lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis
2019-10-28WeiDaiJiaLiangDaiMaoHuaTangMuShiYeShuoFang
Wei Dai, Jia-Liang Dai, Mao-Hua Tang, Mu-Shi Ye, Shuo Fang
Abstract BACKGROUND Hepatocellular carcinoma (HCC) has become a great threat for people's health.Many long noncoding RNAs are involved in the pathogenesis of HCC. SNHG15,as a tissue specific long noncoding RNAs, has been studied in many human cancers, except HCC.AIM To explore the regulatory mechanism of SNHG15 in HCC.METHODS In the present research, 101 HCC patient samples, two HCC cell lines and one normal liver cell line were used. RT-qPCR and Western blot analysis were applied to detect SNHG15, miR-490-3p and histone deacetylase 2 (HDAC2)expression. The regulatory mechanism of SNHG15 was investigated using CCK-8, Transwell and luciferase reporter assays.RESULTS Our research showed that up-regulation of SNHG15 was found in HCC and was related to aggressive behaviors in HCC patients. Moreover, knockdown of SNHG15 restrained HCC cell proliferation, migration and invasion. In addition,SNHG15 served as a molecular sponge for miR-490-3p. Further, miR-490-3p directly targets HDAC2. HDAC2 was involved in HCC progression by interacting with the SNHG15/miR-490-3p axis.CONCLUSION In conclusion, long noncoding RNA SNHG15 promotes HCC progression by mediating the miR-490-3p/HDAC2 axis in HCC.
Key words: SNHG15; miR-490-3p; Hepatocellular carcinoma; Histone deacetylase 2;Pathogenesis; Regulatory mechanism
INTRODUCTION
Hepatocellular carcinoma (HCC) is a high-risk, highly harmful malignant tumor.Recently, HCC has jumped to the top three cancers in the Asia-Pacific region including China, posing a threat to human health[1]. Moreover, the pathogenesis of HCC is not fully understood. It has been reported that environmental and dietary factors affect the occurrence of HCC[2]. At present, surgery is still the first choice for the treatment of HCC. However, the recurrence rate of HCC is still high, and the 5-year survival rate is generally below 50%[3]. Thus, exploring the pathogenesis of HCC to screen for effective diagnostic markers and therapeutic targets is very urgent for HCC patients.
Long non-coding RNAs (lncRNAs) are involved in the occurrence of human diseases and cancers[4]. Recently, the presence of increased numbers of lncRNAs have been demonstrated to be involved in HCC progression. For instance, lncRNA HOXDAS1 was overexpressed in HCC and accelerated cell proliferation and cell cycle progression through the MEK/ERK pathway[5]. On the contrary, lncRNA TPTEP1 was down-regulated in HCC and restrained HCC progression by suppressing STAT3 phosphorylation[6].
The specific functions of lncRNA SNHG15 in human cancers have caught our attention. It had been reported that SNHG15 was down-regulated and served as a tumor suppressor in thyroid cancer[7]. However, SNHG15 was up-regulated in prostate cancer and acted as a tumor promoter by mediating miR-338-3p[8]. These results indicate that expression of SNHG15 is tissue specific. In particular, SNHG15 expression was increased in HCC and predicted poor survival in HCC patients[9].However, the specific role of SNHG15 is still unclear and needs to be elucidated.
In addition, it is well-known that lncRNAs exert an effect in human diseases by inhibiting the expression and biological functions of miRNAs[10]. SNHG15 was found to enhance colorectal cancer cell viability through down-regulation of miR-338-3p[11].In this study, miR-490-3p was predicted to have binding sites with SNHG15.Moreover, miR-490-3p sponging by CircSLC3A2 was reported to regulate HCC progression[12]. It had been shown that miR-490-3p expression was reduced in HCC and restrained autophagy[13]. However, whether SNHG15 regulates HCC progression via regulating miR-490-3p remains unknown.
Besides that, histone deacetylase 2 (HDAC2) was predicted as a target of miR-490-3p in this study. Moreover, HDAC2 expression had been reported to be increased in HCC tissues, which was related to adverse prognosis[14]. Additionally, miR-145 was proposed to act as a tumor inhibitor via binding to HDAC2 in liver cancer[15].However, the interaction between miR-490-3p and HDAC2 has not been reported in previous studies. Therefore, their relationship in HCC was investigated in our study.Further, the regulatory mechanism of lncRNA SNHG15 with the miR-490-3p/HDAC2 axis was elucidated in HCC progression. Our results will contribute to better understand the role of lncRNA SNHG15 in HCC progression.
MATERIALS AND METHODS
Clinical tissues
One hundred and one HCC patients in the Affiliated Hospital of Guangdong Medical University, the second Affiliated Hospital of Guangdong Medical University and the Seventh Affiliated Hospital of Sun Yat-sen University participated in this research.The clinical features of the patients were shown in Table 1. Among them, 33 randomly selected HCC tissues and paired adjacent non-neoplastic liver tissues were applied for further experiments. Before the experiment, written informed consent was collected from all HCC patients. Moreover, patients with HCC did not receive any treatment except for surgery. The permission of this study was obtained from the Institutional Ethics Committee of the Affiliated Hospital of Guangdong Medical University, the second Affiliated Hospital of Guangdong Medical University and the Seventh Affiliated Hospital of Sun Yat-sen University.
Cell culture
HCC cell lines HuH-1, HuH-7 and normal human liver cells L-O2 were obtained from the Japanese Collection of Research Bioresources Cell Bank (JCRB, Shanghai, China).The growth conditions of these cells are 5% CO2, 37 °C and culture solution (90%DMEM + 10% fetal bovine serum).
Cell transfection
SNHG15 siRNA (si-SNH G15), pcDNA3.1-SNHG15 vectors, HDAC2 siRNA (si-HDAC2) and miR-490-3p mimics or inhibitor were purchased from Genepharma(Shanghai, China). Next, the siRNAs, vectors, mimics or inhibitors (20 nM) were transfected into HuH-1 or L-O2 cells using Lipofectamine 2000 (Invitrogen).Untreated cells were used as the controls.
RT-qPCR
Total RNA isolation was performed with TRIzol reagent (Sigma, United States). The cDNA was reverse transcribed using microRNA reverse transcription kit (TAKARA,Dalian, China). RT-qPCR assay was performing using SYBR Green Master Mix II(TAKARA). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference for RNA or protein. Relative expression of SNHG15, miR-490-3p and HDAC2 were detected with the 2-ΔΔctmethod.
Western blot analysis
Protein samples were lysed using RIPA buffer (Beyotime, Shanghai, China). Next,10% SDS-PAGE was used to separate protein. Protein samples were transferred to PVDF membranes and blocked with 5% nonfat milk. Next, protein samples were incubated with HDAC2 and GAPDH primary antibodies (Abcam, Shanghai, China)overnight at 4 °C. After washing, secondary antibodies (Abcam, United States) were incubated with the protein samples for 1 h. Finally, ECL kit (Beyotime) was used to assess protein bands.
CCK-8 assay
Transfected HuH-1 cells were incubated for 24 h (at 37 °C, 5% CO2) in 96-well plates.Next, HuH-1 cells (3 × 103/well) were incubated in DMEM medium for 24, 48, 72 and 96 h. CCK-8 (Dojindo, Kumamoto, Japan) was added and incubated with the cells for 4 h. Finally, the optical density at 450 nm was observed by a microplate reader(Molecular Devices) to assess cell viability.
Transwell assay
First, cell invasion was detected in the upper chamber with Matrigel (BD Biosciences,United States). After 30 min, Transwell upper chamber was added with HuH-1 or LO2 cell suspension (5 × 103cells/well). Next, lower chamber was added with DMEM medium (10% fetal bovine serum). After 24 h, 0.1% crystal violet was applied to stain the moved cells. For cell migration, 5 × 103transfected cells were placed in the upperchambers without Matrigel. Observation and photographing were performed by a light microscope.
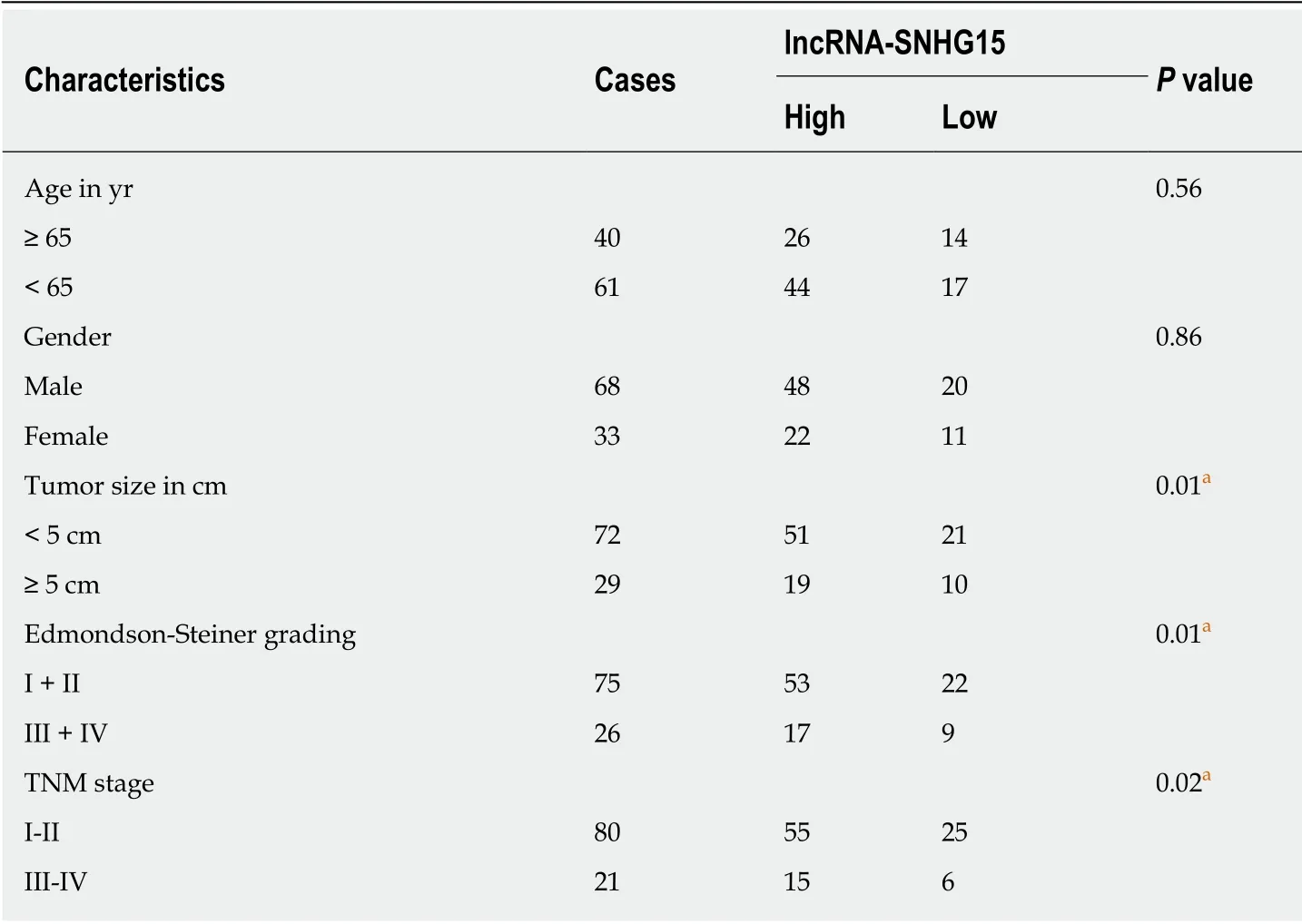
Table 1 Relationship between lncRNA-SNHG15 expression and the clinic-pathological characteristics in hepatocellular carcinoma patients
Dual luciferase reporter assay
Reporter plasmids of SNHG15 (wt-SNHG15 and mut-SNHG15) and HDAC2 (wt-HDAC2 and mut-HDAC2) were cloned into empty pGL3 vectors (GenePharma,Shanghai, China. Then, the above reporter vectors were transfected into HuH-1 cells with miR-490-3p mimics. After 48 h, luciferase activities were examined by dualluciferase reporter assay system (Promega, United States). HuH-1 cells with empty pGL3 vectors were used as the control.
Statistical analysis
Data were analyzed by SPSS 17.0 or Graphpad Prism 6, which are shown as mean ±standard deviation. One-way ANOVA and Student's t-test were employed to compare differences among multiple groups. Chi-squared test was used to analyze the association between SNHG15 and clinical features in HCC patients. P < 0.05 indicated significant difference.
RESULTS
Dysregulation of SNHG15 in HCC
First, a difference in SNHG15 expression was detected in HCC tissues. We found higher SNHG15 expression in HCC tissues than normal tissues (Figure 1A). Similarly,up-regulation of SNHG15 was found in HuH-1 and HuH-7 cells compared to L-O2 cells (Figure 1B). The expression of SNHG15 in HuH-1 cells is higher than HuH-7 cells. Therefore, HuH-1 cells were used for further experiments. In addition, the association between HCC clinical features and abnormal expression of SNHG15 was analyzed. Tumor size, TNM stage and degrees of differentiation in HCC under Edmondson-Steiner grading system were associated with abnormal SNHG15 expression in HCC patients (Table 1). These results suggested that SNHG15 may affect tumorigenesis of HCC.
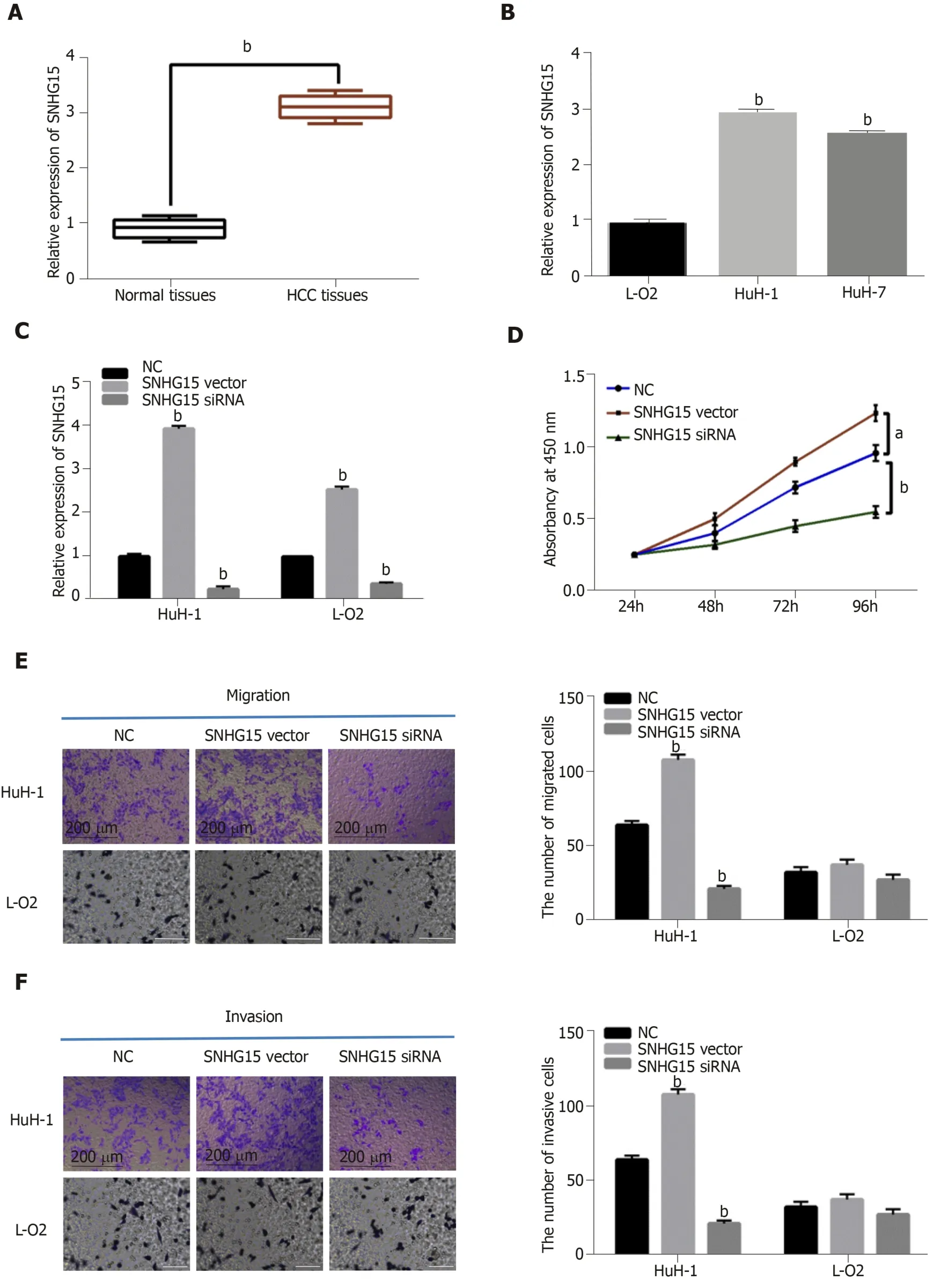
Figure 1 The dysregulation of lncRNA-SNHG15 in hepatocellular carcinoma. A: SNHG15 expression in 33 randomly selected hepatocellular carcinoma tissues and normal tissues; B: SNHG15 expression in HuH-1, HuH-7 and L-O2 cell lines; C: lncRNA-SNHG15 expression in HuH-1 and L-O2 cells containing SNHG15 siRNA or vector; D: Cell proliferation in HuH-1 and L-O2 cells with SNHG15 siRNA or vector; E: Cell migration in HuH-1 cells with SNHG15 siRNA or vector; F: Cell invasion in HuH-1 cells with SNHG15 siRNA or vector. Untreated cells were used as control (NC). aP < 0.05, bP < 0.01. SNHG15: Small nucleolar RNA host gene 15; HuH-1:Hepatocellular carcinoma cell line; HuH-7: Hepatocellular carcinoma cell line; L-O2: Normal human liver cell lin3; siRNA: Small interfering RNA.
Next, SNHG15 siRNA or SNHG15 overexpression vector was transfected into HuH-1 cells to explore its role in HCC. The transfection efficiency was confirmed by RT-qPCR (Figure 1C). CCK-8 assay suggested that knockdown of SNHG15 suppressed proliferation of HuH-1 cells, while up-regulation of SNHG15 accelerated cell proliferation in HuH-1 cells (Figure 1D). In addition, the Transwell assay showed that cell migration was also promoted by up-regulation of SNHG15 and restrained by knockdown of SNHG15 (Figure 1E). We also found that SNHG15 siRNA decreased its expression in L-O2 cells, whereas SNHG15 overexpression vector enhanced its expression in L-O2 cells (Figure 1C). However, up-regulation or down-regulation of SNHG15 had little effect on cell migration and invasion in normal human liver cells LO2 (Figure 1E and F). These findings indicated the potential carcinogenesis of SNHG15 in HCC.
Direct interaction of SNHG15 and miR-490-3p in HCC
To explore the regulatory mechanism of SNHG15, its target miRNA was searched in starBASEv2.0 database (http://starbase.sysu.edu). It predicted that SNHG15 has potential binding sites with miR-490-3p (Figure 2A). Luciferase reporter assay showed that miR-490-3p mimics decreased SNHG15-wt luciferase activity, but not SNHG15-mut (Figure 2B). Next, lower miR-490-3p expression was identified in HCC tissues compared to normal tissues (Figure 2C). Furthermore, SNHG15 was negatively associated with miR-490-3p expression in HCC tissues (Figure 2D). In HuH-1 cells,knockdown of SNHG15 enhanced miR-490-3p expression, while up-regulation of SNHG15 reduced miR-490-3p expression (Figure 2E). Interestingly, down-regulation or overexpression of miR-490-3p could also inversely regulate SNHG15 expression in HuH-1 cells (Figure 2F). To further explain their interaction, the SNHG15 vector was transfected into HuH-1 cells containing miR-490-3p mimics. Moreover, the increased miR-490-3p expression mediated by its mimics was weakened by up-regulation of SNHG15 (Figure 2G). Functionally, miR-490-3p induced inhibition of cell proliferation was also restored by SNHG15 up-regulation (Figure 2H). Similarly, up-regulation of SNHG15 also weakened the suppressive effect of miR-490-3p on migration and invasion of HCC cells (Figure 2I and 2J). Based on the results, we hypothesized that SNHG15 may accelerate HCC progression via molecular sponging of miR-490-3p.
miR-490-3p directly targets HDAC2
TargetScan (http://www.targetscan.org) predicted that miR-490-3p has a binding site to HDAC2 (Figure 3A). Dual-luciferase reporter assay indicated that miR-490-3p mimics reduced HDAC2-wt luciferase activity, but not HDAC2-mut (Figure 3B).Moreover, miR-490-3p mimics were found to decrease HDAC2 expression, while a miR-490-3p inhibitor up-regulated HDAC2 in HuH-1 cells (Figure 3C and D). These studies indicated that miR-490-3p directly targets HDAC2. In addition, the dysregulation of HDAC2 was identified in HCC tissues. Furthermore, HDAC2 was up-regulated in HCC tissues compared with normal tissues (Figure 3E), and miR-490-3p had an inverse correlation with HDAC2 expression in HCC tissues (Figure 3F). On the contrary, a positive correlation between SNHG15 and HDAC2 expression was identified in HCC tissues (Figure 3G). According to these results, we suspected that HDAC2 may be involved in HCC progression by affecting the SNHG15/miR-490-3p axis.
HDAC2 regulated HCC progression through mediating SNHG15/miR-490-3p
Finally, HuH-1 cells containing HDAC2 siRNA were transfected with a SNHG15 vector or a miR-490-3p inhibitor to further explain their interaction. First, we found that mRNA and protein expression of HDAC2 was down-regulated by HDAC2 siRNA. However, the decreased expression of HDAC2 was reversed by the miR-490-3p inhibitor or SNHG15 vector (Figure 4A and B). Functionally, the inhibition of cell proliferation induced by HDAC2 siRNA was restored by miR-490-3p downregulation or SNHG15 up-regulation (Figure 4C). Similarly, the reverse effects of the miR-490-3p inhibitor or SNHG15 vector on migration and invasion were also identified in HuH-1 cells with HDAC2 siRNA (Figure 4D and 4E). These results showed that the SNHG15/miR-490-3p axis exerts an effect in the development of HCC progression by interacting with HDAC2.
DISCUSSION
As potential therapeutic targets, many lncRNAs have been found to regulate HCC progression. For example, lncRNA-MNX1-AS1 was found to accelerate HCC development via regulating the miR-218-5p/COMMD8 axis[16]. In our research,lncRNA SNHG15 also served as an oncogene in HCC. In particular, up-regulation of SNHG15 was identified in HCC, which was related to adverse clinical outcomes in HCC patients. Functionally, knockdown of SNHG15 decreased migration, invasion and proliferation of HCC cells. At the same time, SNHG15 was found to accelerate HCC progression by targeting the miR-490-3p/HDAC2 axis, indicating that SNHG15 may be a therapeutic target for HCC patients.
Consistent with our results, up-regulation of SNHG15 was also detected in colorectal carcinoma and lung cancer[17,18]. Functionally, up-regulated expression of SNHG15 accelerated proliferation and invasion of gastric cancer cells[19]. In addition,Kong et al[20]reported that SNHG15 facilitated human breast cancer cell migration by sponging miR-211-3p. The same effect of SNHG15 was also identified in HCC, which was consistent with previous studies. Besides that, Zhang et al[9]demonstrated that abnormal expression of SNHG15 was related to TNM stage and vein invasion in HCC patients. Similarly, tumor size, Edmondson-Steiner grading and TNM stage were also related to SNHG15 expression in our research. Further, SNHG15 was confirmed as a molecular sponge for miR-490-3p in this study, which has not been found in previous studies.
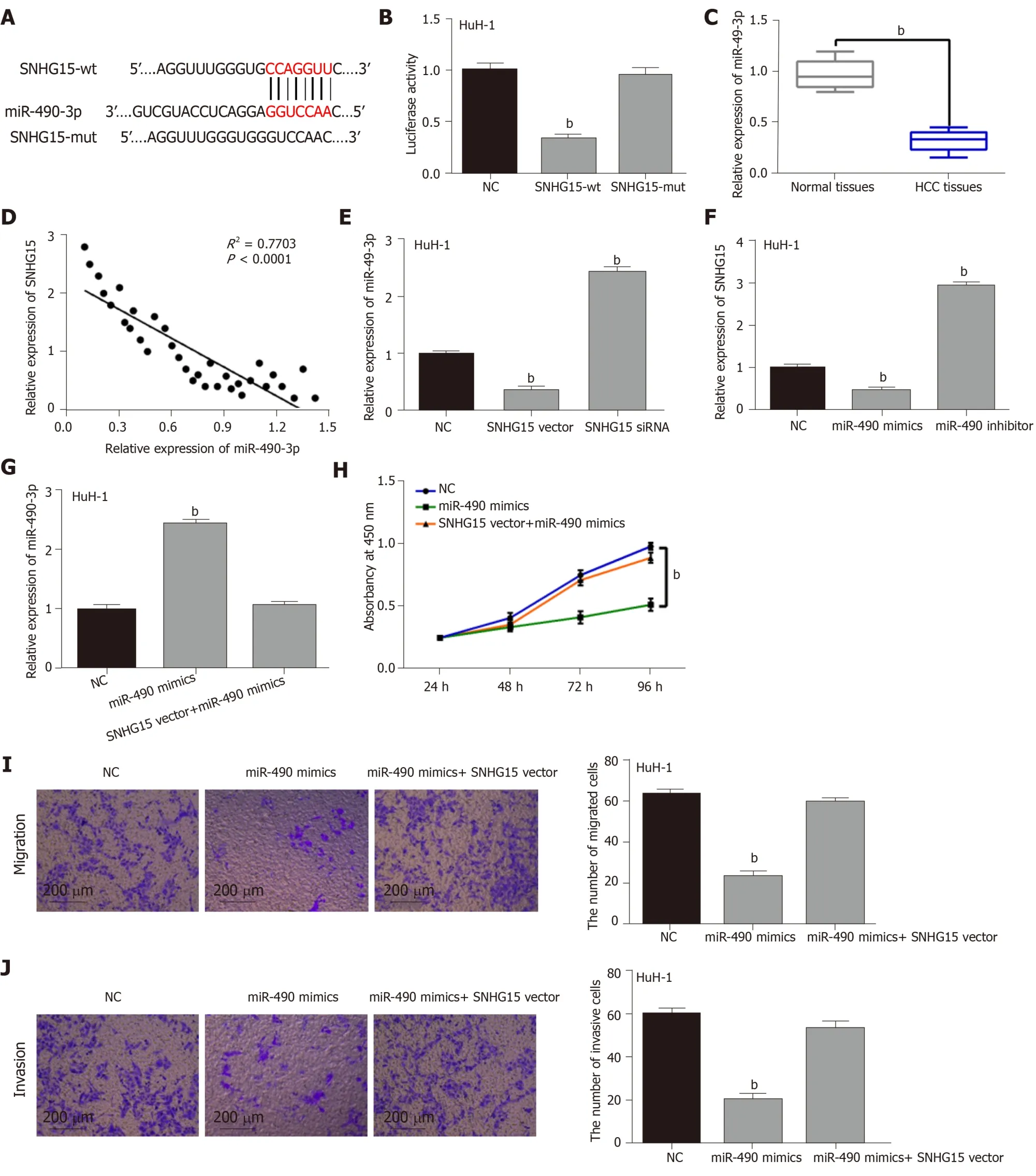
Figure 2 Direct interaction of lncRNA-SNHG15 and miR-490-3p. A: The binding sites between lncRNA-SNHG15 with miR-490-3p; B: Luciferase reporter assay; C:miR-490-3p expression in 33 randomly selected HCC tissues and normal tissues; D: SNHG15 had negative correlation with miR-490-3p expression in 33 randomly selected HCC tissues; E: miR-490-3p expression regulated by SNHG15 siRNA or vector in HuH-1 cells; F: SNHG15 expression in HuH-1 cells containing miR-490-3p mimics or inhibitor; G: miR-490-3p expression in cells with miR-490-3p mimics or miR-490-3p mimics + SNHG15 vector. H: Cell proliferation in HuH-1 cells containing miR-490-3p mimics or miR-490-3p mimics + SNHG15 vector; I: Cell migration in HuH-1 cells containing miR-490-3p mimics or miR-490-3p mimics + SNHG15 vector;J: Cell invasion in HuH-1 cells containing miR-490-3p mimics or miR-490-3p mimics + SNHG15 vector. Untreated cells were used as control (NC). bP < 0.01.SNHG15: Small nucleolar RNA host gene 15; miR-490-3p: Hsa-miR-490-3p; HCC: Hepatocellular carcinoma; siRNA: Small interfering RNA; HuH-1: Hepatocellular carcinoma cell line.
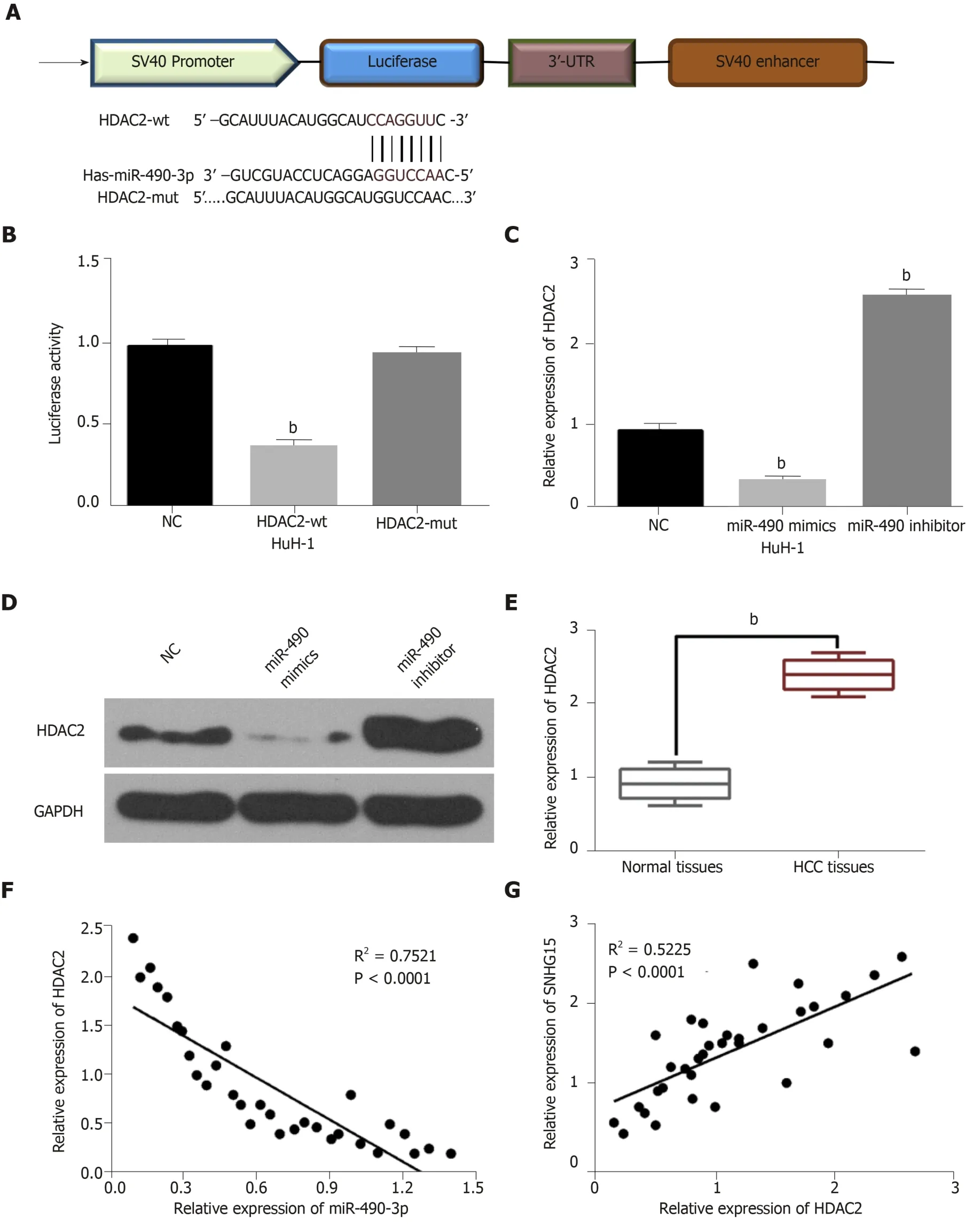
Figure 3 miR-490-3p directly targets HDAC2. A: The binding sites between miR-490-3p and HDAC2; B: Luciferase reporter assay; C: miR-490-3p regulated HDAC2 expression in HuH-1 cells; D: miR-490-3p regulated HDAC2 expression in HuH-1 cells; E: HDAC2 expression in 33 randomly selected hepatocellular carcinoma tissues and normal tissues; F: HDAC2 was negatively correlated with miR-490-3p in 33 randomly selected HCC tissues; G: lncRNA-SNHG15 was positively correlated with HDAC2 in 33 randomly selected HCC tissues. Untreated cells were used as control (NC). bP < 0.01. miR-490-3p: Hsa-miR-490-3p; HDAC2: Histone deacetylase 2; HuH-1: Hepatocellular carcinoma cell line; HCC: Hepatocellular carcinoma; SNHG15: Small nucleolar RNA host gene 15.
The dysregulation of miR-490-3p has been identified in the tumorigenesis of human cancers. Down-regulation of miR-490-3p had been found in ovarian epithelial carcinoma and colorectal cancer[21,22]. In this research, miR-490-3p was also downregulated in HCC. Furthermore, miR-490-3p overexpression was found to decrease migration, invasion and proliferation of HCC cells. Consistently, the inhibitory role of miR-490-3p had been also identified in lung cancer and colorectal cancer[23,24]. miR-490-3p promoted viability and epithelial to mesenchymal transition of HCC cells[25], which also supports the accuracy of our results. In addition, reciprocal suppression between SNHG15 and miR-490-3p was identified in HCC. Furthermore, SNHG15 upregulation abolished the suppressive effect of miR-490-3p in HCC progression. These findings imply that SNHG15 accelerated HCC development by sponging miR-490-3p.
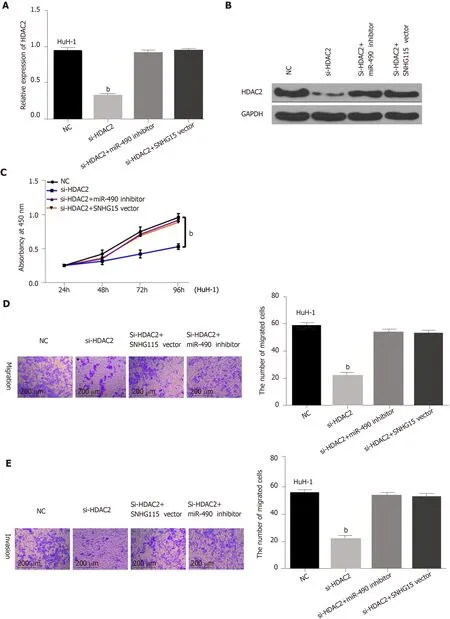
Figure 4 HDAC2 regulated hepatocellular carcinoma progression through mediating SNHG15/miR-490-3p. A: HDAC2 expression in HuH-1 cells containing HDAC2 siRNA, si-HDAC2 + miR-490-3p inhibitor or si-HDAC2 + SNHG15 vector; B: HDAC2 expression in HuH-1 cells containing HDAC2 siRNA, si-HDAC2 + miR-490-3p inhibitor or si-HDAC2 + SNHG15 vector; C: Cell proliferation in HuH-1 cells containing HDAC2 siRNA, si-HDAC2 + miR-490-3p inhibitor or si-HDAC2 +SNHG15 vector; D: Cell migration in HuH-1 cells containing HDAC2 siRNA, si-HDAC2 + miR-490-3p inhibitor or si-HDAC2 + SNHG15 vector; E: Cell invasion in HuH-1 cells containing HDAC2 siRNA, si-HDAC2 + miR-490-3p inhibitor or si-HDAC2 + SNHG15 vector. Untreated cells were set as control (NC). bP < 0.01. SNHG15:Small nucleolar RNA host gene 15; miR-490-3p: Hsa-miR-490-3p; HDAC2: Histone deacetylase 2; HuH-1: Human hepatoma cell line; siRNA: Small interfering RNA;si-HDAC2: HDAC2 siRNA.
We explored the downstream mechanism of miR-490-3p in HCC. We found that miR-490-3p directly targets HDAC2. Moreover, up-regulation of HDAC2 was found in HCC, and a negative association between their expressions was detected in HCC.Meanwhile, a positive correlation between the expression of SNHG15 and HDAC2 was observed in HCC. Previous studies suggested that HDAC2 was up-regulated in breast cancer and colorectal cancer, acting as an oncogene[26,27]. In our research,knockdown of HDAC2 was also found to inhibit HCC progression, serving as a tumor promoter. More importantly, down-regulation of miR-490-3p or up-regulation of SNHG15 was identified to recover the inhibitory effect of HDAC2 silencing in HCC.Taken together, we for the first time demonstrated that HDAC2 regulated by the SNHG15/miR-490-3p axis promoted the tumorigenesis of HCC.
ARTICLE HIGHLIGHTS
Research background
Among all cancers, hepatocellular carcinoma (HCC) related mortality is one of the highest and has seen a dramatic increase in annual global incidence rate. Many recent studies have demonstrated how transcriptional regulation affects HCC. Long non-coding RNAs (lncRNA)play a role in the initiation and progression of HCC, such as maintenance of cell growth, evasion of apoptosis, promotion of invasion and metastasis, stemness maintenance and epithelial to mesenchymal transition.
Research motivation
To discover biomarkers for the diagnosis and treatment of HCC.Research objectives To investigate the underlying mechanisms of lncRNA-SNHG15 in HCC.
Research methods
lncRNA-SNHG15 expression was observed by qRT-PCR assay in HCC tissue and cell lines.Clinicopathological characteristics were collected, arranged and combined with expression analysis of HCC to evaluate the functions of lncRNA-SNHG15. Moreover, cell function assays and western blot were performed to explore the functions of lncRNA-SNHG15 and targets regulated by lncRNA-SNHG15 in HCC cell lines.
Research results
We found that lncRNA-SNHG15 was increased in HCC tissues and cell lines and exhibited a significantly positive relationship with tumor sizes, TNM stage and Edmondson-Steiner grading.Cell experiments showed SNHG15 increased the proliferation and invasion capacity of HCC cell lines, and miR-490-3p/histone deacetylase 2 may be the target regulated by lncRNA-SNHG15 in HCC cells.
Research conclusions
Our study demonstrated that lncRNA-SNHG15 can significantly promote cell growth, migration and invasion of HCC. Furthermore, it can work through miR-490-3p/histone deacetylase 2.Therefore, our study provides some molecular mechanism and three new biomarkers for HCC.
Research perspectives
In the future, research may reveal the important role of lncRNA-SNHG15 that enhances the sensitivity of HCC detection and further develop its application in anti-cancer treatments.
杂志排行
World Journal of Gastroenterology的其它文章
- Role of ion channels in gastrointestinal cancer
- Targeted therapies in metastatic gastric cancer: Current knowledge and future perspective
- Sirtuin 1 alleviates endoplasmic reticulum stress-mediated apoptosis of intestinal epithelial cells in ulcerative colitis
- Up-regulated Wnt1-inducible signaling pathway protein 1 correlates with poor prognosis and drug resistance by reducing DNA repair in gastric cancer
- Hepatitis C virus clearance and less liver damage in patients with high cholesterol, low-density lipoprotein cholesterol and APOE ε4 allele
- Nomogram to predict prolonged postoperative ileus after gastrectomy in gastric cancer
