精神分裂症相关单核苷酸多态性调控microRNA功能研究进展
2019-08-26梁文权侯豫赵存友
梁文权,侯豫,赵存友
精神分裂症相关单核苷酸多态性调控microRNA功能研究进展
梁文权1,侯豫2,赵存友1
1. 南方医科大学基础医学院医学遗传学教研室,广州 510515 2. 陆军总医院附属八一儿童医院神经发育科,北京 100700
MicroRNA (miRNAs)是一类长约22nt且对基因表达具有广泛的调控作用的非编码RNA,并在神经元增殖、分化和发育成熟的过程中扮演重要角色。近年来全基因组关联研究(genome-wide association studies, GWAS)发现了众多的精神分裂症相关单核苷酸多态性(single nucleotide polymorphisms, SNPs)位点多位于非编码区,表明miRNA在精神分裂症的发病过程中存在重要作用。本文综述了精神分裂症相关SNP与miRNA可能发生相互作用的4种机制(SNP位于miRNA基因、SNP位于miRNA宿主基因、SNP位于miRNA种子序列和SNP位于miRNA结合位点),为研究miRNA在精神分裂症发生发展中的作用提供参考。
全基因组关联研究;单核苷酸多态性;微小RNA;精神分裂症
精神分裂症是多基因变异引发的复杂性遗传病,也是最严重的精神类疾病之一[1]。该疾病通常在青壮年时期发病,致残率高,医疗资源消耗大,不仅影响患者的劳动能力,而且给其家属及社会造成严重负担,因此探索行之有效的预防和治疗措施,已经成为一项全球性的社会和经济问题。既往的遗传学研究发现了若干与精神分裂症相关的基因或位点,尤其近年来的全基因组关联研究(genome-wide association study, GWAS)手段更是发现了大量与该疾病相关的单核苷酸多态性(single nucleotide polymorphisms, SNPs)。然而,这些遗传变异只有少数位于蛋白编码区,更多的位于非编码区,这使得研究者从基因表达调控角度研究疾病相关非编码区的变异成为可能。已有多项报道发现与精神分裂症相关的SNP位于microRNA (miRNA)的靶基因或结合位点,miRNA通过对上述基因或位点的负调控作用介导了精神分裂症的发生和发展。
miRNA是一类长约22 nt的非编码RNA,通过与靶基因mRNA 3ʹUTR区特异位点的结合使靶基因mRNA发生降解或翻译阻遏[2,3]。当位于靶基因3ʹUTR区的SNP位于miRNA的结合位点时,这会使miRNA与靶基因的结合能力发生改变,从而造成靶基因的表达或翻译发生改变[4]。人类mRNA 3ʹUTR拥有超过45 000个保守的miRNA结合位点,约60%的人类蛋白质编码基因拥有至少一个保守的miRNA结合位点[5], 这些保守位点的碱基变异会影响到miRNA的调控功能。近年来采用GWAS手段发现的大量与精神分裂症相关的SNPs多位于非编码区,为从miRNA角度研究精神分裂症的发病机制提供了线索。自2008年第1篇关于精神分裂症GWAS分析的文章发表以来[6],通过GWAS分析已发现许多与精神分裂症相关的SNP[7~10]。对欧洲人群通过GWAS研究,发现基因内含子区的SNP rs1625579与精神分裂症相关[11],在汉族人群中也得到了验证[12]。之后的研究支持为精神分裂症危险因素[13],并且基因上的GWAS SNP rs1625579导致的等位基因表达不均一性[14]。Ning等[15]通过连锁不平衡分析发现在中国汉族核心家系中基因座上的SNPs (rs1877670,rs3750192和rs7009708)构建的单倍型与精神分裂症显著性关联。越来越多的研究发现miRNA在神经发育过程中调控基因表达,表明miRNA的表达改变可能会导致诸如精神分裂症等精神疾病的发生。Perkins等[16]发现在精神分裂症病人前额叶皮质表达下降,基因位于22q11.2微缺失上,携带该微缺失的病人有1/4被确诊为精神分裂症患 者[17]。基因芯片研究发现在精神分裂症患者的尸检组织中颞上回和背外侧前额叶中miRNAs的表达水平普遍增加[18]。遭遇童年创伤的精神分裂症患者血液样本中显著下降,该miRNA在产前应激的大鼠海马组织中同样发现表达显著下降在[19]。研究发现和或、和对精神分裂症具有较好的预测作用[20]。这些证据表明,精神分裂症患者的miRNA表达谱发生改变是精神分裂症发生发展的一个重要因素。
非编码miRNA的调控功能为人们研究GWAS中发现的众多非编码区SNP的可能调控机制提供了新的线索。本文将从以下4个方面阐述与精神分裂症相关的SNP通过miRNA实现对基因表达的调控作用(图1):(1) SNP位于miRNA基因:通过影响miRNA的表达量从而使下游的靶基因的表达量发生改变;(2) SNP位于miRNA的宿主基因:通过调控宿主基因的表达从而调控miRNA的表达量;(3) SNP位于miRNA的种子序列内:影响miRNA与mRNA结合的稳定性,使mRNA的稳定性或翻译受阻;(4) SNP位于miRNA靶基因的结合位点内:通过改变miRNA与靶基因的结合能力从而调控靶基因的表达。
1 SNP位于miRNA基因
Ripke等[11]发现位于下游8 kb的rs1625579与精神分裂症相关(=1.6×10–11)。另有研究发现在北美人群中携带TT基因型的精神分裂症发病年龄较携带G基因的发病年龄提前,影像学研究发现携带TT基因型的人群脑白质密度减少、海马体积缩小以及侧脑室增大[14]。研究表明,在脑组织中表达丰富[21],与神经元的分化与突触可塑性相关,rs1625579及其连锁的SNP (rs1198588、rs2660304和rs2802535)调控的表达量。Siegert等[22]研究发现拥有上述SNPs的minor-allele的神经类似细胞(neuron-like cells)的表达量上升,的上调使和的表达量下调导致突触囊泡释放功能异常[22]。
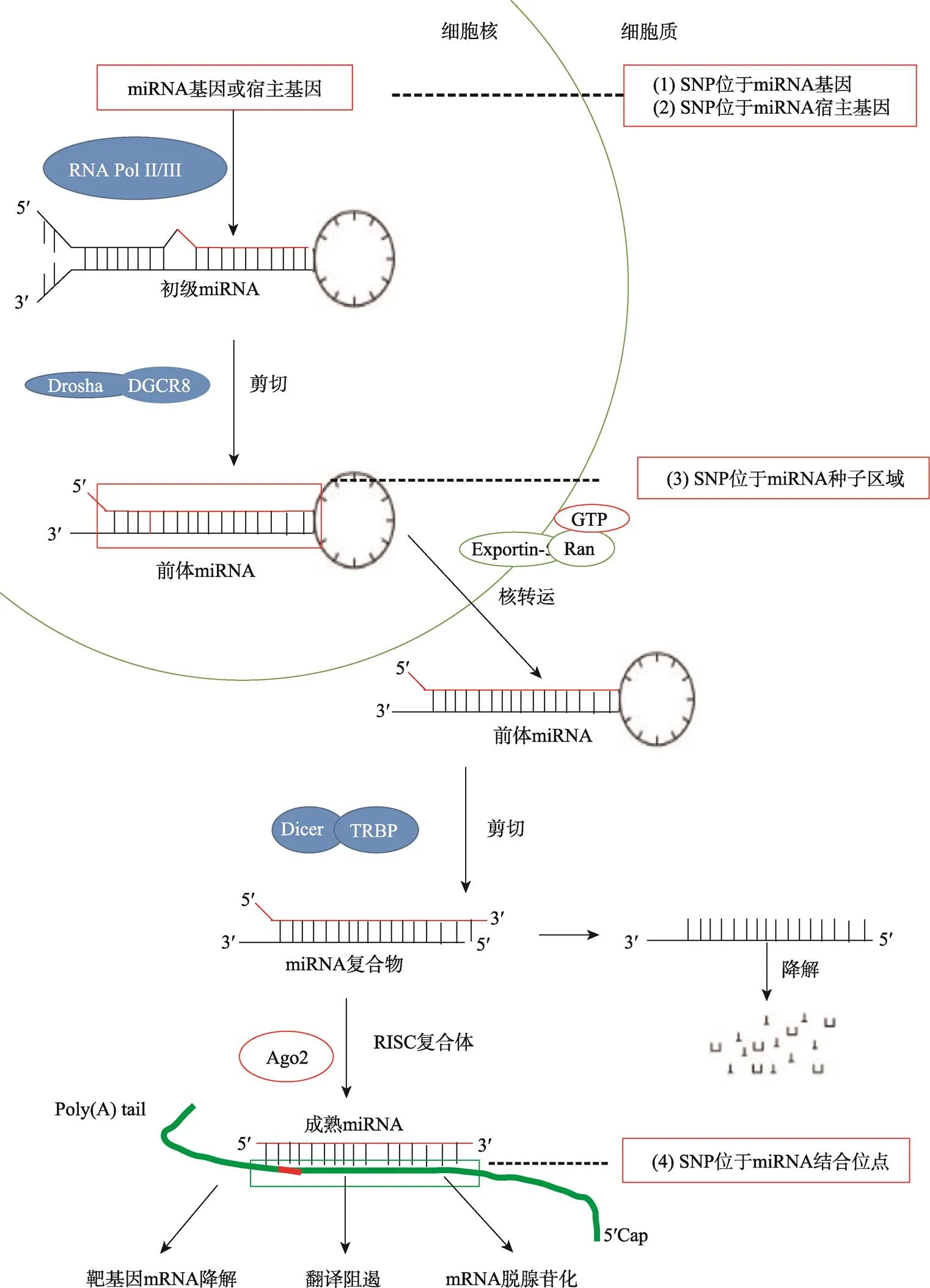
图1 SNP通过miRNA实现对基因表达的调控
miRNA加工通路首先在细胞核内经RNA聚合酶Ⅱ或Ⅲ作用下产生初级miRNA转录本(pri-miRNA),之后被Drosha-DGCR8 复合物裂解形成具有发夹结构的前体miRNA(pre-miRNA),并由Exportin-5-Ran-GTP复合物从细胞核中输出。在细胞质中,RNase Dicer和双链RNA结合蛋白TRBP复合物共同作用将pre-miRNA发夹切割到成熟长度。成熟miRNA的功能链与Argonaute (Ago2)蛋白一起加载到RNA诱导沉默复合物(RISC)中,引导RISC到靶向mRNA,进而通过mRNA裂解、翻译抑制或脱腺苷化沉默靶mRNA。在这一过程中,SNP通过miRNA实现对基因表达调控作用的4种主要机制是:(1) SNP位于miRNA基因;(2) SNP位于miRNA宿主基因;(3) SNP位于miRNA种子区域;(4) SNP位于miRNA结合位点。根据参考文献[27]修改绘制。
Budach等[23]研究发现SNP位于miRNA的启动子中,提示SNP基因型的变化将对miRNA的表达量产生影响[23]。miRNA-eQTL多富集于miRNA的前体、启动子、增强子和转录因子结合区域,并且与其宿主基因的mRNA的eQTL存在大量交集。目前只有少量几篇miRNA的eQTL数据发表,包括Parts等[24]发表的脂肪细胞的miRNA-eQTL、Siddle等[25]发表的树突状细胞的miRNA-eQTL和Huan 等[26]发现在全血中的miRNA-eQTL。上述研究表明位于miRNA的SNP可以调控miRNA的表达量,从而影响miRNA的下游调控通路,而使罹患精神分裂症的风险上升。
2 SNP位于miRNA宿主基因
约40%的哺乳动物miRNA位于其宿主基因的内含子区[28],有证据表明位于宿主基因内含子区的miRNA的成熟并不需要Drosha酶的参与[29]。有研究表明被miRNA沉默的基因与miRNA的宿主基因在功能上具有拮抗作用,而miRNA的表达量与宿主基因的表达量呈正相关[30];该研究还发现通过沉默与其宿主基因具有拮抗功能的和从而使对轴突生长的刺激作用增强[30]。另外有研究检测了和与其宿主基因在海马神经元成熟过程中的动态表达变化,发现mRNA的3ʹUTR具有的结合位点,mimics转染大鼠B35成神经细胞瘤细胞发现的表达水平显著降低,而转染观察不到该种现 象[31]。亦有研究报道多巴胺受体基因也是靶向基因,的缺失或的下调可导致22q11.2微缺失小鼠出现精神病性症状[32]。利用序列特异性miRNA海绵在发育期小鼠皮层中阻遏作用发现神经元极性的丢失和皮层内神经元数目的减少,相反的,在皮层中过表达使神经元的数目增加并且呈现多极性[33]。Ferrari等[34]发现6个SNPs (rs906175、rs2725391、rs969413、rs2659030、rs9319617和rs1048775)的致病基因型能降低的表达量。上述研究提示,位于miRNA宿主基因的SNP可能影响宿主基因的表达,从而使miRNA的表达异常;或者位于宿主基因的SNP影响miRNA的剪切成熟过程,从而使miRNA的表达异常;又或者位于宿主基因的SNP会使miRNA的转录调控机制异常,导致miRNA表达异常。不管如何,miRNA的表达异常,都有可能使罹患疾病的风险率上升。
3 SNP位于miRNA的种子序列内
在成熟miRNA序列中5ʹ端2~8位的碱基序列被称为miRNA的种子序列,这一种子序列在miRNA与mRNA的识别与互补配对中扮演重要角色[35]。据统计,超过500个SNPs位于miRNA的种子序列,研究人员发现位于miRNA种子序列中心位置的SNP比位于种子序列外围区域的少,因此miRNA种子序列中心区域在进化上更保守[36]。研究表明即使是在miRNA种子序列的单个SNP变异也将导致大于50%的该miRNA靶基因频谱变化[36]。目前还缺乏SNP位于miRNA种子序列导致精神分裂症的直接证据,然而已有文章报道了这种SNP在其他疾病中的重要作用。Mencıʹa等[37]发现在内耳毛细胞表达的种子序列的点突变将导致常染色体显性遗传的渐进性失聪,种子序列的SNP (+13 G>A)将使下游靶基因、、、和表达发生改变。有研究表明在种子序列的SNP (+57C>T)能够导致EDICT (endothelial dystrophy, congenital cataract, and stromal thinning)综合征[38]。Jiang等[39]研究发现有617个SNPs位于牛源的331个成熟miRNAs的种子序列内,位于种子序列的SNP (rs109462250)因为结合位点的改变影响的表达,通过双荧光验证了rs109462250的42198 087 G>A突变使与的结合能力降低。Chai等[40]发现位于种子序列的SNP(rs331295049 A17G)使的表达上调(miR-378/G),并且该SNP改变了的二级结构,使其对原有85%靶基因失去调控作用,但同时又产生了700多个新的靶基因。上述研究表明,位于miRNA种子序列的SNP具有十分重要的生物学作用,然而现在缺乏SNP位于miRNA种子序列内导致精神分裂症患病风险上升的相关报道。
4 SNP位于miRNA结合位点内
据报道,大约5%的miRNA识别元件(miR recognition element, MRE)中具有SNP,约3%的miRNA识别元件种子序列((miR recognition element seed sites, MRESS)具有SNP,同时研究表明约38%的MRESS-SNP位于8mer MRESSs[41]。
位于靶基因mRNA 3ʹUTR区miRNA结合位点内的SNP将会影响miRNA与mRNA的结合,从而使miRNA对mRNA的调控过程受阻。近年来,人们已经发现若干个位于miRNA结合位点的SNP对精神分裂症的发生发展具有影响。Hou等[42]通过对GWAS精神分裂症相关变异位点的检索发现,位于基因3ʹUTR区rs4702-G可通过增加的结合能力而参与FURIN的表达下调,而FURIN的表达量降低将导致成熟脑源性神经营养因子(brain derived neurotrophic factor,BDNF)的生成减少,后者的降低已经报道与精神分裂症的发生相关。Gong等[4]发现在精神分裂症患者中rs10759-C将降低miR-124与RGS4 mRNA结合的最小自由能(minimum free energy, MFE),从而使与mRNA的结合更加稳定,显著降低RGS4的表达量。Rossi等[43]发现rs11122396位于3ʹ UTR区,且位于的结合位点,通过双荧光素酶报告载体证实了rs11122396对结合mRNA的影响,且他们认为SNP对miRNA的影响是有等位基因差异的,即SNP只对miRNA结合其中一条染色体转录的mRNA有影响,对另外一条染色体转录的mRNA无影响,简称等位基因差异性调控。Hauberg等[23]发现rs11191548能影响与mRNA的结合,从而影响NT5C2的表达,且通过双荧光素酶报告载体验证; 值得注意的是是所有保守miRNA中对精神分裂症易感基因有明显调节作用的miRNA之一[44]。研究证明位于3ʹ-UTR的SNP(rs1060120)将影响对H3F3B的转录后调控,并且可能参与的精神分裂症的发生[45]。John等[46]在精神分裂症病人样本中验证了35个位于精神分裂症相关基因3ʹ-UTR miRNA结合位点上的SNPs,研究发现位于3ʹ-UTR的rs7430与精神分裂症具有显著相关性(= 0.01; OR (95%) 1.24 (1.04~1.48))。位于3ʹ-UTR的rs550067317将影响miR-137对EFNB2的调控能力[47]。上述研究表明位于miRNA结合区域的SNP将会使miRNA与mRNA的结合能发生变化,从而影响miRNA与RNA的结合,使miRNA对mRNA的调控发生异常。
5 SNP对miRNA调控过程影响的预测方法
研究SNP与miRNA的相互作用,可能需要利用生物信息学方法对miRNA的靶基因以及SNP与miRNA的关系进行预测。因为miRNA能调控数百个靶基因,所以研究miRNA最大的困难是预测miRNA的靶基因,现有的miRNA靶基因预测软件或网站如表1。
现有的预测软件或网站只能从生物信息学方面预测位于miRNA结合位点或种子序列的SNP对miRNA结合mRNA的影响,具体的功能还需要构建荧光报告载体验证。同样,人们也可以通过过表达miRNA结合ChIP-seq的方式获得miRNA结合的mRNA的序列信息,然而其生物学功能仍需构建荧光报告载体和基因敲降实验进行验证。还可以通过CRISPR/Cas9获得不同基因型的细胞,从而验证miRNA对位于mRNA 3ʹ UTR的不同基因型SNP的作用或验证不同基因型对miRNA表达或剪切成熟过程的影响或位于miRNA种子序列不同基因型SNP对其下游通路的影响。最后可以构建转基因动物模型从而验证不同基因型SNP罹患精神分裂症的风险。

表1 miRNA靶基因预测软件或网站
6 结语与展望
miRNA在大脑中的广泛性表达和在精神分裂症中的表达紊乱,提示miRNA与精神分裂症存在相关性。近年来多篇GWAS的研究结果为人们解析精神分裂症的致病机制提供了许多可能的具有生物学功能的遗传变异资源,然而在后GWAS时代如何去验证GWAS报道的SNP却十分困难,因为虽然有部分SNP位于蛋白质编码区,然而也有一部分位于非编码区,如何去验证位于非编码区的SNP的生物学功能至今是一个难题。已有研究证明miRNA与GWAS-SNPs存在相互作用,提示位于非编码区域的GWAS-SNPs可能在转录调控或转录后调控的过程中具有重要的生物学作用,例如位于宿主基因的SNPs可能影响miRNA的表达从而改变下游靶基因的表达,位于miRNA基因上的SNPs直接导致miRNA表达异常参与了精神分裂症的发生与发展,位于种子序列的SNPs可能导致mRNA表达谱的改变从而导致精神分裂症的发生,位于结合位点的SNPs可能是miRNA与靶基因的转录后调控过程发生异常导致精神分裂症的发生。
精神分裂症作为一种重性精神疾病,在我国“十三五”脑计划提出的大背景下必将受到更多的关注,精神分裂症作为环境与遗传因素共同作用的多基因遗传病,或许miRNA结合SNP的研究是阐明精神分裂症病理过程的更好的切入点,为以后临床诊断和治疗精神分裂症提供理论基础。尽管GWAS研究已经发现许多与精神分裂症相关的SNP,但如何验证其功能仍将是未来的一个研究热点,此外将miRNA与精神分裂症相关SNP结合在一起,从遗传学和表观遗传学角度阐明精神分裂症的发病机制也将会是未来研究的一个重要发展方向。
[1] Plomin R, Owen MJ, Mcguffin P. The genetic basis of complex human behaviors., 1994, 264(5166): 1733–1739.
[2] O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-Myc-regulated microRNAs modulate E2F1 expression., 2005, 435(7043): 839–843.
[3] Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution., 2005, 123(6): 1133–1146.
[4] Gong Y, Wu CN, Xu J, Feng G, Xing QH, Fu W, Li C, He L, Zhao XZ. Polymorphisms in microRNA target sites influence susceptibility to schizophrenia by altering the binding of miRNAs to their targets., 2013, 23(10): 1182–1189.
[5] Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs., 2009, 19(1): 92–105.
[6] Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL.. Genomewide association for schizophrenia in the CATIE study: results of stage 1., 2008, 13(6): 570–584.
[7] Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia- associated genetic loci., 2014, 511(7510): 421– 427.
[8] Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, Yi Q, Li C, Li X, Shen J, Song Z, Ji W, Wang M, Zhou J, Chen B, Liu Y, Wang J, Wang P, Yang P, Wang Q, Feng G, Liu B, Sun W, Li B, He G, Li W, Wan C, Xu Q, Li W, Wen Z, Liu K, Huang F, Ji J, Ripke S, Yue W, Sullivan PF, O'Donovan MC, Shi Y. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia., 2017, 49(11): 1576–1583.
[9] Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Byrne EM, Dannlowski U, Eley TC, Hayward C, Martin NG, McIntosh AM, Plomin R, Porteous DJ, Wray NR, Caballero A, Geschwind DH, Huckins LM, Ruderfer DM, Santiago E, Sklar P, Stahl EA, Won H, Agerbo E, Als TD, Andreassen OA, B?kvad-Hansen M, Mortensen PB, Pedersen CB, B?rglum AD, Bybjerg-Grauholm J, Djurovic S, Durmishi N, Pedersen MG, Golimbet V, Grove J, Hougaard DM, Mattheisen M, Molden E, Mors O, Nordentoft M, Pejovic-Milovancevic M, Sigurdsson E, Silagadze T, Hansen CS, Stefansson K, Stefansson H, Steinberg S, Tosato S, Werge T,; GERAD Consortium; CRESTAR Consortium, Collier DA, Rujescu D, Kirov G, Owen MJ, O'Donovan MC, Walters JTR. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection., 2018, 50(3): 381–389.
[10] International Schizophrenia Consortium, Purcell SM, Wray N R, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder., 2009, 460(7256): 748–752.
[11] Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci., 2011, 43(10): 969–976.
[12] Zhang P, Bian Y, Liu N, Tang Y, Pan C, Hu Y, Tang Z. The SNP rs1625579 in miR-137 gene and risk of schizophrenia in Chinese population: a meta-analysis., 2016, 67: 26–32.
[13] Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, Lakatos A, Miles MF, Lander A, Vawter MP, Xie X. Identifying gene regulatory networks in schizophrenia., 2010, 53(3): 839–847.
[14] Green MJ, Cairns MJ, Wu J, Dragovic M, Jablensky A, Tooney PA, Scott RJ, Carr VJ. Australian Schizophrenia Research Bank. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia., 2013, 18(7): 774–780.
[15] Ning QL, Ma XD, Jiao LZ, Niu XR, Li JP, Wang B, Zhang H, Ma J. A family-base association study of the EGR3 gene polymorphisims and schizophrenia.,2012, 34(3): 307–314.宁启兰, 马旭东, 焦李子, 牛晓蓉, 李建鹏, 王彬, 张欢, 马捷. 基于核心家系的EGR3基因与精神分裂症的关联研究. 遗传, 2012, 34(3): 307–314.
[16] Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, Parker JS, Jin J, Hammond SM. MicroRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder., 2007, 8(2): R27.
[17] Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome., 1999, 56(10): 940–945.
[18] Sellier C, Hwang VJ, Dandekar R, Durbin-Johnson B, Charlet-Berguerand N, Ander BP, Sharp FR, Angkustsiri K, Simon TJ, Tassone F. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome., 2014, 9(8): e103884.
[19] Cattane N, Mora C, Lopizzo N, Lopizzo N, Borsini A, Maj C, Pedrini L, Rossi R, Riva MA, Pariante CM, Cattaneo A. Identification of a miRNAs signature associated with exposure to stress early in life and enhanced vulnerability for schizophrenia: New insights for the key role of miR- 125b-1-3p in neurodevelopmental processes., 2019, 205: 63–75.
[20] He K, Guo C, Guo M, Tong S, Zhang Q, Sun H, He L, Shi Y. Identification of serum microRNAs as diagnostic biomarkers for schizophrenia., 2019, 156: 23.
[21] Guella I, Sequeira A, Rollins B, Morgan L, Torri F, van Erp TG, Myers RM, Barchas JD, Schatzberg AF, Watson SJ, Akil H, Bunney WE, Potkin SG, Macciardi F, Vawter MP. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex., 2013, 47(9): 1215–1221.
[22] Siegert S, Seo J, Kwon EJ, Rudenko A, Cho S, Wang W, Flood Z, Martorell AJ, Ericsson M, Mungenast AE, Tsai LH. The schizophrenia risk gene product miR-137 alters presynaptic plasticity., 2015, 18(7): 1008– 1016.
[23] Hauberg ME, Holm-Nielsen MH, Mattheisen M, Askou AL, Grove J, Børglum AD, Corydon TJ. Schizophrenia risk variants affecting microRNA function and site-specific regulation of NT5C2 by miR-206., 2016, 26(9): 1522–1526.
[24] Parts L, Hedman Å K, Keildson S, Knights AJ, Abreu- Goodger C, van de Bunt M, Guerra-Assunção JA, Bartonicek N, van Dongen S, Mägi R, Nisbet J, Barrett A, Rantalainen M, Nica AC, Quail MA, Small KS, Glass D, Enright AJ, Winn J; MuTHER Consortium, Deloukas P, Dermitzakis ET, McCarthy MI, Spector TD, Durbin R, Lindgren CM. Extent, causes, and consequences of small RNA expression variation in human adipose tissue., 2012, 8(5): e1002704.
[25] Siddle KJ, Deschamps M, Tailleux L, Nédélec Y, Pothlichet J, Lugo-Villarino G, Libri V, Gicquel B, Neyrolles O, Laval G, Patin E, Barreiro LB, Quintana-Murci L. A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection., 2014, 24(5): 850–859.
[26] Huan T, Rong J, Liu C, Zhang X, Tanriverdi K, Joehanes R, Chen BH, Murabito JM, Yao C, Courchesne P, Munson PJ, O'Donnell CJ, Cox N, Johnson AD, Larson MG, Levy D, Freedman JE. Genome-wide identification of microRNA expression quantitative trait loci., 2015, 6: 6601.
[27] Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation., 2009, 11(3): 228–234.
[28] Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes., 2005, 11(3): 241–247.
[29] Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing., 2007, 448(7149): 83–86.
[30] Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene., 2008, 36(16): 5232–5241.
[31] Kos A, Olde LoohuisNF, Wieczorek ML, Glennon JC, Martens GJ, Kolk SM, Aschrafi A. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase., 2012, 7(2): e31022.
[32] Chun S, Du F, Westmoreland JJ, Han SB, Wang YD, Eddins D, Bayazitov IT, Devaraju P, Yu J, Mellado Lagarde MM, Anderson K, Zakharenko SS. Thalamic miR-338-3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion., 2017, 23(1): 39–48.
[33] Kos A, de Mooij-Malsen AJ, van Bokhoven H, van Bokhoven H, Kaplan BB, Martens GJ, Kolk SM, Aschrafi A. MicroRNA-338 modulates cortical neuronal placement and polarity., 2017, 14(7): 905–913.
[34] Ferrari R, Grassi M, Salvi E, Borroni B, Palluzzi F, Pepe D, D'Avila F, Padovani A, Archetti S, Rainero I, Rubino E, Pinessi L, Benussi L, Binetti G, Ghidoni R, Galimberti D, Scarpini E, Serpente M, Rossi G, Giaccone G, Tagliavini F, Nacmias B, Piaceri I, Bagnoli S, Bruni AC, Maletta RG, Bernardi L, Postiglione A, Milan G, Franceschi M, Puca AA, Novelli V, Barlassina C, Glorioso N, Manunta P, Singleton A, Cusi D, Hardy J, Momeni P. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia., 2015, 36(10): 2904.e13 –26.
[35] Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets., 2005, 120(1): 15–20.
[36] Bhattacharya A, Cui Y. Knowledge-based analysis of functional impacts of mutations in microRNA seed regions., 2015, 40(4): 791–798.
[37] Mencia A, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss., 2009, 41(5): 609–613.
[38] Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome., 2012, 53(1): 348–353.
[39] Jiang Q, Zhao H, Li R, Zhang Y, Liu Y, Wang J, Wang X, Ju Z, Liu W, Hou M, Huang J. In silico genome-wide miRNA-QTL-SNPs analyses identify a functional SNP associated with mastitis in Holsteins., 2019, 20(1): 46.
[40] Chai J, Chen L, Luo Z, Zhang T, Chen L, Lou P, Sun W, Long X, Lan J, Wang J, Pu H, Qiu J, Shuai S, Guo Z. Spontaneous single nucleotide polymorphism in porcine microRNA-378 seed region leads to functional alteration., 2018, 82(7): 1081–1089.
[41] Richardson K, Lai CQ, Parnell LD, Lee YC, Ordovas JM. A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS., 2011, 12: 504.
[42] Hou Y, Liang W, Zhang J, Li Q, Ou H, Wang Z, Li S, Huang X, Zhao C. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338-3p reduces BDNF production., 2018, 199: 176–180.
[43] Rossi M, Kilpinen H, Muona M, Surakka I, Ingle C, Lahtinen J, Hennah W, Ripatti S, Hovatta I. Allele-specific regulation of DISC1 expression by miR-135b-5p., 2014, 22(6): 840–843.
[44] Hauberg ME, Roussos P, Grove J, Børglum AD, Mattheisen M, Schizophrenia Working Group of the Psychiatric Genomics Consortium. Analyzing the Role of MicroRNAs in schizophrenia in the context of common genetic risk variants., 2016, 73(4): 369–377.
[45] Manley W, Moreau MP, Azaro M, Siecinski SK, Davis G, Buyske S, Vieland V, Bassett AS, Brzustowicz L. Validation of a microRNA target site polymorphism in H3F3B that is potentially associated with a broad schizophrenia phenotype., 2018, 13(3): e0194233.
[46] John J, Bhatia T, Kukshal P, Chandna P, Nimgaonkar VL, Deshpande SN, Thelma BK. Association study of MiRSNPs with schizophrenia, tardive dyskinesia and cognition., 2016, 174(1–3): 29–34.
[47] Wu S, Zhang R, Nie F, Wang X, Jiang C, Liu M, Valenzuela RK, Liu W, Shi Y, Ma J. MicroRNA-137 Inhibits EFNB2 expression affected by a genetic variant and is expressed aberrantly in peripheral blood of schizophrenia patients., 2016, 12: 133–142.
[48] Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs., 2015, 4.
[49] Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites., 2010, 11(8): R90.
[50] Sticht C, De La Torre C, Parveen A, Gretz N. MiRWalk: an online resource for prediction of microRNA binding sites., 2018, 13(10): e0206239.
[51] Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function., 2019, 47(D1): D155–D162.
[52] Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes., 2004, 10(10): 1507–1517.
[53] Gong J, Liu C, Liu W, Wu Y, Ma Z, Chen H, Guo AY An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools., 2015, 2015: bav029.
[54] Deveci M, Catalyürek UV, Toland AE. MrSNP: software to detect SNP effects on microRNA binding., 2014, 15: 73.
[55] Liu C, Zhang F, Li T, Lu M, Wang L, Yue W, Zhang D. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs., 2012, 13: 661.
[56] Zorc M, Obsteter J, Dovc P, Kunej T. Genetic variability of MicroRNA genes in 15 animal species., 2015, 3: 51–56.
Schizophrenia-associated single nucleotide polymorphisms affecting microRNA function
Wenquan Liang1, Yu Hou2, Cunyou Zhao1
MicroRNAs (miRNAs) compose a class of non-coding transcripts with a mean length of 22 nucleotides, and play critical roles in regulating gene expression in the process of development, proliferation and differentiation of neurons. Recent genome-wide association studies (GWAS) find most of schizophrenia-associated single nucleotide polymorphisms (SNPs) locating in the non-coding regions, providing functional implications of miRNAs in the development of schizophrenia. In this review, we highlight the interplays between GWAS-SNPs and miRNAs in four perspectives: SNP in miRNA gene; miRNA located in the host gene; SNP located in the miRNA’s seed sequence; SNP located in the miRNA’s binding site. We also speculate on the future research on the role of miRNA in the development of schizophrenia.
GWAS; SNP; miRNA; schizophrenia
2019-05-08;
2019-07-07
广东省科技计划项目(编号:2019B030316032),广州市科技计划项目(编号:201804010259)和国家自然科学基金项目(编号:81601175,81671333)资助[Supported by the Guangdong Science and Technology Foundation (No. 2019B030316032), the Guangzhou Science and Technology Foundation (No. 201804010259) and the National Natural Science Foundation of China (Nos. 81601175, 81671333)]
梁文权,硕士在读研究生,专业方向:表观遗传。E-mail: wen1390229749@163.com
赵存友,博士,教授,研究方向:表观遗传。E-mail: cyzhao@smu.edu.cn
10.16288/j.yczz.19-126
2019/8/5 20:59:49
URI: http://kns.cnki.net/kcms/detail/11.1913.R.20190805.2059.003.html
(责任编委: 夏昆)
