Cloning and Bioinformatics Analysis of tyeA Gene of Vibrio alginolyticus
2019-11-20*
*
1. Fisheries College, Guangdong Ocean University, Zhanjiang 524088, China; 2. Key Laboratory of Pathogenic Biology and Epidemiology of Aquatic Economic Animals of Guangdong Province, Zhanjiang 524088, China; 3. Key Laboratory of Aquatic Economic Animal Disease Control of Guangdong Provincial Department of Education, Zhanjiang 524088, China
Abstract [Objectives] This study aimed to clone the tyeA gene of Vibrio alginolyticus HY9901 strain and analyze its sequence by bioinformatics. [Methods] By referring to the entire genome sequence of V. alginolyticus on GenBank, specific primers were designed. According to the principle of PCR amplification, the target gene tyeA was amplified. By means of bioinformatics, the sequence of tyeA was further analyzed, and the phylogenetic tree of tyeA genes of Vibrio spp. and the corresponding subunit three- dimensional structure models were constructed. [Results] The length of the tyeA gene of V. alginolyticus strain HY9901 is 285 bp, and its theoretical molecular weight is 10.98 kD. According to prediction, there is no signal peptide or transmembrane region at the N- terminus of the sequence, and the amino acid sequence contains two casein kinase II phosphorylation sites. The results of protein subcellular localization prediction show that the TyeA protein is located in the cell membrane. The protein is unstable and non- hydrophilic. The tertiary structure of TyeA protein of V. alginolyticus is similar to that of Yersinia sp. According to prediction, TyeA has a major functional domain Pfam. In terms of secondary structure, alpha helix, random coil, extended strand and beta turn account for 85.11%, 7.45%, 4.26% and 3.19%, respectively. The homology of TyeA between V. alginolyticus and Vibrio parahaemolyticus is up to 83%, so they are classified into one cluster. [Conclusions]This study will help to further understand the regulation mechanism of type III secretion system in V. alginolyticus.
Key words Vibrio alginolyticus, tyeA gene, Gene cloning, Bioinformatics analysis
1 Introduction
Vibrioalginolyticusis a type of gram- negative bacterium, without spores or cysts, halophilic anaerobic. According to research, it is ubiquitous in seawater and estuaries in various parts of the world[1].Among theVibriospp. in seawater,V.alginolyticusaccounts for the largest proportion[2].V.alginolyticusis highly parasitic in marine animals[3]and is associated with diseases of marine animals, including fishes, shellfishes, shrimps and coral reefs[4-5].V.alginolyticusis a zoonotic pathogenic bacterium. In recent years, it has brought huge farming pressure and economic losses to the aquaculture industry. At the same time, the bacterium can cause symptoms such as food poisoning and diarrhea in humans[6-7].Type III secretion system (T3SS) is a strictly controlled virulence mechanism, and many gram- negative bacteria use it to colonize in hosts[8].T3SS consists of a cytoplasmic bulb, a matrix spanning the inner and outer membranes of the cell and an extracellular needle. When infecting the host,T3SS secretes virulence protein extracellularly, to the surface of the host cell, or even into the host cell, ultimately leading to death of the host cell[9].It has been found that TyeA is a regulator of T3SS.A large number of studies have shown that the YopN- TyeA heterodimer composed of TyeA and YopN plays a particularly important role in regulating the secretion of effector proteins[10-12]. WhenYersiniaspp. infect the host, the pre- assembled YSC- YOP type III secretion system injects the YOP effector into the host immune cells. The YopN- TyeA heterodimer is the center that controls the targeting activity of YSC- YOP[13-14].It has also been found that YopN- TyeA heterodimer plays a particularly important role in regulating and controlling the secretion and translocation of YSC- YOP component substrates[15].
In order to further study the regulation mechanism of the secretion of T3SS effector protein byV.alginolyticus, this paper analyzed the sequence and expressed thetyeAgene ofV.alginolyticus, and analyzed the secondary structure, tertiary structure and physicochemical properties of TyeA protein, so as to provide a theoretical basis for the subsequent research on the regulatory function oftyeAgene inV.alginolyticusand the development of related subunit vaccines.
2 Materials and methods
2.1 Strains and vectorTheV.alginolyticusstrain HY9901 was obtained from the virulent strain preserved in the Key Laboratory of Pathogenic Biology and Epidemiology of Aquatic Economic Animals of Guangdong Province; the Escherichia coil strain DH5α was provided by the Key Laboratory of Pathogenic Biology and Epidemiology of Aquatic Economic Animals of Guangdong Province; and the cloning vector pMD18- T was purchased from TaKaRa Company.
2.2 ReagentsThe used reagents mainly included RxTaq DNA polymerase (Takara, Japan), bacterial genomic DNA extraction kit (TransGen, China), DNA gel recovery kit (TransGen, China), standard molecular weight DNA Marker (TaKaRa, Japan) and TSB medium. Other reagents of analytical grade were imported or domestically purchased. The antibiotic ampicillin (Ap) had a mass concentration of 100 μg/mL.
2.3 Instruments and equipmentPCR instrument (Bio- Rad, Canda; TaKaRa, Japan); constant- pressure and constant- current electrophoresis apparatus (Liuyi, China); benchtop high- speed refrigerated centrifuge (Eppendorf, America);CHA- S electrothermal constant- temperature incubator (Boxun, China); ultra- clean workbench (Huilong, China).
2.4 Extraction of total DNA fromV.alginolyticusstrain HY9901The extraction steps of total DNA fromV.alginolyticuswere as follows:
(i) Expanding culture. TheV.alginolyticusstrain preserved at -80℃ was taken out, inoculated into TSB (2%) liquid medium and subjected to shaking culture at 28℃ for 18 h.
(ii) Enrichment. Six portions, 1 mL for each, of the bacterial solution were transferred into six centrifuge tubes and centrifuged at 10 000 r/min for 5 min. After discarding the supernatant, the precipitate in the six tubes was mixed in a centrifuge tube and concentrated.
(iii) Vortexing, resuspension and neutralization. According to the instruction of the kit, the centrifuge tube was added with 250 μL of vortex liquid, vortexed until the precipitate disappeared, added with 250 μL of resuspension, inverted 4-6 times, added with 350 μL of neutralization solution, inverted 4-6 times, and centrifuged at 10 000 r/min for 5 min in success.
(iv) Washing of spin column. The supernatant was transferred to the spin column, centrifuged at 10 000 r/min for 1 min, added with 500 μL of washing solution, and centrifuged at 10 000 r/min for 30 s. The above operation was repeated twice. Finally, the empty column was centrifuged at 10 000 r/min for 1 min.
(v) Elution of DNA. After the operation above, the empty column was added with 20 μL of elution solution, incubated for 2 min at room temperature, and centrifuged at 14 000 r/min for 2 min. The eluate was collected, incubated for 2 min at room temperature, and centrifuged at 14 000 r/min for 2 min.
(vi) Preservation. After completing the above steps, the bacterial genomic DNA was successfully extracted, and it was store in a refrigerator at -20℃ for later use.
2.5 Cloning of thetyeAgeneBased on the entire gene sequence ofV.alginolyticuson Genbank, a pair of primers was designed. The upstream primer P1 was 5′- ATGGCTTATCAAGTTTCTA- 3′, and the downstream primer P2 was 5′- TCAATCCAACTCATCTTCC- 3′.
The PCR reaction conditions were as follows: pre- denaturation at 95℃ for 5 min; 95℃ for 30 s, (60, 64, 65 and 67)℃ for 30 s, 72℃ for 60 s, 33 cycles, extension at 72℃ for 10 min. A small amount of the PCR product was examined by 1% agarose gel electrophoresis.
2.6 Purification of the PCR productThe remaining PCR product was purified according to the instruction of the DNA gel recovery kit.
2.7 Ligation and transformation of PCR productThe PCR product was ligated into the pMD18- T vector overnight at 4℃ in a refrigerator.E.coliDH5α competent cells were then transformed. The main steps of transformation were as follows.
(i) Adding 5 μL of ligation product to 50 μL of recipient bacterium; (ii) Ice bath for 30 min, water bath (42℃) for 90 s, and placing on ice for 3-5 min; (iii) Adding 1 000 μL of resistant LB medium (pcDNA3.1 resistance of Amp), and placing on a shaker at 37℃, 200 r/min for 1 h; (iv) Centrifuging at 3 000 r/min for 2 min at room temperature, discarding the excess supernatant and keeping 100 μL, suspending the remained supernatant gently, coating to an Ap+/LB plate containing IPTG/X- Gal; (v) Placing for 30 min and inverting at 37℃ for 10-12 h.
2.8 Selection and sequencing of positive clonesScreening was performed on Ap+/LB plates containing IPTG/X- Gal. Positive clones were picked and sent to Shanghai Sangon Bioengineering Technology Service Co., Ltd. for sequencing.
2.9 Bioinformatics analysis of DLD ofV.alginolyticusstrain HY9901[16-17]Homology alignment and similarity analysis of genetic sequences were performed using NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Homologous alignment analysis of amino acid sequences was performed using DNAMAN. Using ExPASyProteomics Server software (http://web.expasy.org/protparam/), the physicochemical properties ofV.alginolyticusTyeA protein were analyzed, the amino acid sequence was deduced, and the open reading frame (ORF), molecular weight calculated value (Mw) and theoretical isoelectric point (pI) were determined. By using SignalP 4.0 Server software (http://www.cbs.dtu.dk/services/SignalP), the sequence of the signal peptide was predicted. By using TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM), the transmembrane domain was predicted. By using SoftBerry- Psite (http://linux1.softberry.com/berry.phtml? Topic=psite&group= programs & subgroup=proloc),the distribution of functional sites in the amino acid sequence was predicted. The promoter was analyzed using Promoter 2.0 software. Subcellular localization was predicted using PSORT II Prediction (http://psort.hgc.jp/form2.html). The phylogenetic tree was constructed by neighbor- joining using Clastal 2.0 and MEGA 5.0 software. The three- dimensional structure of the TyeA protein ofV.alginolyticusstrain HY9901 was constructed using the SWISS- MODEL (http://www.swissmodel.expasy.org/) program. The pathways involved were analyzed using the Kegg software (http://www.genome.jp/kegg/).The STRING database(http://string.embl.de/) was used for searching for interactions between protein networks.
3 Results
3.1 Cloning oftyeAgeneAccording to the whole gene sequence ofV.alginolyticuson Genbank, specific primers were designed for the 3′ and 5′ termini. Using the designed primers, PCR amplification was performed. The PCR product was examined by 1% agarose gel electrophoresis, and a clear, bright and concentrated band, in size of 285 bp, was obtained, in agreement with the theoretical result, indicating successful amplification (Fig.1). Positive colonies were analyzed by sequencing. The results show that thetyeAgene has a 285 bp open reading frame (ORF) encoding 94 amino acids. The accession number is MN328351 in GenBank.

Note: M, DL500 DNA molecular marker; 1-5, PCR product.
Fig.1 Cloning of thetyeAgene
3.2 Physicochemical properties of TyeA proteinThe TyeA protein ofV.alginolyticusstrain HY 9901 was analyzed using ExPASy software. The results showed that the total number of atoms is 1 532 and the molecular structural formula is C489H759N125O156S3. The theoretical molecular weight of the protein is 10.981 33 kD and the theoretical pI value is 4.11. The instability coefficient is 46.69, greater than the threshold of 40, suggesting that the protein is unstable. The fat coefficient is 102.77.Thetotal average hydrophilic coefficient is -0.023 5, suggesting that the protein is non- hydrophilic. The total number of acidic amino acid residues (Asp+Glu) is 23, and the total number of basic amino acid residues (Arg+Lys) is 8. The N- terminus is methionine (Met).The half- lives of the protein expressed in yeast andE.coliare greater than 20 and 10 h, respectively. The half- life of the protein expressed in mammalian reticulocytesinvitrois 30 h.
3.3 Sequence analysis of TyeA proteinThe structure of signal peptide at the N terminus was predicted using the SignalP 4.0 Server program and no signal peptide was found. The prediction using the TMHMM Server v.2.0 program shows that there was no transmembrane region. The prediction using the SoftBerry- Psite (predict protein) program shows that the amino acid sequence contains two casein kinase II phosphorylation sites (Fig.2). The results of protein subcellular localization prediction show that TyeA is located in the cell membrane, indicating that the protein encoded by this gene is a cell membrane protein.
3.4 Homology and evolution analysis of TyeABLAST analysis found that TyeA ofV.alginolyticushas high homology with those of other species, and its homology with the TyeA ofVibrioparahaemolyticusis as high as 83%. The results of multiple- sequence similarity comparison indicate that TyeA inVibriospp. is highly conserved (Fig.3).Using the neighbor- joining method of MEGA 5.0, the phylogenetic tree of TyeA proteins of some bacterial species was constructed. The results show that the TyeA protein ofV.alginolyticusstrain HY9901 and the TyeA protein ofV.parahaemolyticusare classified into one cluster, indicating that they are closely related (Fig.4).

Note: "*" represents a terminator, and "__" marks casein kinase II phosphorylation sites.
Fig.2 Sequence analysis of thetyeAgene
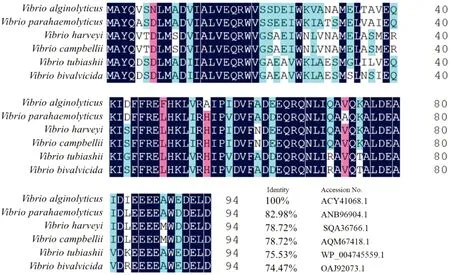
Fig.3 Similarity comparison between TyeA amino acid sequences
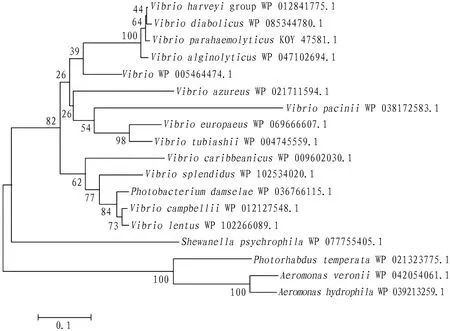
Fig.4 Phylogenetic tree for TyeA amino acid sequences
3.5 Prediction of functional domains and secondary structure of TyeAUsing the SMART program, the functional domain of the TyeA protein was predicted. It was found that TyeA has a major functional domain Pfam (Fig.5).The secondary structure of the TyeA protein ofV.alginolyticusstrain HY9901 was predicted by SOPMA software. The results show that in the secondary structure of TyeA, alpha helix, random coli, extended strand and beta turn account for 85.11%, 7.45%, 4.26% and 3.19%, respectively (Fig.6).

Fig.5 Domain prediction of TyeA protein
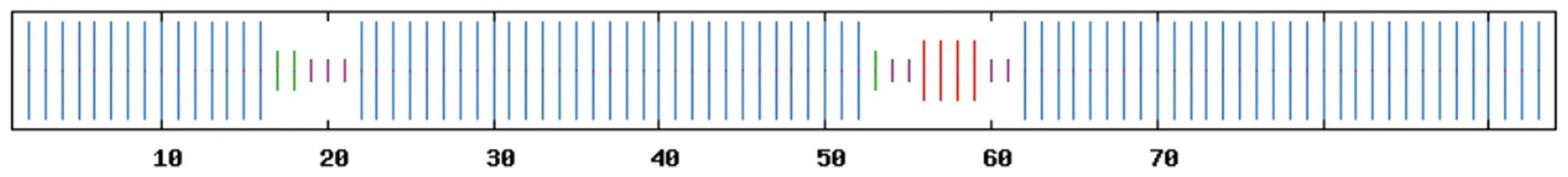
Note: Blue, alpha- helix; purple, random coil; red, extended strand; green, beta- turn.
Fig.6 Prediction map of secondary structure of TyeA protein
3.6 Subunit structure of TyeAThe TyeA amino acid sequence was submitted to the SWISS- MODEL program, and many similar homologous sequences were obtained by searching the database from thetyeAgene sequence. One or some of them were selected as templates. After comparison, adjustment and energy minimization, the tertiary structure model of a single subunit of TyeA was obtained (Fig.7). By comparison, it was found that the TyeA single subunit tertiary structure model ofV.alginolyticusis similar to that ofYersiniasp. (Yersiniasp.:QMEAN, -1.24; Cβ, 1.11;ALL atom, 1.51; resolution, 1.30; torsion, -2.1;V.alginolyticus: QMEAN, -0.17; Cβ, 1.11; ALL atom, 1.53; resolution, 1.19; torsion, -1.01).
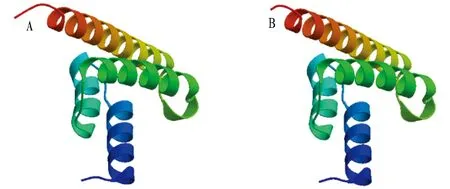
Note: A.Vibrioalginolyticusstrain HY9901; B.Yersiniasp.
Fig.7 Tertiary structure models of subunits of TyeA proteins inVibrioalginolyticusstrain HY9901 and Yersinia sp.
4 Conclusions and discussion
4.1 ConclusionsThis study successfully cloned thetyeAgene ofV.alginolyticusstrain HY9901, of which the bioinformatics analysis was carried out. The results show that the TyeA protein ofV.alginolyticusstrain HY9901 is a non- hydrophilic unstable protein consisting of 94 amino acids with a size of 10.98 kDa; and its secondary structure contains four conformations of α- helix, β- turn, random coil and extended chain. The homology comparison of amino acid sequences shows that thetyeAgene ofV.alginolyticusstrain HY9901 has the highest homology withVibrioharveyi, up to 99%, has the similar homologies withV.parahaemolyticusandVibriodiabolicus(77%), and has a lower homology withVibriocampbellii(77%).The analysis results of phylogenetic tree are consistent with the homology analysis results oftyeAnucleotide sequences, indicating thattyeAnucleotide sequence is highly conserved among the same species. The homology oftyeAnucleotide sequence increases as the relationship becomes close. It can also be speculated that the TyeA protein ofV.alginolyticusstrain HY9901 may have a similar function or effect as TyeA ofV.harveyi.
4.2 DiscussionThe homology and evolutionary analysis of TyeA revealed that TyeA ofV.alginolyticushas high homology with TyeA of other species, indicating that thetyeAgene is highly conserved. The tertiary structure model constructed by using SWISS- MODEL software shows that the TyeA subunit tertiary structure ofV.alginolyticusis similar to that ofYersiniasp. ThetyeAgene is a very important regulatory gene in the effector secretion of the type III secretion system inYersiniaspp. A large number of studies have found that YopN- TyeA heterodimer plays a particularly important role in regulating and controlling the secretion and translocation of YSC- YOP component substrates[8-10].
The Yopn- TyeA complex found in the study constitutes a "plug" that blocks secretion, preventing the secretion of effector proteins under non- secretory conditions[18].The Yopn- TyeA complex can sense calcium ion concentration, thus playing an important role in the switch of the Yops secretion channel.Invitro, the secretion of the YOP effector protein is blocked in the presence of calcium and triggered by the removal of calcium[19].In the body, the level of calcium is sufficient to prevent the secretion of YOP until the bacteria are exposed to eukaryotic cells. Such a regulatory mechanism avoids excessive secretion of effector proteins. A strain ofYersiniapestislacking the expression of YopN, TyeA, SycN, YscB or LcrG all can selectively secrete Yops effector proteins regardless of the presence or absence of calcium[20]. Therefore, the function of the Yopn- TyeA complex protein is to prevent the secretion of the Yop effector protein until the activation signal of T3SS is encountered.
So far, how the YopN- TyeA complex blocks the exit channel on the T3SS unit is still not known. But it has been found that the TyeA homologous proteins PCR1 and PcrG (LcrG inYersinia) inPseudomonasaeruginosaassemble with the intact intimal protein PcrD (YscV) to block entry of the substrate into the secretory channel[21].
At present, there are few reports on the regulation of effector proteins in the type III secretion system by thetyeAgene inV.alginolyticus. This experiment provides a basis for the subsequent research and development. In the future, thetyeAandvopNgenes inV.alginolyticuswill be knocked out to verify the regulation of related proteins and further study whether TyeA and VopN also present in complex form and their regulation mechanism on effector proteins, thereby further understanding the regulation mechanism of type III secretion system inV.alginolyticus.
杂志排行
Asian Agricultural Research的其它文章
- Research on Intelligent Monitoring System Based on Raspberry Pi 3
- Application of New Modified Bentonite in Decolorization of Simulated Printing and Dyeing Wastewater
- Protection and Renovation Strategies for Ancient Town Architectural Space Design from the Perspective of Place Spirit Theory: A Case Study of Hekou Ancient Town, Yanshan County of Jiangxi Province
- Coordinated Development Level of Economic Society and Ecological Environment in Binzhou City
- Teaching Reform of Building Construction Course under the Background of New Digital Technology
- Effects of Different Breeds, Number of Squabs and Temperature on Production Performance of Pigeons
