Up-regulation of tumor necrosis factor-α pathway survival genes and of the receptor TNFR2 in gastric cancer
2019-04-24AnaFlviaTeixeiraRossiliaCocenzoContieroFernandadaSilvaManoelCaetanobioEduardoSeverinoAnaElizabeteSilva
Ana Flávia Teixeira Rossi,Júlia Cocenzo Contiero,Fernanda da Silva Manoel-Caetano,Fábio Eduardo Severino,Ana Elizabete Silva
Abstract
Key words: Gastric cancer;Tumor necrosis factor-α signaling;TNFR1;TNFR2;Cellular survival;MicroRNAs
INTRODUCTION
As the sixth most common cancer and the fifth leading cause of cancer-related death worldwide,gastric cancer (GC) is currently one of the most relevant neoplasms[1,2].Despite the geographical variabilities,it has a high incidence especially in Asia and Eastern Europe,and also in developing countries of South America[3].In Brazil,it is estimated 13540 new cases of this cancer in men and 7750 in women in the year 2019,occupying the fourth in incidence among men and the sixth among women[4].Intestinal-type adenocarcinoma is the most common type of GC and is the result of a multi-step process that begins with chronic gastritis and can progress to atrophic gastritis,intestinal metaplasia,dysplasia and cancer[5].This process can originate from chronic inflammation,mainly as a consequence ofHelicobacter pylori(H.pylori)infection[6].This Gram-negative bacterium promotes oxidative stress that induces DNA damage and triggers a repair response by stimulating growth and survival factors as well as regulatory cytokines that can initiate the carcinogenic process[7].
Tumor necrosis factor (TNF)-α is a pleiotropic cytokine important to ensure tissue homeostasis[8,9].Deregulation of the TNF-α signaling can affect cellular responses,causing inflammatory diseases and cancer[10].It is one of the main pro-inflammatory mediator produced during the inflammatory response[11],causes diverse cellular responses such as apoptosis,cell survival,angiogenesis,and metastasis[11,12].In cancer,TNF-α can have both pro- and anti-tumoral effects based on interaction with its receptors TNFR1 and TNFR2[13].Only TNFR1 possesses an intracellular death domain and thus can induce both apoptotic signaling and transcription of cell survival genes[14].Although its role is less well understood,TNFR2 activation largely results in NF-κB stimulation and cell proliferation[15].
In gastric tissues,increased expression of TNF-α has been observed in the normal mucosa as well as in chronic gastritis,intestinal metaplasia and dysplasia[16],active chronic gastritis[17]and GC[18],this factor plays an important role in progression of the lesion cascade that leads to GC.
Recently,we reported up-regulation of TNF-α mRNA and protein and other inflammatory mediators in chronic gastritis associated withH.pylori;we also observed deregulation of microRNAs (miRNAs) that interact with genes encoding cytokines.Interestingly,after bacterial eradication treatment,expression ofTNFmRNA was reduced and that of several miRNAs,such as miR-103a and miR-181c,was increased[19].Considering that TNF-α regulates cellular processes as apoptosis and cell survival according to binding to its TNFR1 and TNFR2 receptors,we used GC fresh tissue samples to investigate the mRNA expression of these receptors and important downstream genes involved in TNF-α signaling pathway [TNF,TNFR1(TNFRSF1A),TNFR2(TNFRSF1B),TRADD,TRAF2,CFLIP(CFLAR),NFKB1,NFKB2,CASP8andCASP3],and potentially related miRNAs (miR-19a,miR-34a,miR-103a,miR-130a and miR-181c) chosen from public database search[20-29].In addition,we evaluated the relationship between miRNA and mRNAviaconstruction of an interaction network.We observe that TNF-α signaling is deregulated in GC through up-regulation ofTNFR2and downstream survival genes,which may favor the cell survival,and conversely,down-regulation of TNFR1 andCASP3involved in apoptosis evasion.
MATERIALS AND METHODS
Clinical samples
This study was approved by local Research Ethics Committee (CEP - IBILCE/UNESP,number 1.336.892),and written informed consent was obtained from all individuals in previous studies[30,31].
A total of 30 fresh tissues samples of gastric adenocarcinoma were collected from gastric biopsies or surgical resection of the 30 patients attended at the Hospital de Base,São José do Rio Preto,SP,Brazil,without previous chemotherapy and radiotherapy,as previously described[30,31].The samples were diagnosed according Lauren classification as diffuse and intestinal- type[32].In addition,four fresh tissue samples were collected from gastric biopsies from individuals with histologically normal gastric mucosa (H.pylori-negative).These normal tissue samples were used as a pool for calibration in the reverse transcription (RT)-qPCR analysis.The demographic and clinical pathological data are presented in Table 1.For this study,total DNA and RNA stored and extracted using TRIzol®reagent (InvitrogenTM,Carlsbad,Califórnia,United States) in previous works were used[30,31].
Molecular diagnosis of H.pylori infection
Molecular diagnosis ofH.pyloriwas performed by nested PCR in DNA samples to evaluate the presence of theHSP60gene according to the protocol described by Singhet al[33].A 501-bp fragment was only observed inH.pylori-positive samples.Negative controls,without DNA and withH.pylori-negative DNA,were used in all reactions.
Quantification of mRNA and miRNA expression by RT-qPCR
Complementary DNA (cDNA) was synthesized from mRNA and miRNA using a High Capacity cDNA Archive Kit (Applied Biosystems,California,United States) and TaqMan®MicroRNA Reverse Transcription Kit (Applied Biosystems,California,United States),respectively,according to the manufacturer’s protocol.
Quantitative polymerase chain reaction (qPCR) was performed using aStepOnePlus Real Time PCR System 2.2.3 (Applied Biosystems,California,United States) with TaqMan®for target genesTNF(Hs01113624_g1),TNFR1 (TNFRSF1A)(Hs01042313_m1),TNFR2 (TNFRSF1B) (Hs00961749_m1),TRADD(Hs00182558_m1),TRAF2(Hs00184192_m1),CFLIP (CFLAR)(Hs00153439_m1),NFKB1(Hs 00765730_m1),NFKB2(Hs01028901_g1),CASP8(Hs01116281_m1) andCASP3(Hs00234387_m1),and for target miRNAs hsa-miR-19a-3p (MIMAT0000073;ID 000395),hsa-miR-34a-3p (MIMAT0004557;ID 002316),hsa-miR-103a-3p(MIMAT0000101;ID 000439),hsa-miR-130a-3p (MIMAT0000425;ID 000454) and hsamiR-181c-5p (MIMAT0000258;ID 000482)(Applied Biosystems,California,United States).All reactions were performed in triplicate with a final volume of 10 µL using GoTaq®Probe qPCR Master Mix 2X (Promega,Wisconsin,United States).Relative quantification (RQ) of mRNA and miRNA expression was calculated by the 2(-ΔΔCt)method[34]using a pool of normal mucosa samples as a calibrator.Reference genesACTB(Catalog#:4352935E) andGAPDH(Catalog#:4352934E) and endogenous RNU6B (ID 001093) and RNU48 (ID 001006) were adopted for normalization of mRNA and miRNA quantification,respectively,as validated in our previous studies[17,28].qPCR experiments followed MIQE guidelines[35],and RQ values are expressed as the median of gene and miRNA expression for GC in relation to that of the normal mucosa pool.

Table 1 Characterization of individuals with normal mucosa and gastric cancer
In silico analysis for prediction of miRNA targets and the miRNA-mRNA interaction network
In silicoanalysis was performed using public databases for predicted and validated target genes of the five miRNAs evaluated.The databases used were as follows:TarBarse[20],miRWalk 2.0[21],miRTarbase[22],miRDB (MirTar2 v4.0)[23,24],microRNA.org[25],PITA 0 0 ALL[26],TargetScan[27],RNA-22[28]and miRmap[29].Only target genes predicted by at least three databases were considered.
Data were integrated using bioinformatic methods,and protein annotations were then used to construct protein:protein interaction networks (PPI).The PPI networks were generated using MetasearchSTRINGplatform v10.5[36].Data visualization and integration between PPI and miRNA:target gene networks were performed usingCytoscapev3.1.1[37].
Statistical analysis
The D’Agostino and Pearson normality test was used to evaluate the distribution of the data,which did not present normal distribution,therefore non-parametric tests were used.Alterations in expression of genes or miRNAs in the GC group in relation to a pool of normal mucosa were evaluated by the Wilcoxon Signed Rank test[38].The Mann-Whitney test was employed to investigate associations ofH.pyloriinfection,adenocarcinoma histological type,gender and age with mRNA and miRNA expression in GC group.Correlation analysis between mRNA expression and between miRNA/mRNA expression was performed by Spearman's correlation.Values ofP< 0.05 were considered statistically significant.
RESULTS
Molecular diagnosis for H.pylori
Among the 30 GC samples diagnosed forH.pyloriinfection,50% (15) were positive for the presence of this bacterium.All four samples of normal gastric mucosa had the diagnosis confirmed asH.pylori-negative (Table 1).
Expression of TNF-α pathway genes
GC samples showed significantly up-regulated mRNA expression ofTNF(RQ=3.78,P< 0.001),TNFR2(RQ=1.98,P< 0.001),TRADD(RQ=2.14,P=0.004),TRAF2(RQ=3.70,P< 0.001),CFLIP(RQ=2.03,P< 0.001),NFKB2(RQ=2.16,P< 0.001),andCASP8(RQ=2.58,P< 0.001) in comparison with normal mucosa,whereasTNFR1(RQ=0.66,P=0.037) andCASP3(RQ=0.29,P< 0.0001) were down-regulated (Figure 1).No any significant change inNFKB1mRNA expression was found in GC samples.
Correlation analysis between the mRNA expression of genes involved in the TNF-α pathway showed an intricate network of positive correlations in GC (Table 2).TNFAcorrelated with expression of all other genes,with exception ofTNFR1,while this showed a strong correlation only withTNFR2.Conversely,TNFR2correlated positively with expression of all other genes.The strongest correlation was observed betweenCFLIPandCASP8(r=0.91,P< 0.001) (Table 2).
Expression of miRNAs related to TNF-α pathway
In general,expression of miRNAs (miR-19a,miR-103a,miR-130a and miR-181c)were not deregulated in GC (Figure 2),an exception is the significant up-regulation of miR-34a (RQ=1.39,P=0.017).
No association between miRNA expression was found for gender,age orH.pyloriinfection in GC samples.However,association analysis showed that expression of miR-103a (P=0.035) and miR-130a (P=0.011) was significantly increased in the diffuse-type of GC (RQ=1.53 and 4.83,respectively) compared to the intestinal-type(RQ=0.55 and 0.52,respectively).In addition,expression of miR-103a was downregulated when comparing only the intestinal- type of GC samples with normal mucosa (RQ=0.55,P=0.037) (Figure 3).When we evaluated the influence of these factors on mRNA expression of TNF-α pathway genes,no association was found.
miRNA–mRNA correlation and interaction networks
A miRNA:mRNA interaction network integrating miRNAs with the proteins encoded by genes of the TNF-α pathway was constructed based onin silicoanalysis using public databases (Figure 4).The obtained protein-protein interaction (PPI) network reinforced the significant correlation gene:gene found between the genes of the TNF-α pathway in GC (Table 2) samples.We also performed a correlation analysis between the transcripts levels of miRNAs:mRNA of TNF-α pathway in GC samples,but no negative correlation was observed (data not shown).However,the miRNA:mRNA interaction network shows several possible relationships between mRNA and miRNAs evaluated.
This interaction network generated in this study suggests that all evaluated miRNAs can act on the TNF-α pathway to influence cellular responses withTNFas a predicted or validated target.Furthermore,it highlights miR-19a and miR-103a,which can regulate the greatest number of genes,includingTNF,TNFR1,TNFR2,CFLIP,TRADD,CASP3andCASP8.
DISCUSSION
This is the first study to investigate the expression of genes that participate in the TNF-α pathway and its relationship to miRNA expression in GC.Our results show that this pathway is deregulated in GC may promote cell survival through TNFα/TNFR2/NF-κB and to prevent apoptosis.We observed up-regulation ofTNF,TNFR2receptor,downstream cell survival genes asTRAF2,TRADD,CFLIPandNFKB2,beyondCASP8and miR-34a,and down-regulation ofTNFR1andCASP3,indicating predominant expression of anti-apoptotic in relation to pro-apoptotic mediators in GC tissues.In addition,the miRNA:mRNA interaction network generated indicates that miR-19a,miR-34a,miR-103a,miR-130a and miR-181c may target genes of the TNF-α signaling,such asTNFA,TNFR1andTNFR2.

Figure 1 Relative expression of tumor necrosis factor-α pathway genes in gastric cancer.
TNF-α pathway genes and their receptors may have opposite effects on tumorigenesis depending on the developmental stage and tumor type[11].In GC,the present study showed increased expression ofTNFAso that this cytokine has a promoting effect that is necessary even when such interleukins as IL-1β and IL-6 are present in the tumor tissue[8].Considering the role of TNFA receptors,Oshimaet al[8]showed that TNFR1-mediated stimulation of TNF-α signaling in bone marrowderived cells from knockout mice induces tumor-promoting factors,asNoxo1andGna14,in tumor cells.In contrast,anti-tumor effects of this receptor were observed in CD4+T cells isolated from mice,as the absence of TNFR1 signaling promoted angiogenesis and carcinogenesis[39].Regarding TNFR2,induction of its expression in a colorectal cancer cell line results in a significant increase in cellular proliferation,promoting tumor growthviathe PI3K/AKT pathway[40].Similar to the results of the present study,Al-Lamkiet al[41]found a significant increase in TNFR2,but not in TNFR1,in clear cell renal carcinoma and reported that this increase was correlated with a high degree of malignancy and activation of NF-κB and VEGFR2,causing cell cycle entry.Furthermore,blocking TNFR2 by short hairpin RNA (shRNA) in Lewis lung carcinoma cell culture leads to increased TNF-mediated apoptosis and decreased angiogenesis,which may result in tumor regression[42].Therefore,TNFR2 is possibly the major receptor related to tumor progression triggered by the TNF-α pathway in GC.
In the cell proliferation pathway (Figure 5),after binding of TNF-α to TNFR2,TRAF2 associates with the intracellular portion of the receptor.As the key mediator of this signaling pathway,TRAF2 promotes the binding of anti-apoptotic proteins(cIAP1 and cIAP2) to TNFR2[43-45].This TNFR2/TRAF2 interaction evokes transcriptional activation of genes related to cell survival,such as NF-κB[15],which induces transcription of anti-apoptotic genes including cFLIP,TRAF1 and TRAF2[46,47]to prevent cell death.In our study,we observed up-regulation ofTRAF2,NFKB2andCFLIPmRNAs in GC.These findings are consistent with the positive correlations observed betweenTNFR2mRNA and anti-apoptotic genes (CFLIPandTRAF2) as well as activation of cell proliferation (NFKB1andNFKB2).All these results together corroborate the predominance of cell survival pathway gene expression mediated by TNF-α/TNFR2/NF-κB in GC.
However,TNFR1mRNA was down-regulated in GC samples,indicating that the apoptosis pathway may be impaired.This result is also consistent with increased expression ofTRADD,TRAF2andCFLIPand decreased expression ofCASP3.Interaction between TRAF2 and TNFR1,viathe TRADD adapter protein[48],inhibits the ability of this receptor to induce apoptosis[49]by impairing formation of the deathinducing signaling complex (DISC),also called Complex II (Figure 5).When the apoptotic pathway is activated,TNFR1 and the adapter proteins TRADD,TRAF2 and RIP1 associated with it undergo conformational changes that result in TRADD dissociation from the receptor and release of the death domain.TRADD together with FADD and pro-caspase-8 form the DISC[50,51],which activate caspase-8viaproteolytic cleavage of its precursor,consequently resulting in activation of the caspase-3 effector and apoptosis[14].In contrast,cFLIP,which is induced by NF-κB,inhibits the activity of the DISC due to its homology with caspase-8[50].cFLIP interacts directly with procaspase-8 to form a heterodimer that suppresses precursor cleavage and consequentactivation of the caspase[52].Therefore,when NF-κB is activated,cells are resistant to apoptosis triggered by TNFR1 through promotion of cFLIP expression and inhibition of pro-caspase-8 binding to FADD[50].In the GC samples evaluated in this study,we found increased expression ofCASP8mRNA,which may result from factors generated in inflammatory conditions such as nitric oxide[53]and interferon-Υ[54]that are up-regulated in stomach cancer[55,56].However,the cFLIP-mediated apoptotic signaling blockade results in low expression ofCASP3and consequent reduction of cell death.
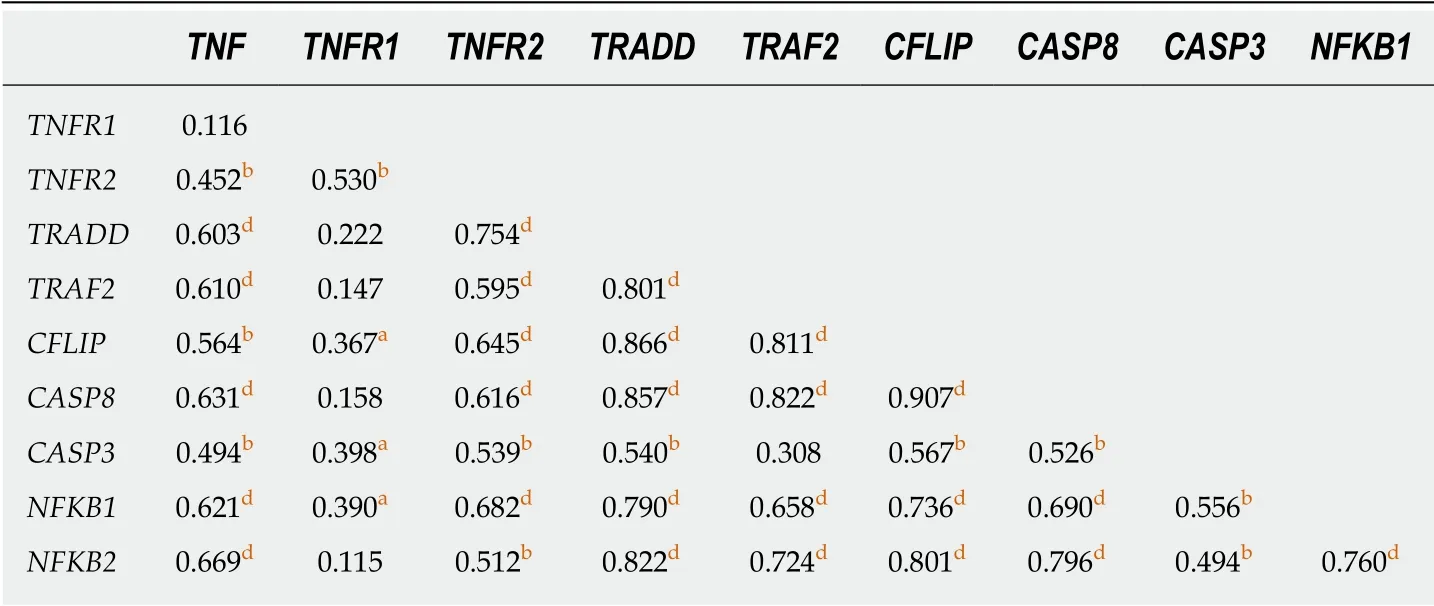
Table 2 Correlation analysis between mRNA expression and tumor necrosis factor-α pathway genes in gastric cancer
Deregulation of global gene expression is a key characteristic during malignant progression,and miRNAs are important regulators of this process,including in GC[57].Thus,we consulted public databases and selected miRNAs miR-19a,miR-34a,miR-103a,miR-130a and miR-181c,which target genes of the TNF-α pathway,as a change in their expression may impact regulation of this pathway.In general,the expression of miRNAs was not deregulated in GC,though the miR-34a was up-regulated.
Several studies highlight the tumor-suppressor role of miR-34a,whereby reduced expression is associated with an advanced clinical stage,lymph node metastasis[58],a lower survival rate[59],and a higher recurrence rate[60]in patients with GC.However,our results corroborate an oncogenic action of the miR-34a because a significant increase in its expression observed in GC[61]and association with a lower overall survival[62].Nonetheless,as a recent meta-analysis did not find a significant association between expression of this miRNA and overall survival[63],further studies are needed to clarify the role of miR-34a in GC.
In our study,expression of the oncogenic miRNAs miR-19a,miR-103a and miR-130a was not deregulated in GC samples.When we stratified the samples by histological type,significantly higher expression of both miR-103a and miR-130a was detected in diffuse-type GC than in the intestinal-type.Furthermore,expression of miR-103a was down-regulated when considered only the intestinal type of GC in relation to normal mucosa.Although miR-103a has already been confirmed as a highly sensitive and specific biomarker of the diffuse type[64],the action of miR-130a in gastric carcinogenesis remains unsure.All these miRNAs may have both oncogene and tumor-suppressor roles in different stages of neoplastic development and tumor type.Up-regulation of these miRNAs has been associated with metastasis,proliferation and cell invasion in GC[65-67].Conversely,some studies have shown decreased expression of miR-103a and miR-130a,correlating these findings with GC tumorigenesis,cell migration,invasion and metastasis[68,69].
Regarding miR-181-c,our results showed no alteration of its expression in GC samples.However,in our previous study,we observed reduced expression of this miRNA in biopsies from chronic gastritis patients,but after bacterial eradication therapy,a significant increase of its expression was observed.Thus suggesting that the inflammatory process induced byH.pylorican regulate its expression[19].Few studies have evaluated this miRNA in GC and the results are discordant.Recently,Zabagliaet al[70]observed lower expression of miR-181c in GC patients compared with control and chronic gastritis groups.On the other hand,Cuiet al[71]correlated reduced expression of this miRNA with a higher survival rate of GC patients.Therefore,the role of this miRNA in the gastric carcinogenesis needs to be further investigated.
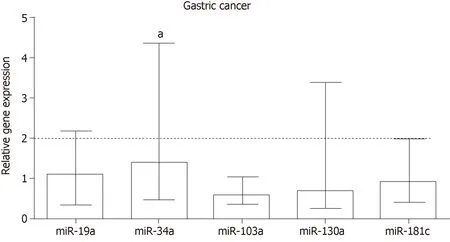
Figure 2 Relative expression of miRNAs miR-19a,miR-34a,miR-103a,miR-130a and miR-181c in gastric cancer.
H.pyloriinfection is an important factor associated with gastric lesions,and GC may result in deregulation of expression of various genes and miRNAs that function in defense mechanisms,inflammatory and immune responses and cellular kinetics[19,72].Although,in the present study,mRNA and miRNA expression were not influenced byH.pyloriinfection,in a previous study[19],we observed thatH.pyloriinfection deregulates miRNA expression in chronic gastritis patients,suggesting that it may act differently at distinct stages of gastric tumorigenesis,most likely being more important in the early stages of carcinogenesis.
In addition,we analyzed correlations and the miRNA:mRNA interaction network involving miR-19a,miR-34a,miR-103a,miR-130a and miR-181c,which can regulate the TNF-α pathway.Although no negative correlation between miRNA and mRNA expression was observed in GC,we did identify an intricate network of relationships between TNF-α pathway genes and miRNAs,especiallyTNFandTNFR2,which are predicted or validated targets of some of these miRNAs in other types of cancer,highlight miR-19a and miR-103a.Therefore both miRNAs are strong candidates for functional studies in GC cell line.
TNF-α is a validated target of miR-19a-3p[73],miR-34a-3p[74]and miR-130a-3p[75]in esophageal squamous cell carcinoma,macrophages and cervical cancer,respectively.Moreover,a negative feedback betweenTNFand miR-130a exists.In cervical cancer lines treated with TNF-α,an increase in the nuclear level of the p50 subunit of NF-κB occurs,which results in increased expression of miR-130a and consequently reduced expression ofTNF[75].TNF-α is also a predicted target of miR-103a-3p and miR-181c-5p (Figure 4) and is inversely correlated inH.pylori-positive chronic gastritis patients,both before and after eradication treatment[19].Absence of a negative correlation between miRNA and mRNA expression in GC samples does not invalidate the relationship shown in our miRNA:mRNA interaction network building from bioinformatics data,since the regulation of gene expression by miRNAs can be influenced by several factors,as infectious agents[19],DNA methylation[76]and probably by distinct stages of carcinogenesis.
As observed in our interaction network (Figure 4),TNF-α is not the only gene targeted by miRNAs.TNFR2can be regulated by miR-19a,miR-103a and miR-130a,whereas miR-19a also regulatesTNFR1.Other downstream genes are also predicted targets of these miRNAs,includingTRADD,CFLIP,CASP8andCASP3,highlighting that miR-19a and miR-103a target the largest number of genes of the TNF-α pathway.Therefore,deregulated expression of these miRNAs may influence several genes and disrupt cell processes such as survival and apoptosis.
Our study shows up-regulated expression of important genes of the TNF-α signaling pathway related to cell survival in GC mediated by TNFR2 receptor,and down-regulation of pro-apoptoticCASP3gene and the TNFR1 receptor.This predominant expression of anti-apoptotic in relation to pro-apoptotic mediators may favor cell survival and apoptosis evasion,highlighting the pro-tumor effect of TNF-α on GC.Furthermore,this signaling pathway can be regulated by the action of miRNAs,as shown by our interaction network,may influence the development and tumoral progression.However,further studies blocking TNFR2 should be conducted for negatively influence the expression of cell survival genes.

Figure 3 Relative expression of miRNAs miR-19a,miR-34a,miR-103a,miR-130a and miR-181c in intestinal and diffuse types of gastric cancer.
ARTICLE HIGHLIGHTS
Research background
Tumor necrosis factor (TNF)-α is a proinflammatory cytokine with opposite effects according activation of its TNFR1 and TNFR2 receptors,can provide signals for activation,differentiation,survival cell,invasion and propagation of cancer cells or induce apoptosis.This way,TNF-α signaling pathway perform several biological functions,as well as modulate the immune response and inflammation,so deregulation of this pathway has been implicated with inflammatory diseases and cancer.Therefore,studies are needed to better understand the relationships of this signaling network and the protumorigenic or antitumorigenic effects,as well as the tumor development and progression in different types of neoplasms.
Research motivation
We previously evaluated the effect of eradication therapy of Helicobacter pylori (H.pylori) on expression levels of cytokines genes and the expression of miRNAs involved with regulation of inflammatory process.We observed up-regulation of TNF-α mRNA and protein and other inflammatory mediators in samples of chronic gastritis patients infected by H.pylori.Moreover,we also observed deregulation of miRNAs that interact with genes encoding cytokines.Interestingly,after bacterial eradication treatment,the expression levels of TNF mRNA were reduced and that of several miRNAs,such as miR-103a and miR-181c,were increased.Considering that TNF-α can have both pro- and anti-tumoral effects activating processes as apoptosis and cell survival depending on the interaction with its TNFR1 and TNFR2 receptors,we proposed the present study.Therefore,we decided to quantify the transcript levels of the TNF-α signaling pathway genes and of TNFR1 and TNFR2 receptors,as well as the involvement of miRNAs that may participate in the regulation of this signaling pathway,in fresh tissues of gastric cancer (GC) patients.
Research objectives
Considering that while the TNF-α/TNFR1 binding promotes the activation of the apoptosis cascade and the TNF-α/TNFR2 leads to activation of the cell survival pathway,the main objective of this study was to investigate which of the two receptors are most expressed in GC,and downstream genes of TNF-α signaling pathway,thus favoring the cell survival or apoptosis processes.A secondary objective was to evaluate the relationship between miRNA and mRNAviaconstruction of an interaction network.The results may highlight important genes that regulate cell proliferation and possible molecular targets that act on gastric carcinogenesis.
Research methods
Sensitive and validated techniques were employed for RNA quantification of the genes and miRNAs in normal and tumor tissues.For this purpose,we used TaqMan gene and miRNA expression assays (Applied Biosystems,Foster City,CA,United States) with specific probes for each gene and miRNA for relative quantification.The reactions were analyzed using the StepOnePlus real-time PCR System (Applied Biosystems,Foster City,CA,United States).Molecular diagnosis ofH.pyloriwas performed by nested PCR for geneHSP60.In addition,we also used a bioinformatic tool ‘miRNA Data Integration Portal’ (http://ophid.utoronto.ca/mirDIP/) to build an miRNA:mRNA interaction network by using Cytoscape software (version 3.1.1).
Research results
Ours results showed up-regulation ofTNFR2receptor and its ligandTNF,as well as downstream genes of cellular survival asTRADD,TRAF2,CFLIP,andNFKB2,besidesCASP8and miR-34a in GC tissues,whereas mediators of apoptosis such asTNFR1andCASP3were down-regulated.Although we did not observe changes in the expression of most miRNAs,the miRNA:mRNA interactions network suggests a mechanism of regulation mainly by miR-19a and miR-103a,which targets a greater number of genes of the TNF-α pathway.
Research conclusions
Our findings highlight the TNFR2 receptor as up-regulated in GC tissue,as well as its TNFA ligand,favoring the pro-tumoral effect of this cytokine and transcription of cell survival genesviaTNFA/TNFR2 /NF-kB,and down-regulation of TNFR1 andCASP3related to apoptosis evasion.Moreover,this pathway can be modulated by an intricate regulatory network of miRNA:mRNA.
Research perspectives
The antitumor effect of TNF-α has been investigated as a form of cancer therapy associated or not with chemotherapeutic agents;however,its toxicity and adverse effects have limited its application.Another strategy for cancer therapy is to block the TNF receptors,so may increase the effectiveness of the TNF-α treatment and decrease its systemic toxicity.Therefore,in vitrofunctional studies may better elucidate the role of these receptors in gastric carcinogenesis.
杂志排行
World Journal of Gastrointestinal Oncology的其它文章
- Identification of genomic features associated with immunotherapy response in gastrointestinal cancers
- Predictive factors of histological response of colorectal liver metastases after neoadjuvant chemotherapy
- Iodine-125 implantation with transjugular intrahepatic portosystemic shunt for main portal vein tumor thrombus
- Validated model for prediction of recurrent hepatocellular carcinoma after liver transplantation in Asian population
- Significance of HER2 protein expression and HER2 gene amplification in colorectal adenocarcinomas
