Z,FU J L,et al.CsMYB3 and CsRuby1 form an ‘activator-and- repressor loop for the regulation of anthocyanin biosynthesis in citrus[J].Plant and Cell Physiology,2020,61(2):318-330.
[79] HUANG D,WANG X,TANG Z Z,et al.Subfunctionalization of the Ruby2-Ruby1 gene cluster during the domestication of citrus[J].Nature Plants,2018,4(11):930-941.
