Regional Differences of the Urinary Proteomes in Healthy Chinese Individuals
2019-10-15JianqiangWuWeiweiQinLiPanXiaorongWangBiaoZhangGuangliangShanYouheGao
Jianqiang Wu , Weiwei Qin, Li Pan, Xiaorong Wang,Biao Zhang, Guangliang Shan*, Youhe Gao*
1Medical Research Center, Peking Union Medical College Hospital, Peking Union Medical College & Chinese Academy of Medical Sciences, Beijing 100730, China
2Department of Pathophysiology, 4Department of Epidemiology and Statistics, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences & School of Basic Medicine,Peking Union Medical College, Beijing 100005, China
3Department of Biochemistry and Molecular Biology, Gene Engineering Drug and Biotechnology Beijing Key Laboratory, Beijing Normal University, Beijing 100875, China
Key words: urine; proteomics; biobank; biomarkers; regional differences
Objective Urine is a promising biomarker source for clinical proteomics studies. Regional physiological differences are common in multi-center clinical studies. In this study, we investigate whether significant differences are present in the urinary proteomes of individuals from different regions in China.Methods In this study, morning urine samples were collected from healthy urban residents in three regions of China (Haikou, Xi’an and Xining) and urinary proteins were preserved using a membrane-based method(Urimem). The urine proteomes of 27 normal samples were analyzed using LC-MS/MS and compared among three regions. Functional annotation of the differential proteins among the three areas was analyzed using the DAVID online database, and pathway enrichment of the differential urinary proteins was analyzed using KEGG.Results We identified 1898 proteins from Urimem samples using label-free proteome quantification, of which 56 urine proteins were differentially expressed among the three regions (P < 0.05). Hierarchical clustering analysis showed that inter-regional differences caused less significant changes in the urine proteome than intersex differences. After gender stratification, 16 differential proteins were identified in male samples and 84 differential proteins were identified in female samples. Among these differential proteins, several proteins have been previously reported as urinary disease biomarkers.Conclusions Urimem will facilitate urinary protein storage for large-scale urine sample collection. Regional differences are a confounding factor influencing the urine proteome and should be considered in future multicenter biomarker studies.
BIOMARKERS are measurable changes associated with specific pathophysiological conditions. Unlike blood, which is controlled by homeostatic mechanisms, urine reflects systemic changes in the body and can be a more promising source of biomarkers.[1-2]Currently, in addition to its application in urogenital diseases, urinary proteomics has been used in systematic diseases, such as cancers, cardiovascular diseases, diabetes, and mental disorders, to identify urinary disease biomarkers.[3]More importantly, urine has potential uses in detecting small and early pathological changes in the very early stages of disease.[4-7]For example, several urinary proteins change significantly even before a tumor mass is palpable, indicating that urine is an excellent source of biomarkers for early cancer detection.[6]Another study suggested that early changes in urinary proteins can be detected in a lung fibrosis model, which may lead to early treatment and a better prognosis.[7]
In the last decade, advances in proteomics techniques have accelerated the pace of biomarker research. Urinary proteome changes, some of which have been reported to be associated with disease,have been found in various systematic diseases both in animal models and patients. However, candidate biomarkers or biomarker panels need to be validated in large-scale, multi-center patient cohorts. Previous studies showed that several factors can influence the urine proteome, such as gender, age, exercise and medicine.[2,8-11]Regional differences are a common phenomenon in multi-center clinical studies. These differences may consist of genetic background, food consumption, lifestyle and environmental factors. However, whether there are significant differences in the urinary proteome among different regions remains unknown.
In this study, morning urine samples were collected from healthy Han residents in three urban areas of China (Haikou, Xi’an and Xining). These three cities are the capitals of Hainan, Shaanxi and Qinghai Provinces, respectively. Urimem is a method that can store urine proteins simply and economically.[12]In the current study, a large number of urine samples were collected at sites in the three regions, and urine proteins were stored using Urimem. Then, 27 Urimem samples were selected and their donors were matched by age,sex and ethnicity. The proteomes of these 27 normal samples were compared based on region to investigate the regional differences in the urinary proteome.
SUBJECTS AND METHODS
Participants
The China National Health Survey (CNHS) was conducted to evaluate physiological constants and health conditions of the Chinese population. Research on the normal urine proteome of the healthy Chinese population is a subproject of CNHS. A total of 754 healthy volunteers between 10 and 80 years of age were enrolled in this study. Urine samples from 283 healthy urban residents (164 males and 119 females) in Haikou, Hainan Province, China, were collected on December 27-29, 2013; urine samples from 255 healthy urban residents (121 males and 134 females) in Xi’an,Shaanxi Province, China, were collected on July 8-10,2014; and urine samples from 216 healthy urban residents (128 males and 88 females) in Xining, Qinghai Province, China, were collected on August 15-17,2015.
This study was approved by the local Ethics Committee of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (No. 029-2013),and signed informed consent was obtained from all participants.
Data collection
A questionnaire was administered to obtain information from the participants, including sex, age, ethnicity, personal medical history, education level, physical activity, smoking status and drinking status.
Blood pressure was measured on a seated participant using an Omron digital blood pressure measuring device. After a donor had fasted overnight, a venous blood sample was collected at the time of the interview. Blood samples were processed within 4 h of collection, shipped to a laboratory in Beijing, and stored at -80°C until analysis.
Blood biochemical and immune indicators, including total cholesterol (TC), triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), immunoglobulin G (IgG),immunoglobulin A (IgA), immunoglobulin M (IgM),alanine aminotransferase (ALT), serum total protein(TP), serum albumin (ALB), gamma glutamyl transferase (GGT), alkaline phosphatase (ALP), aspartate aminotransferase (AST), calcium (Ca), phosphorous (P),creatinine, urea nitrogen, fasting venous blood glucose(Glu) and uric acid (UA), were assessed at the Peking Union Medical College Hospital and Chinese PLA General Hospital.
Urinary protein preservation on a membrane
Approximately 50-100 ml of morning midstream urine was collected from each healthy participant. The urinary protein concentration was determined using routine urine tests. The urinary proteins were preserved on nitrocellulose (NC) membranes using a filtering device as previously reported in our study.[12]NC membranes were used to absorb the urine proteins in the current study because their reproducibility is similar to that of protein isolation, and proteins can be easily eluted from these membranes.[13]
Briefly, after urine collection, 20 ml of each urine sample was mixed with 10 ml of phosphate buffer (1 mol//L Na2HPO4, 1 mol//L NaH2PO4, pH=6). The diluted urine sample was then added to a vacuum filter bottle(10-cm2filter area). In this device, a polyvinylidene fluoride (PVDF) membrane (0.45 µm HV, Millipore,Tullagreen, Carrigtwohill, Ireland) with ultra-low protein-binding capacity was used to filter cell debris from the urine. Another 0.2-µm NC membrane (GE Healthcare, Freiburg im Breisgau, Germany) was used to absorb urine proteins. Three sheets of filter paper were wetted with pure water and placed below the NC membrane. The vacuum filter bottle was connected to a vacuum pump, and the urine in the bottle passed through the PVDF membrane, while the urine protein in the flow-through was saved on the NC membrane.Then, the protein-bound NC membrane was dried with a hair dryer at room temperature and sealed in a sterile plastic bag with a label. The medical record number,sex, age, date of urine collection and blood pressure were recorded on the label. Urine samples from each participant were preserved on 2 or 3 NC membranes(Urimem).
Elution of urinary proteins from the membranes
To avoid potential interference from confounding factors, Urimem membranes for preservation of urinary protein from healthy urban Han Chinese adults aged between 30 and 36 with no chronic or metabolic diseases (hypertension, diabetes, hepatitis and cancers)and no drug treatment within 2 weeks were selected.Additionally, blood biochemical indicators from each participant were reviewed to select healthy individuals whose blood indicators fell within the reference ranges for Chinese populations. To rule out potential kidney diseases, participants without severe proteinuria were selected after a routine urine protein test. Finally, 9 participants (5 males and 4 females) from each geographic area were randomly selected for further proteome analysis.
Urinary proteins were eluted from the membranes according to a previously described method.[14]The NC membrane with bound proteins was cut into small pieces and placed in a 2-ml Eppendorf tube. Then,1.7 ml of acetone and 0.2 ml of 5% NH4HCO3were added to the tube and vigorously vortexed for 30 s.The tubes were heated at 55°C for 60 min and were strongly vortexed for 30 s every 20 min. After the membrane was fully dissolved, the liquid was gently mixed using a rotary mixer at 4°C for 2 h to precipitate the protein. Then, the mixture was centrifuged at 12 000 xgfor 15 min at 18°C. After the supernatant was removed, the tube was placed upside down on filter paper for 5 min at room temperature. The precipitate was re-suspended in lysis buffer (8 mol/L urea,2 mol/L thiourea, 50 mmol/L Tris and 25 mmol/L DTT),and the protein concentration was determined using Bradford method.
Tryptic digestion and LC-MS/MS analysi s
A mass of 200 µg of urinary proteins eluted from each Urimem sample was digested with trypsin (Trypsin Gold, mass spectrometry grade, Promega) using the filter-aided sample preparation method with 10-kDa filtration devices (Pall, NY, USA) at 37°C overnight. The peptide mixtures were then desalted using Oasis HLB cartridges (Waters, Milford, MA, USA), dried by vacuum evaporation and re-dissolved in 0.1% formic acid.
For the LC-MS/MS analysis, 1 µg of peptides from each individual sample was loaded onto a trap column (75 µm×2 cm, 3 mm, C18, 100 Å) and was separated on a reverse-phase analytical column(50 µm×150 mm, 2 µm, C18, 100 Å) using the EASYnLC 1200 HPLC system (Thermo Fisher Scientific,Waltham, MA, USA). The elution for the analytical column was performed over 60 min at a flow rate of 300 µl/min. Then, the eluted peptides were analyzed using an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). MS data were acquired in the high-sensitivity mode using the following parameters: data-dependent MS/MS scans per full scan with top-speed mode (3 s), MS scans at a resolution of 120 000 and MS/MS scans at a resolution of 30 000 in Orbitrap, 30% HCD collision energy, chargestate screening (+2 to +7), dynamic exclusion (exclusion duration 30 s) and a maximum injection time of 45 ms.
Label-free quantification
Label-free quantitation of the proteomic data was performed using Progenesis LC-MS software (version 4.1, Nonlinear, Newcastle upon Tyne, UK) according to the procedure described by Haucket al.[15]Twenty-seven sample features were automatically aligned according to their retention times to maximally overlay all the two-dimensional feature maps (m/z and retention time). Features with charge states of +2 to+4 were selected for further analysis. After alignment and feature exclusion, the samples were divided into three groups (Haikou, Xi’an and Xining) using a between-subject design, and the raw abundances of all features were normalized. Normalization corrected for factors resulting from experimental variation and was automatically calculated over all features in all samples.
All MS/MS spectra were exported from the Progenesis software, and the data were searched against the SwissProt Human database (08/2013, containing 20 267 sequences) using Mascot software (version 2.5.1, Matrix Science, London, UK). The parent ion tolerance was set at 10 ppm, and the fragment ion mass tolerance was set to 0.05 Da. A maximum of two missed cleavage sites in the trypsin digestion was allowed. Carbamidomethylation of cysteines was set as a fixed modification, and the oxidation of methionine was considered a variable modification. For quantification, all peptides with an ion score ≥20 andP<0.01 were included, and the total cumulative abundance of a specific protein was calculated by summing the abundances of all peptides allocated to it. Calculations of the proteinPvalue (one-wayANOVA) were then performed on the sum of the normalized abundances across all runs.ANOVAvalues ofP<0.05 and additionally regulation of ≥2-fold were regarded as significant for all further results.
Functional and pathway analyses
Functional annotation of the differential proteins among the three areas was analyzed using the DAVID online database.[16]Protein classification was performed using gene ontology (GO) analysis for molecular functions and biological processes. Differentially expressed proteins were also uploaded into the Kyoto Encyclopedia of Genes and Genomes (KEGG) for pathway analysis.[17]To identify differential proteins which had been annotated as urinary candidate biomarkers,all significant changed proteins were searched against the Urinary Protein Biomarker Database.[3]
Statistical analysis
Statistical analyses were performed with GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA,USA; www.graphpad.com) and SPSS 19.0 software (IBM Corporation, Armonk, NY, USA). The clinical characteristics data were expressed as the means ± SDs. Comparisons among the three groups were conducted using a one-wayANOVAfollowed by multiple comparisons with the least significant differences (LSD) test. Group differences resulting inP-values<0.05 were considered statistically significant. Using the method as previously reported,[18]the coefficients of variation (CVs) of the samples in each region were calculated to show the level of sample variation in each geographic area.
RESULTS
Characteristics of the participants from the three regions of China
In this study, we recruited healthy urban residents from the capitals of three Chinese provinces (Hainan,Shaanxi, and Qinghai). Haikou is the capital of Hainan Province, located in southern China (longitude:110.35 degrees, latitude: 20.02 degrees, elevation:8 m); Xi’an is the capital of Shaanxi Province, located in central China (longitude: 108.95 degrees, latitude:34.27 degrees, elevation: 415 m); and Xining is the capital of Qinghai Province, located in northwest China(longitude: 101.74 degrees, latitude: 36.56 degrees,elevation: 2250 m). The latitudes of Xi’an and Xining are similar, but significantly higher than that of Haikou.The elevations of these three cities are significantly different.
Detailed information of laboratory biochemical analyses on the 27 participants is listed in Table 1.Age, sex, blood pressure and most blood biochemical indicators were similar among the healthy participants from the three areas (P>0.05). However, TP, ALB, TC,LDL-C, Glu and Ca were significantly different (P<0.05)in the blood of the participants from these three cities(Figure 1). Serum protein concentrations, including TP and ALB, were lower in Xi’an residents than those in residents from the other cities (P<0.001). The concentrations of serum lipids, including TC and LDL-C,were higher in Haikou residents than those in residents from Xi’an (P<0.05). Additionally, Glu level was higher in Xining residents than that in residents from Haikou(P<0.01). Finally, the serum concentration of Ca was significantly lower in residents from Xi’an than that in residents from the other cities (P<0.01).
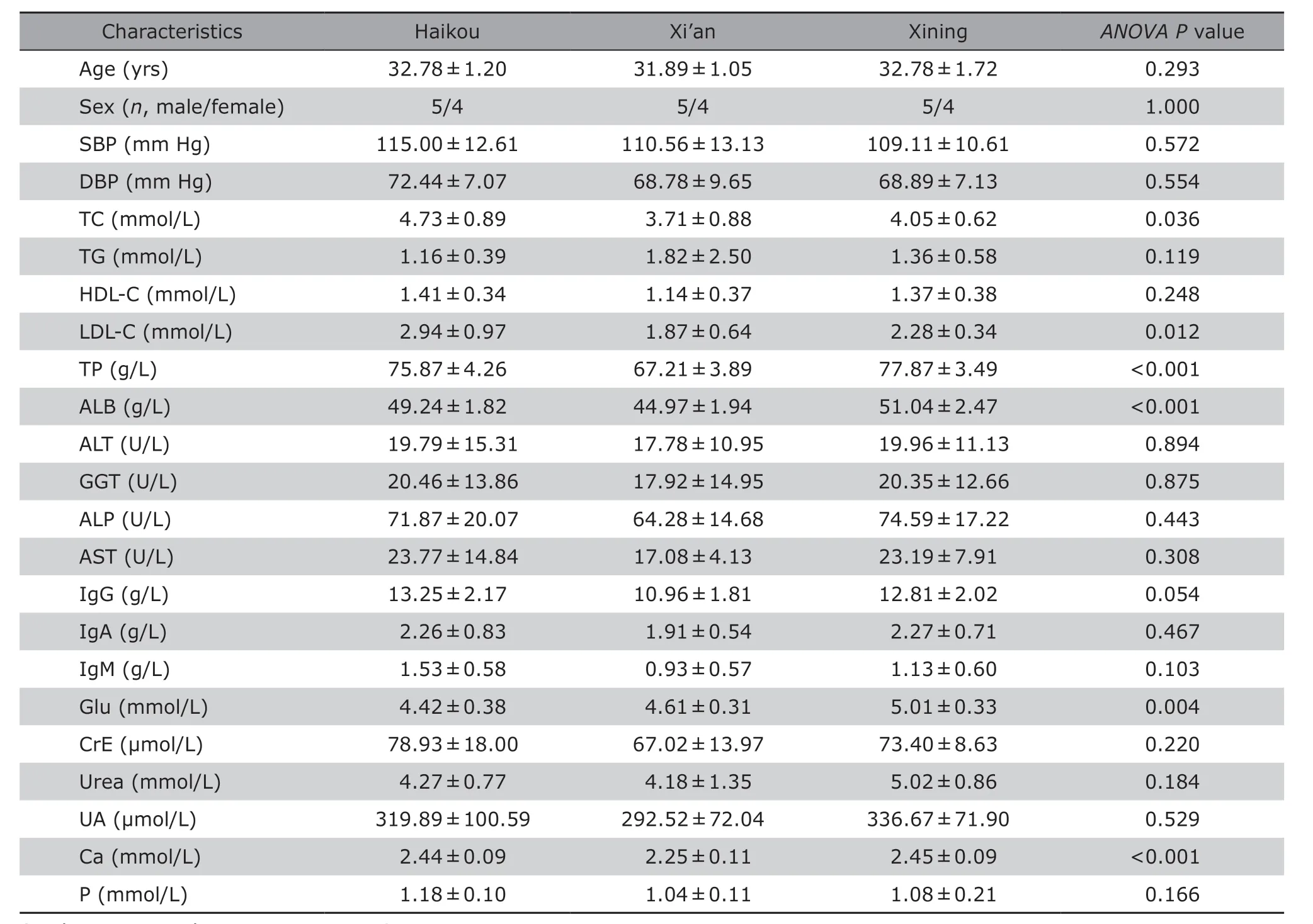
Table 1. Characteristics of the 27 participants according to location§ (n = 9)
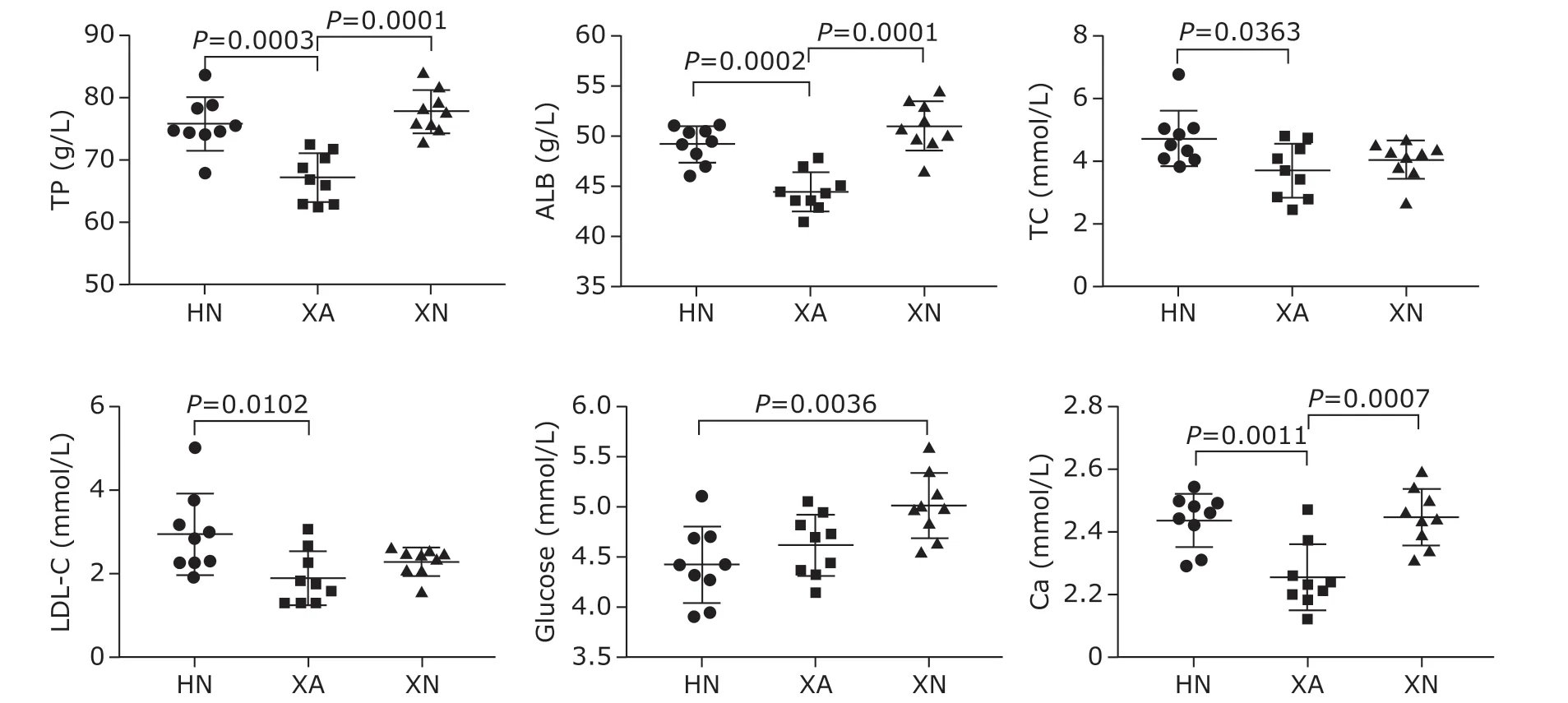
Figure 1. Blood biochemical indicators that differed among the three areas of China.
Regional differences in the normal human urine proteome
To investigate the regional differences in the normal urine proteome, Urimem samples from 27 healthy residents from three Chinese regions were analyzed by LC-MS/MS. Finally, a total of 1898 urine proteins were identified from the Urimem samples using label-free identification (Supplementary Table S1).
To show the level of sample variation in each geographic area, CVs of the samples in each region were calculated. As shown in Figure 2, the CV values of the samples from Haikou (median CV=0.60) were slightly higher than those of the samples from Xi’an (median CV=0.54) and Xining (median CV=0.49), suggesting that the individual variation of urine proteome samples in Haikou is more significant. In addition, a total of 56 differential proteins were identified among the three regions (≥2 unique peptides,ANOVAP<0.05, and fold change ≥2; Supplementary Table S2).
Sex is a confounding factor that may influence the urinary proteome. Between the male and female samples, a total of 53 urine proteins showed changes greater than 2-fold and hadP-values<0.05 (Supplementary Table S3). As shown in Figure 3, a hierarchical clustering analysis of all identified proteins was performed to comprehensively define inter-regional and inter-sex differences in the urinary proteome.The results showed that all female samples were clustered into one group and that several samples in the same area could be clustered together. These results suggested that male and female urinary proteomes exhibited different patterns, and inter-regional differences were smaller than inter-sex differences. Then,the male and female samples were separated, and the differential proteins were re-analyzed among the three regions. A total of 16 differential urine proteins were identified in male samples, whereas 84 differential proteins were identified in female samples. Moreover,there were 49 urine proteins that were differentially expressed between Xi’an and Haikou, 42 between Xi’an and Xining, and 35 between Haikou and Xining (Supplementary Table S4). Additionally, we observed that 2 differential proteins in male samples and 14 differential proteins in female samples were among the 56 previous identified differential proteins. These results showed that inter-gender variation to some degree affects the inter-regional analysis and that difference among the three regions are more significant in female samples than in male samples.

Figure 2. The coefficient of variation (CV) values of sample variation in each geographic area.
Some disease biomarkers are differentially expressed in the three regions
All significantly changed proteins were searched against the Urinary Protein Biomarker Database. The differential urine proteins among the three areas included proteins that have been reported as urinary disease biomarkers or biomarker candidates in clinical studies (Table 1).
Functional analysis of differential proteins
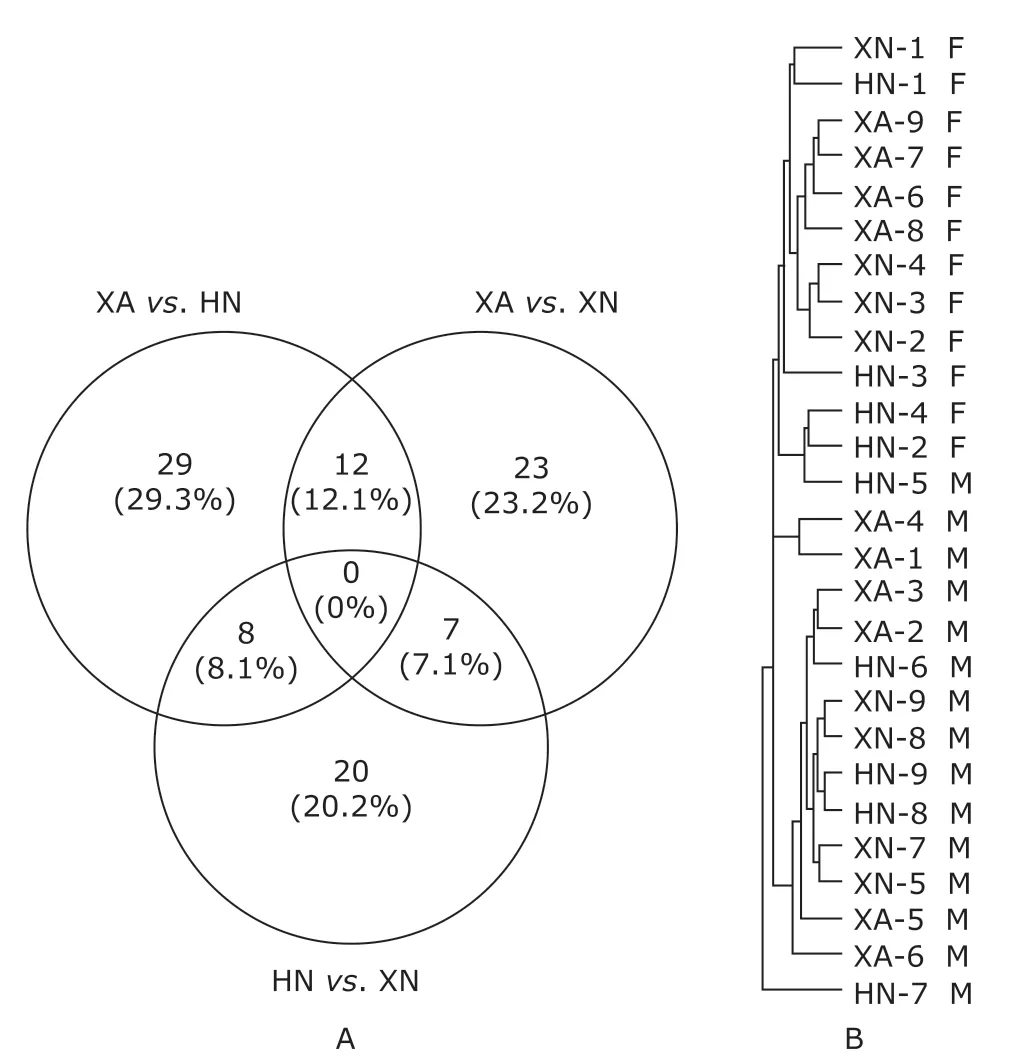
Figure 3. The effects of inter-regional and inter-sex differences on the urine proteome.
Functional annotation of the differential proteins was performed using DAVID. Forty-nine differential proteins between Xi’an and Haikou, 42 differential proteins between Xi’an and Xining, and 35 differential proteins between Haikou and Xining were uploaded into the database. In the biological process category,inflammatory response (P=3.7E-3), tricarboxylic acid cycle (P=2.8E-3), cellular protein metabolic process(P=4.1E-3) and gluconeogenesis (P=6.5E-3) were overrepresented in differential proteins of Xi’anvs.Haikou; carbohydrate metabolic process (P=8.2E-3),response to estradiol (P=0.020), cyclooxygenase pathway (P=0.024) and response to hyperoxia(P=0.040) were overrepresented in differential proteins of Xi’anvs. Xining; and response to reactive oxygen species (P=2.5E-3), leukocyte cell-cell adhesion (P=1.0E-3), response to heat (P=3.8E-3),response to hypoxia (P=4.2E-3), cellular oxidant detoxification (P=7.8E-3) and surfactant homeostasis(P=0.015) were overrepresented in differential proteins of Xiningvs. Haikou.
Then, KEGG was used for pathway enrichment analysis of the differential urinary proteins. The pathways for metabolic pathways (P=1.0E-5), biosynthesis of antibiotics (P=1.4E-4), Citrate cycle (TCA cycle)(P=6.0E-3) and carbon metabolism (P=9.5E-3) were significantly enriched in differential proteins of Xi’anvs. Haikou. The pathways for metabolic pathways(P=2.4E-3), lysosome (P=6.4E-3) and arachidonic acid metabolism (P=0.016) were significantly enriched in differential proteins of Xi’anvs. Xining. The pathways for leukocyte transendothelial migration (P=0.039), lysosome (P=0.043) and nitrogen metabolism (P=0.046) were significantly enriched in differential proteins of Xiningvs. Haikou.
DISCUSSION
In our present study, we investigated whether regional differences can significantly influence the human urinary proteome. To clarify the potential effect of regional differences on urine proteomics, we performed comparative proteomics analysis of Urimem samples from three areas in China using label-free quantification. Our results indicate that some urine proteins were differentially expressed among Xi’an, Hainan and Xining. These proteins included several proteins identified as disease biomarkers or biomarker candidates in previous clinical studies.
As shown in Figure 1, several blood biochemical indicators were significantly different in the blood of the participants from the three regions, consistent with the results of several previous studies. For example,the serum TP and ALB levels of healthy population in Xi’an were significantly different from those in other cities in China.[19]Another study suggested that several serum indicators, including blood urea nitrogen,creatinine, and UA, changed with increasing elevationin four areas of Yunnan Province.[20]In another recent study, significant differences in serum immunological indexes and the coagulation index of healthy people between Xi’an and Xining were reported, including higher expression of IgG, IgM and IgA in Xining than in Xi’an.[21]Our results in Table 1 are consistent with this study. Because blood and Urimem samples come from the same participants, and the urinary proteome contains a number of plasma proteins, changes in the blood composition are likely to have a potential impact on the urinary proteome.

Table 1. The effects of regional differences on urinary disease biomarkers
In the first analysis, some differential urine proteins were identified among the three areas. Because sex has been reported to influence the urine proteome,[22]we compared the effects of inter-regional and inter-sex differences on the urinary proteome. The results indicated that the inter-regional differences were smaller than the inter-gender differences. Then,the male and female samples were separated, and differential analysis was repeated. The differential urine proteins among the three areas included proteins reported as urinary disease biomarkers in previous clinical studies. For example, catalase protects cells from the toxic effects of hydrogen peroxide and is involved in oxidative damage. This oxidative stress biomarker was upregulated in the urine samples of diabetic patients.[23]Epidermal growth factor receptor kinase substrate 8-like protein 2 and syntenin-1 regulate migration, growth, proliferation, and cell cycle progression in a variety of cancers. These two proteins were reported to be differentially expressed in the urine samples of bladder cancer patients and to be potential cancer biomarkers.[24]Thus, in multi-center biomarker studies, it is difficult to definitively determine whether potential disease biomarkers are truly associated with disease states or regional differences. Therefore, regional differences are a confounding factor influencing the urine proteome and should be considered in future multi-center biomarker studies.
Previous studies demonstrated that some intrinsic factors (such as sex and age) and extrinsic influences(such as diet, exercise, and environmental factors)may influence the urine proteome, and intrinsic factors likely cause more significant changes to the urine proteome than extrinsic influences.[2]Regional differences are attributable to genetic background, food consumption, lifestyle and environmental factors. In this study, the participants were matched by age, sex and ethnicity. Thus, regional differences in our study were likely due to extrinsic influences. The most significantly different environmental factor among the three regions was elevation (Haikou, 8 m; Xi’an, 415 m; Xining,2250 m). The exposure of healthy subjects to high elevation can cause hypoxia. Functional analysis of differential proteins among the three regions showed that several differential proteins between Xining and Xi’an were involved in the ‘response to hyperoxia’ process, and some differential proteins between Xining and Haikou were involved in the process of ‘response to hypoxia’. Tissue hypoxia can cause oxidative stress and changes in white blood cell and lung surfactants that can affect respiratory function. Thus, several of differential proteins between Xining and Haikou are also associated with biological processes related to hypoxia, such as ‘reactive oxygen species’, ‘cellular oxidant detoxification’, ‘leukocyte cell-cell adhesion’ and‘surfactant homeostasis’. This result is consistent with the greater altitude change between Xining and Haikou.
Another significantly different factor among the three regions is food consumption. Residents in Haikou tend to eat rice and more fresh fruit and vegetables.Residents in Xi’an and Xining tend to eat pasta and consume less fresh fruit and vegetables. Moreover,residents in Xining tend to consume more animal meat and milk. Pathway analysis of differential proteins between Xi’an and Haikou showed that the carbon metabolism pathway was enriched. By contrast, the nitrogen metabolism pathway was enriched in differential proteins between Xining and Haikou. These pathways difference may be attributed to the greater consumption of meat and milk by Xining residents.
The system of preserving urinary proteins on membranes using a vacuum filter bottle was first described in our previous study.[12]The feasibility and validity of Urimem were subsequently proven in flowup studies.[13-14,25]The findings showed that for protein absorption, the reproducibility of NC membrane was similar to that of PVDF membranes. The proteome coverage of urinary proteins absorbed onto NC membranes has also been evaluated using LC-MS/MS analysis. Recently, membrane-based urine protein absorption using a syringe-push system was described as a simple rapid method for preparing urine for proteomics analysis.[26]These studies demonstrated that dry membranes can be stored at room temperature for an extended period of time and remain stable for further proteomic analysis. The long-term stability of membranes with dried absorbed urine proteins allows specimen collection from areas without freezers and overcomes sample shipping problems, such as protein stability during sample shipping and shipping expenses due to weight. Membrane-based methods for urine protein preservation are quick and suitable for batch sample processing in large-scale clinical studies. Additionally, membranes with different properties can be used to selectively retain different substances from urine, such as DNA, metabolites and microRNAs.[27]Massive sample storage (biobanking) in a simple and economical manner is the foundation of future largescale clinical precision medicine studies, and this membrane-based method may be one good option for urine sample processing and storage.[28]Urimem is a membrane that can preserve urinary proteins simply and economically. Using six filtering devices, urine samples from about 100 individuals can be collected and preserved within one day in this study. Urimem is a timesaving and suitable tool for large-scale and rapid sample preservation, especially in places without centrifugation or refrigeration facilities. The extensive use of Urimem in and out of hospitals will accelerate biobank development and future urine biomarker studies using proteomics approaches.
In this study, proteome differences in the normal urine samples of urban residents from three Chinese regions. Some urinary proteins were differentially expressed in different regions, and several proteins were reported as disease biomarkers in previous clinical studies. The potential application of our findings in biomarker discovery is that regional differences should be taken into consideration in biomarker identification and validation, particularly in multi-region and multi-center clinical studies. The object of this study was to assess the presence of significant differences in the urinary proteomes of individuals from different regions. Due to the low throughput of MS, only 27 Urimem samples are used for the proteomics analysis, and the results should be confirmed in future studies with a larger sample size and additional populations in other regions.
The Supporting Informationis available free of charge on the CMSJ website at doi: 10.24920/003504.
Table S1.Total urine proteins identified from the Urimem samples using label-free identification.
Table S2.Differentially expressed urine proteins among the three areas.
Table S3.Gender differences in the urine proteome.
Table S4.Differentially expressed urine proteins among the three areas after gender stratification.
杂志排行
Chinese Medical Sciences Journal的其它文章
- Research on the Antitumor Compounds from Cephalotaxus Hainanensis
- Artificial Musk R&D and Manufacturing
- Management of an Adult with Goodpasture’s Syndrome Following Brain Trauma with Extracorporeal Membrane Oxygenation: A Case Report
- Transvaginal Reduction of a Heterotopic Cornual Pregnancy with Conservation of Intrauterine Pregnancy
- Research Progress on Diagnosis and Treatment of Chronic Osteomyelitis
- Association between GSTT1 Homozygous Deletion and Risk of Pancreatic Cancer: A Meta Analysis
