Identification of CDK6 and RHOU in Serum Exosome as Biomarkers for the Invasiveness of Non-functioning Pituitary Adenoma
2019-10-15ShanYuXiaoshuangWangKaicanCaoXinjieBaoJiaYu
Shan Yu, Xiaoshuang Wang, Kaican Cao, Xinjie Bao, Jia Yu*
1State Key Laboratory of Medical Molecular Biology & Key Laboratory of RNA and Hematopoietic Regulation & Department of Biochemistry,Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & School of Basic Medicine, Peking Union Medical College, Beijing 100005, China
2Department of Thoracic Surgery, Nanfang Hospital,Guangzhou 510515, China
3Department of Neurosurgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100730, China
Key words: pituitary adenoma; invasiveness; exosome; droplet digital PCR; biomarkers;RHOU; CDK6
Objective To explore circulating biomarkers for screening the invasiveness of non-functioning pituitary adenomas (NF-PAs).Methods The exosomal RNAs were extracted from serum of patients with invasive NF-PA (INF-PA) or noninvasive NF-PA (NNF-PA). Droplet digital PCR was adapted to detect the mRNA expression of candidate genes related to tumor progression or invasion, such as cyclin dependent kinase 6 (CDK6), ras homolog family member U (RHOU), and spire type actin nucleation factor 2 (SPIRE2). Student’s t-test was used to analyze the statistical difference in the mRNA expression of candidate genes between the two groups. Receiver operating characteristic (ROC) curve was used to establish a model for predicting the invasiveness of NF-PAs. The accuracy,sensitivity, specificity and precision of the model were then obtained to evaluate the diagnostic performance.Results CDK6 (0.2600±0.0912 vs. 0.1789±0.0628, t=3.431, P=0.0013) and RHOU mRNA expressions(0.2696±0.1118 vs. 0.1788±0.0857, t=2.946, P=0.0052) were upregulated in INF-PAs patients’ serum exosomes as compared to NNF-PAs. For CDK6, the area under the ROC curve (AUC) was 0.772 (95% CI:0.600-0.943, P=0.005), the accuracy, sensitivity, specificity and precision were 77.27%, 83.33%, 75.00%and 55.56% to predict the invasiveness of NF-PAs. For RHOU, the AUC was 0.757 (95% CI: 0.599-0.915,P=0.007), the accuracy, sensitivity, specificity and precision were 72.73%, 83.33%, 68.75% and 50.00%. In addition, the mRNA levels of CDK6 and RHOU in serum exosomes were significantly positively correlated(r=0.935, P<0.001). After combination of the cut-off scores of the two genes, the accuracy, sensitivity,specificity and precision were 81.82%, 83.33%, 81.25% and 62.50%.Conclusions CDK6 and RHOU mRNA in serum exosomes can be used as markers for predicting invasiveness of NF-PAs. Combination of the two genes performs better in distinguishing INF-PAs from NNF-PAs. These results indicate CDK6 and RHOU play important roles in the invasiveness of NF-PAs, and the established diagnostic method is valuable for directing the clinical screening and postoperative treatment.
PITUITARY adenomas (PAs) are common neoplasms that originate from the adenohypophyseal cells, accounting for 10%-20% of intracranial neoplasms.[1,2]Generally, PAs exhibit clinical manifestations due to excessive hormonal secretion, however, 30% PAs are clinically non-functioning (NF) with no evidence of hormone hypersecretion or no early symptoms until space occupying effect emerges.[3,4]Most NF-PAs are non-invasive (NNF-PAs), which grow slowly, and some patients even need no excision and can carry the benign tumor for life-long time. In contrast, some NFPAs (25%-55%) are invasive, which develop fast and could induce visual dysfunction, headache, hormone hyposecretion, or other neurological complications.[5]More importantly, invasive NF-PAs (INF-PAs) usually invade surrounding blood vessels, bones, sphenoid sinuses and optic chiasm, making them difficult to be resected and prone to relapse within three months.[6]Therefore, patients with INF-PAs relapse constantly or die even if they receive chemotherapy and radiotherapy after surgery.[7]Currently, it is very difficult to distinguish between NNF-PAs and INF-PAs due to the lacking of clinical guidelines, and the only diagnostic option is magnetic resonance imaging (MRI) examination. Thus,it is urgent to explore a novel diagnostic biomarker for predicting invasiveness of NF-PAs.
We had previously found a series of genes showing differential expression pattern in pituitaries between NNF-PA and INF-PA patients by high-throughput RNA-seq and further validated them by quantitative real-time PCR (qRT-PCR) in pituitary tissues (unpublished data). Furthermore, functional enrichment analysis suggested that pathways such as cell cycle and division were involved in tumor invasion. In order to protect the patients with NF-PAs and to predict invasiveness at early stage, we seek to search for circulating biomarkers in body fluids. Blood-brain barrier separates the circulating blood from brain while the adenohypophysis is not influenced, so the substances secreted by the pituitary gland and in blood can be exchanged freely. We then proposed that differentially expressed mRNA molecules in pituitary glands could show similar expression pattern in blood and might be used as diagnostic biomarkers.
One challenge for detecting circulating mRNAs is that they are extremely unstable and abundant ribonucleases exist in blood, however, exosomes are much more stable in body fluids. Exosomes are small vesicles (30-150 nm in diameter) derived from cells that can be detected in all fluids including blood.[8]Exosomes contain proteins and RNAs from their original cells and have been used for clinical diagnosis as well as predicting prognosis in various tumors.[9]Therefore,monitoring mRNAs in plasma or serum exosomes may provide robust circulating biomarkers. Since mRNA level in blood exosomes is fairly low, a unique technique called droplet digital PCR (ddPCR) with high sensitivity was adopted in this study.[10]
Among the highly expressed genes in INF-PAs, we focused on three candidates, cyclin dependent kinase 6 (CDK6), ras homolog family member U (RHOU), and spire type actin nucleation factor 2 (SPIRE2), based on their crucial function in tumors, and investigated their potentials as biomarkers for predicting invasiveness of NF-PAs. The mRNA levels of these genes were analyzed in patients’ serum exosomes by using ddPCR.According to the cut-off score, a diagnostic model for predicting the invasiveness of NF-PAs was finally established. Our results imply that CDK6 and RHOU could act as circulating biomarkers for screening INFPAs.
MATERIALS AND METHODS
Patients and sample collection
Patients undergoing resection of NF-PAs at the Peking Union Medical College Hospital between April and November 2017 were eligible for the study. The diagnosis of NNF-PAs and INF-PAs was made according to MRI examination, intraoperative Knosp classification,combined with postoperative recurrence. In this study,NNF-PAs were defined as Knosp grade 0, 1, or 2, and importantly not involving the cavernous sinus. All INFPAs were defined as Knosp grade 4 and involving the cavernous sinus definitely.[11]This study was approved by the ethical committee of Peking Union Medical College Hospital and all the participants were consent to participating in the analysis.
Exosome extraction from peripheral blood
The peripheral blood of each patient (about 5 ml)was taken in a Vacuette® tube (GREINER BIO-ONE,Germany). After incubated with serum separator clot activator at room temperature for 10 min, the blood sample was centrifuged at 1900×g, 4°C for 10 min.The transferred supernatant was centrifugated at 1600×gfor 10 min at 4°C, and finally the cell-free supernatant was transferred into a new tube.
Exosomes were next extracted from 1 ml serum using Total Exosome Isolation Reagent (from serum)(Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. Approximately (2.5-6.5)×1011exosomes could be obtained from each sample. The pelleted exosomes were resuspended in PBS to examine exosome morphology using transmission electron microscopy with negative staining method and to display the size and quantity of the exosomes using nanoparticle tracking analysis with NanoSight NS300 instrument (Malvern Panalytical, England ).
Exosomal RNA purification and reverse transcription
For total RNA extraction, the exosome pellet was lysed with 1 ml QIAzol Lysis Reagent (QIAGEN BIO-ONE),and proceeded using miRNeasy Micro Kit (QIAGEN B10-ONE). Then aliquots (11 μl) from total RNA were reverse transcribed using SuperScript™ Ⅲ Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions.
ddPCR
The ddPCR was performed following the recommendations of the supplier (Bio-Rad, California, USA). The reaction system contained 1×ddPCR™ Supermix for Probes(no dUTP) (Bio-Rad), 800 nmol/L primer, 250 nmol/L probe (Table 1) and 3 μl of cDNA template in each 20 μl reaction system. After homogenization, the 20 μl ddPCR reaction mixture and 70 μl oil (Bio-Rad)were loaded into a droplet generator cartridge (Bio-Rad) to produce an emulsion about 40 μl in volume by QX200™ Droplet Generator. The emulsion was subsequently transferred to a 96-well PCR plate, heatsealed by a pierceable foil (Bio-Rad) and subjected to thermal cycling under the following conditions: 95°C for 5 min for Taq polymerase activation; 40 cycles of 94°C for 30 s and 60°C for 40 s; then 98°C for 10 min and holding at 12°C. After amplification, the fluorescence intensity was read on a QX200 droplet reader. The result was analyzed with QuantaSoft software[12](version 1.7.4.0917).
Functional enrichment
Functional enrichment analysis was carried out with KOBAS 3.0 and GSEA online. Gene IDs of the interested genes were submitted on KOBAS (http://kobas.cbi.pku.edu.cn/anno_iden.php) with all available database selected. For GSEA enrichment,gene sets were searched in Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
Statistical analysis
Continuous data were expressed as mean±SD.Statistical differences were analyzed by Student’st-test,Chi-squaretest orFishertest using GraphPad Prism (version 6.0). A two-tailedPvalue less than 0.05 was considered as statistically significant. Receiver operating characteristic (ROC) curves and correlation analysis were depicted by IBM SPSS Statistics (version 22.0). ROC curves were used to assess the diagnosing accuracy for differentiating INF-PAs from NNF-PAs. The relative expression value of candidate exosomal mRNA corresponding to the Youden index (sensitivity + specificity -1) was defined as the cut-off score. The areaunder ROC curve (AUC) was calculated to evaluate the diagnostic efficiency.
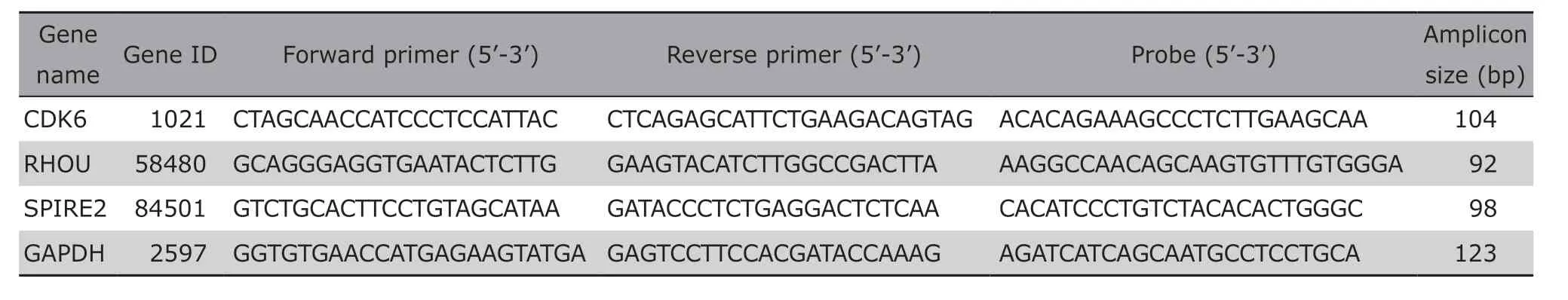
Table 1. Primers and probes for the target and reference genes
RESULTS
Clinical features of NNF-PA and INF-PA patients
Finally, 46 patients were recruited in the study, including 32 NNF-PAs and 14 INF-PAs. No significant difference in clinical features was observed between the NNF-PAs and INF-PAs groups except for tumor size(t=3.7152,P=0.0006; Table 2), which was consistent with the common knowledge that invasive PAs are larger than non-invasive ones at diagnosis. It should be noticed that the famous prognostic markers p53 and Ki-67 cannot distinguish NNF-PAs from INF-PAs either, indicating the demand for developing the novel molecular biomarkers.
Verification of the purified exosomes
Electron microscopy showed that the purified exosome was a tea saucer-like structure of 30-150 nm in size with distinct membrane (Figure 1A). Nanoparticle tracking analysis used to display the size and quantity of the exosomes indicated that nearly all recovered nanoparticles’ diameters were less than 300 nm, with most of them in the range of 100-150 nm (Figure 1B).
CDK6 and RHOU were highly expressed in INFPAs
We next performed ddPCR to detect the mRNA copynumber variations of candidate genes in exosomes.Less than 3 ng exosomal total RNA was purified and subsequently subjected to reverse transcription.To eliminate the significant difference in the total amount of RNA input among different patients which induced by amount of serum exosomes, the copy number of GAPDH was adopted as a reference control to exclude such effects. Of note, we rarely detected SPIRE2 mRNA, suggesting that SPIRE2 was not a good exosome biomarker for NF-PAs. After normalized to GAPDH, both CDK6 and RHOU showed significantly higher relative expression in INF-PAs as compared to NNF-PAs (CDK6: 0.2600±0.0912vs. 0.1789±0.0628,t=3.431,P=0.0013; RHOU: 0.2696±0.1118vs.0.1788±0.0857,t=2.946,P=0.0052; Figure 2),suggesting these two molecules might be used to discriminate between NNF-PAs and INF-PAs.
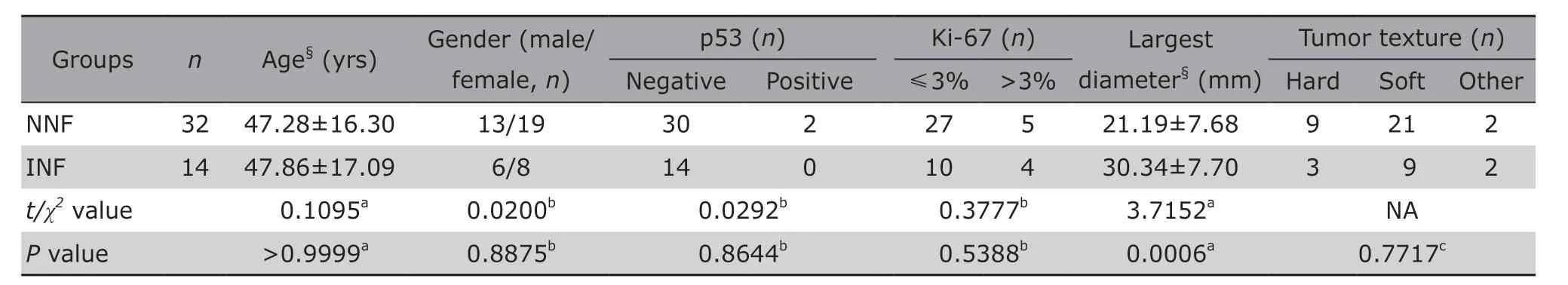
Table 2. Clinicopathological features of NNF-PA and INF-PA patients included in the study

Figure 1. Validation of the exosomes recovered from the serum.
CDK6 and RHOU can be used to discriminate INFPAs from NNF-PAs
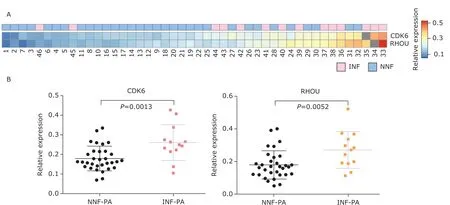
Figure 2. Differential expression of CDK6 and RHOU in exosomes from NF-PAs.
To evaluate the performance of CDK6 and RHOU as diagnostic indexes for INF-PAs, their diagnostic ability was analyzed by ROC curves. The AUC of CDK6 was 0.772 [95% confidence interval (CI): 0.600-0.943,P=0.005], and the AUC of RHOU was 0.757 (95%CI:0.599-0.915,P=0.007), respectively (Figure 3A). We then calculated the cut-off points for CDK6 and RHOU to predict their ability in discriminating the invasiveness of NF-PAs. For CDK6 (cut-off score=0.2146),the accuracy, sensitivity, specificity and precision were 77.27%, 83.33%, 75.00% and 55.56%; similarly,the accuracy, sensitivity, specificity and precision for RHOU (cut-off score=0.1857) were 72.73%, 83.33%,68.75% and 50.00%. These data collectively supported that both CDK6 and RHOU could be used as accurate and sensitive biomarkers to differ INF-PAs from NNF-PAs, although the precision was about 50%.
Additionally, we found that the mRNA levels of CDK6 and RHOU in serum exosomes were significantly positively correlated (Pearson correlation coefficient =0.935) (Figure 3B), suggesting they may be simultaneously involved in relevant biological network to initiate or promote the invasion of NF-PAs.To investigate their potential overlapped pathways,we queried “CDK6 AND RHOU” on KOBAS and GSEA,which turned out that these two genes were mainly co-regulating cell cycle-related processes. The top 20 enrichment pathways on KOBAS are shown in Table 3,including all 9 pathways revealed by GSEA marked as highlighted. The accordance of CDK6 and RHOU suggested that they might be used together for diagnosis. Indeed, after setting the double cut-off scores(0.2146 for CDK6 and 0.1857 for RHOU) as invasiveness threshold, we observed the elevated accuracy(81.82%), sensitivity (83.33%), specificity (81.25%)and precision (62.50%) (Table 4). These results indicated that the combining CDK6 and RHOU were more powerful to distinguish INF-PAs from NNF-PAs.
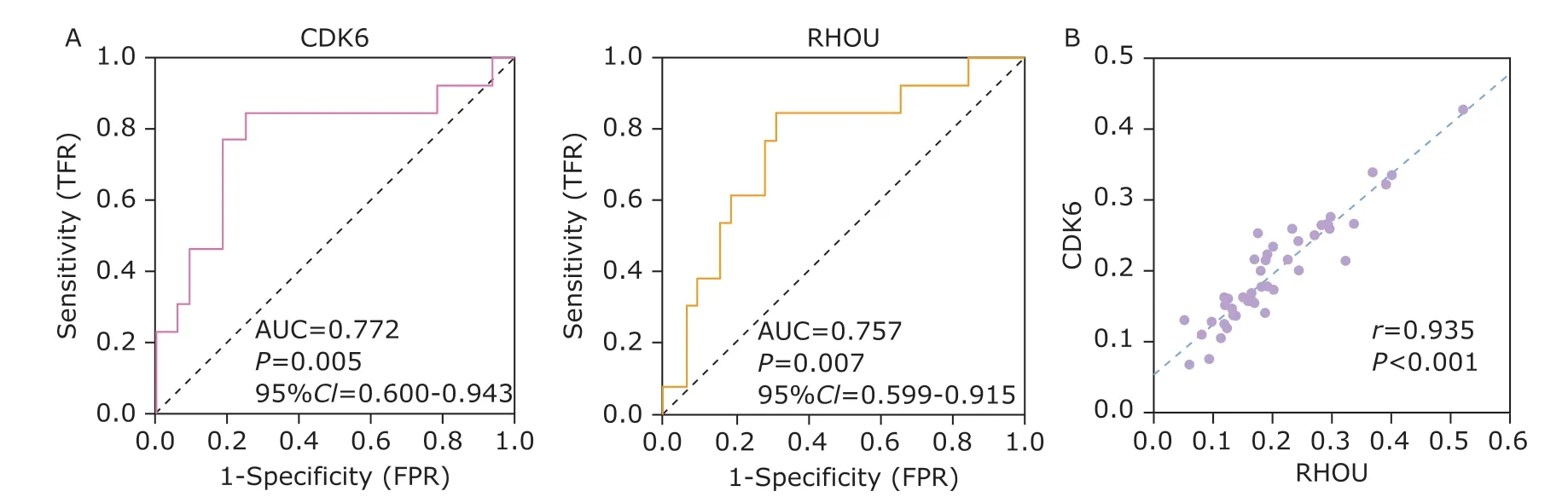
Figure 3. The evaluation of performance using CDK6 or RHOU to discriminate INF-PAs from NNF-PAs.
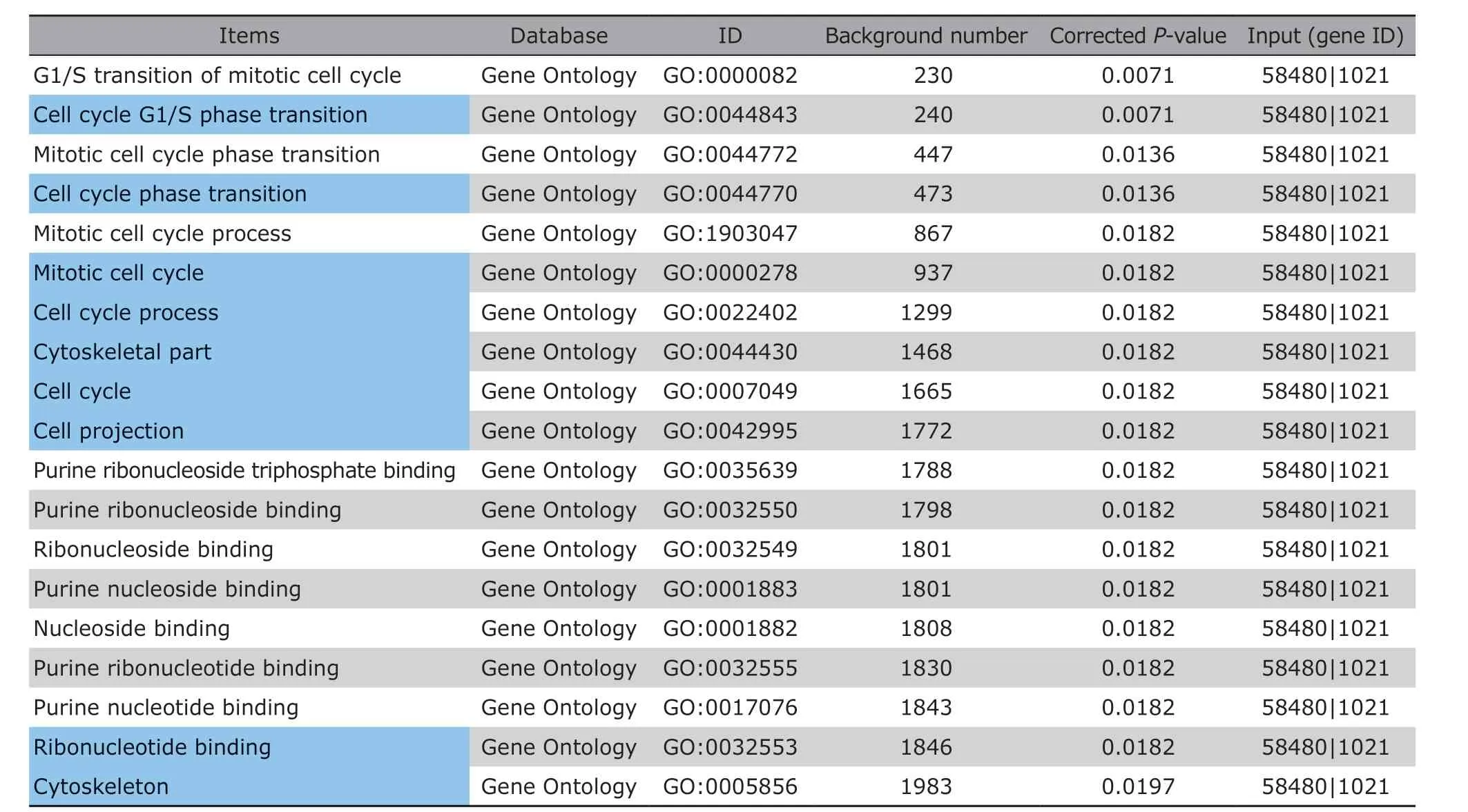
Table 3. Functional enrichments of CDK6 and RHOU
DISCUSSION
It is known that intraoperative observation combined with postoperative recurrence are still the standard to define INF-PAs. No direct correlation was found between clinical features including age and gender, or biomarkers such as p53 and Ki-67 and the invasiveness of NF-PAs in this study. In many different tumors,p53 immunoreactivity or Ki-67 proliferative index >3% facilitates the diagnosis or identification of tumor aggressiveness. However, the inconsistent results for Ki-67 and p53 activity in PAs were observed by different studies.[13,14]In addition, histological features of p53 and Ki-67 were unable to distinguish benign or invasive PAs.[15,16]To develop invasive and robust diagnostic biomarkers, we analyzed the absolute mRNA quantity of several potential cancer related genes in patients’ serum exosomes based on our previous findings. Among these genes, both CDK6 and RHOU weresignificantly up-regulated in the INF-PAs group as compared to NNF-PAs and their variations were significantly positively correlated with each other.
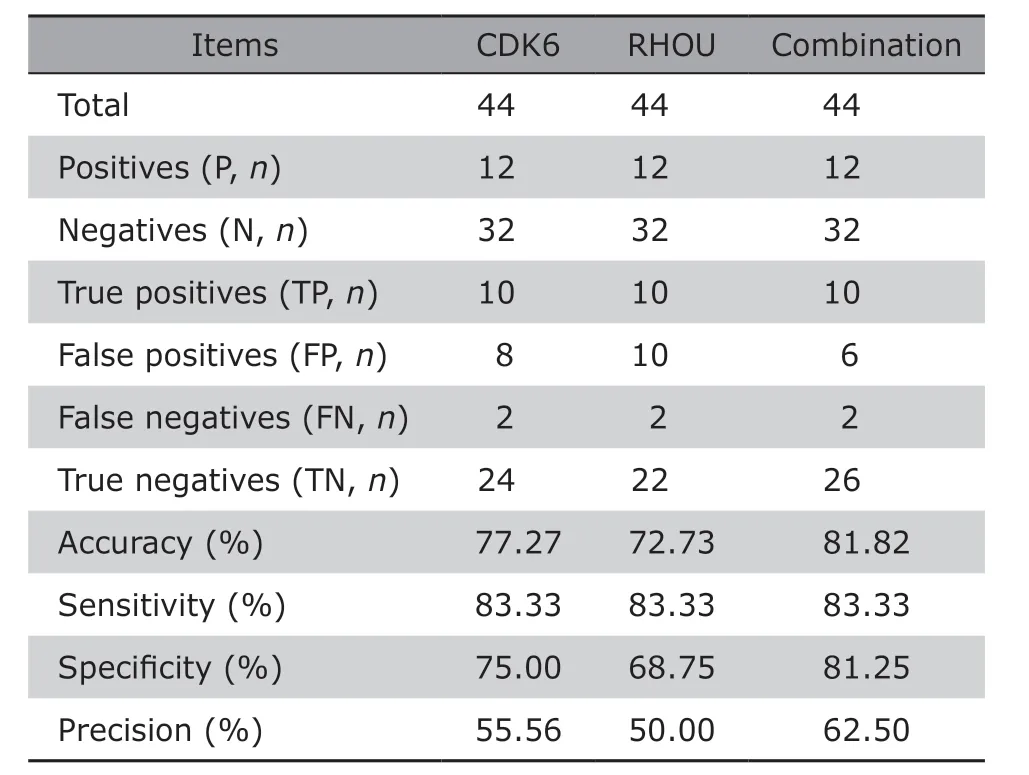
Table 4. Comparisons of the performance using CDK6,RHOU or both to distinguish between NNF-PAs and INFPAs
Cyclin-dependent kinases (CDKs) are a family of serine-threonine kinases which play critical roles in orchestrating G1 phase progression and G1/S transition in cell cycle.[17]It is well studied that CDK6 is an over-expressed oncogenic gene in cancer cells and regulates the progression of many cancers, including medulloblastoma, malignant glioma, glioblastoma,lymphoma, hematopoietic malignancies, and cancers of pancreas, prostate, bladder.[18-25]Rho GTPases affect cell morphology, division, adhesion, motility, and contribute to migration and invasion of cancer cells, such as melanoma cells, breast carcinoma.[26-30]As a member of the Rho family of GTPases, RHOU (ras homolog family member U) has been identified to control cell migration in both multiple myeloma and acute lymphoblastic leukemia.[31,32]
The higher expression of CDK6 and RHOU in INF-PAs indicated their important functions in the invasiveness of NF-PAs, and the underlying mechanism might be applied with other tumors.CDK6 has been recognized to play manifold roles in cancer progression, including promoting cell survival,proliferation and tumor growthviathe retinoblastoma(Rb) protein.[18,22,23]RHOU has been reported as a response gene in Notch signaling in T-ALL cell[32]and in JAK/STAT pathway in multiple myelomas,[31]it can also cooperate with p21 activated kinase (PAK) family[32-35]to increase cell migration and/or adhesion. Notably,enhanced cell proliferation, migration and adhesion could trigger invasion. Besides, the remarkable positive expression correlation of these two genes suggested they might be involved in one regulatory network, for example, they both regulate cell cycle. Canovas Nuneset al.[31]had observed a significant cell cycle arrest at G1/S phase transition in response to RHOU silencing,which further support that RHOU and CDK6 might interact with each other in tumors development (Table 3). In a word, it is speculated that CDK6 and RHOU have both unique and overlapping mechanisms in the invasion of NF-PAs. Further investigations revealing the detail molecular mechanism of both CDK6 and RHOU regulating NF-PAs development may expand our understanding of the pathogenesis and invasiveness of NF-PAs and thus improve postoperative management.
The important roles of CDK6 and RHOU drove us to explore their capacity as biomarkers. Strikingly, using the individual cut-off score of CDK6 or RHOU could already distinguish INF-PAs from NNF-PAs with high sensitivity and accuracy. More importantly, combination of these two biomarkers could significantly improve the specificity and precision to diagnose INF-PAs or NNF-PAs. Further studies with more biomarkers and validation in a larger cohort are required to improve the diagnostic panels.
Taken together, our study identified CDK6 and RHOU can be used as blood-based circulating biomarkers to discriminate between INF-PAs and NNF-PAs,which is also helpful to predict prognosis and develop accurate postoperative treatment for these patients.As single molecule is insufficient to determine tumor subtypes, the combination of several molecules may be a straightforward strategy to establish a better diagnostic system.
Conflicts of interest statement
All authors declare no conflicts of interest.
杂志排行
Chinese Medical Sciences Journal的其它文章
- Research on the Antitumor Compounds from Cephalotaxus Hainanensis
- Artificial Musk R&D and Manufacturing
- Management of an Adult with Goodpasture’s Syndrome Following Brain Trauma with Extracorporeal Membrane Oxygenation: A Case Report
- Transvaginal Reduction of a Heterotopic Cornual Pregnancy with Conservation of Intrauterine Pregnancy
- Research Progress on Diagnosis and Treatment of Chronic Osteomyelitis
- Association between GSTT1 Homozygous Deletion and Risk of Pancreatic Cancer: A Meta Analysis
