PRC1.6复合体表观遗传调控生殖谱系特异性基因的时空表达
2019-04-22孙晓伟李宏阳王健程博
孙晓伟,李宏阳,王健,程博,2
PRC1.6复合体表观遗传调控生殖谱系特异性基因的时空表达
孙晓伟1,李宏阳1,王健1,程博1,2
1. 兰州大学生命科学学院,兰州 730000 2. 兰州大学,教育部细胞活动及逆境适应重点实验室,兰州 730000
多梳抑制复合体1 (polycomb repressive complex 1, PRC1)是一类通过催化和识别染色质表观遗传修饰进而调控基因表达的蛋白复合体,主要参与干细胞干性维持、细胞分化以及细胞周期调控等生理过程,该复合体功能异常影响机体发育或导致癌症发生。在哺乳动物细胞内,PRC1根据组成差异被进一步细分为6种不同的亚型PRC1.1~PRC1.6,它们在靶基因群识别、表观调控机制及生物学功能上存在明显特化。近年来研究发现,PRC1.6复合体在胚胎干细胞及体细胞中对于稳定抑制生殖谱系特异性基因的表达至关重要,同时对于生殖干细胞的干性维持以及精子发生过程中生殖谱系特异性基因的精密调控承担着重要作用。本文在介绍PRC1.6复合体的发现、各组分的分子功能及其参与的生化反应途径的基础上,系统阐述了该复合体在胚胎发育、性腺发育、精子发生等过程中对生殖谱系特异性靶基因群的时空表达发挥的重要调控功能,并探讨了PRC1.6与已知表观遗传调控网络的相互作用,以期为进一步探索生殖谱系基因表达、精子发生的表观遗传调控机制以及男性不育的致病机制提供参考。
多梳抑制复合体1;PRC1.6;转录抑制;生殖谱系特异性基因;时空表达
表观遗传调控是实现真核生物基因选择性表达的主要途径。多梳蛋白家族(polycomb groups,PcGs)是一类重要的表观遗传调控因子,与多种干细胞的干性维持、细胞分化、细胞周期的调控、细胞衰老、X染色体失活等一系列细胞生理活动密切相关[1,2]。它主要通过两类蛋白复合体发挥功能——多梳抑制复合体Ⅰ和Ⅱ (polycomb repressive complex 1/2, PRC1/2)。近年来在高等动物细胞中不断鉴定出组分不同的PRC1亚型,且它们的生物学功能在靶基因群的选择及调控基因表达的作用机制中有所差异。PRC1.6复合体属于其中一类PRC1亚型,主要包含RING1B、PCGF6 (polycomb group ring finger protein 6)、MAXMyc-associated factor X)、MGA (MAX gene associated)、E2F6和L3MBTL2 (lethal (3) malignant brain tumour like 2)等组分。近年来通过对PRC1.6复合体不同核心组分在生物化学、分子生物学、细胞生物学及发育生物学等方面的研究,发现该复合体对于哺乳动物发育过程中多种相关细胞谱系的建成及维持是必需的。本文在介绍该复合体的发现、核心组分的分子生物学功能的基础上,对该复合体在高等动物胚胎发育、性腺发育、精子发生、胚胎干细胞(embryonic stem cells, ESCs)及生殖干细胞(germ line stem cells, GSCs)维持等过程中发挥的生物学功能展开了系统论述。
1 多梳蛋白家族PcGs
PcGs是一大类通过催化和识别组蛋白表观遗传修饰来改变染色质构象、对靶基因进行转录调控的蛋白。第一个多梳蛋白编码基因()是Pamela Lewis于1947年在黑腹果蝇()中发现的,1978年Edward Lewis发现突变导致果蝇发生同源异型转化(如雄性果蝇产生多对性梳的异常发育表型),进一步研究发现多梳蛋白是通过抑制同源异型基因的表达来调控果蝇的体节发育[1,3,4],其功能与发挥基因激活作用的三胸蛋白家族TrxGs(trithorax groups)相拮抗[5,6]。但是近几年也有关于PcGs蛋白激活基因表达的报道[7~9],表明PcGs蛋白在执行转录调控方面的机制可能是非常复杂且多效的。PcGs蛋白主要通过PRC1和PRC2两类蛋白复合体形式发挥作用。其中,EZH2是PRC2中具有催化活性的核心组分,可以催化组蛋白H3第27位赖氨酸的三甲基化(H3K27me3)[10]。其他的PRC2组分参与复合体组装及活性维持[11]。PRC1具有E3泛素连接酶活性,该活性由RING1A/ RING1B及PCGF的环指结构域(ring finger domain)的部分亲水性表面构成[12,13],催化组蛋白H2A第119位赖氨酸的单泛素化(H2AK119ub1)[14]。PRC1和PRC2 调控的靶基因群存在部分重叠,在多数情况下二 者存在相互招募的关系,互助实现对靶基因的转录 调控[15]。
高等动物中PRC1的组分较果蝇要复杂的多,除了对应果蝇PRC1的每个组分都衍生出多个同源蛋白之外,还增添了很多其他组分,且组分之间的组合方式非常复杂多样[16]。目前在哺乳动物中发现PRC1亚型有6类,称为PRC1.1~PRC1.6,其中PRC1.2和PRC1.4因包含有经典的Pc蛋白同系物CBX蛋白而被称作经典PRC1 (canonical PRC1),其余几种亚型因缺乏CBX蛋白而被列为非典型PRC1 (non-canonical PRC1)。本文着重讨论了PRC1.6亚型,关于其他亚型的组成及功能均有较多报道,请读者参考相关综述[17,18]。
2 PRC1.6复合体的发现、核心组分及其分子功能
2.1 PRC1.6复合体的发现
PRC1.6复合体先后被不同的研究团队发现,并被赋予了不同的名称(表1)。2002年,Ogawa等[19]在HeLa及成纤维细胞中利用免疫沉降技术、甘油密度梯度离心并结合质谱检测发现了与E2F6-标签融合蛋白结合的蛋白复合体—E2F6复合物(E2F6.com)。该复合体中除了包含转录因子E2F6/DP-1,还有另一对转录因子异二聚体MAX/MGA;同时发现该复合体在甘油密度梯度分离中与组蛋白甲基转移酶G9a、Eu-HMTase1 (GLP)部分重叠,具有H3K9的甲基转移酶活性[19]。2011年,美国纽约大学医学院Danny Reinberg课题组的Trojer等[20]研究L3MBTL2的功能时再次发现了该复合体,将其命名为PRC1L4 (PRC1-like4)复合体;2012年,同课题组的Gao等[21]通过免疫沉降/质谱实验发现了人源细胞中包含不同PCGF因子(PCGF1~PCGF6)的PRC1复合体,并根据包含PCGF因子的编号将PRC1复合体细分为PRC1.1~PRC1.6复合体亚型,各亚型除了共同组分是PRC1复合体的酶学核心组分RING1B (RING2)外其余组分差异较大。其中PRC1.6组分包括PCGF6、RING1B、RYBP、MAX、MGA、L3MBTL2、E2F6、DP-1/2、HP1γ和HDAC1/2等。2017年,Endoh等[22]在小鼠ESCs中利用带标签的PCGF6蛋白也分离到了类似的复合体并将其称为PCGF6-PRC1。以上4种复合体虽然名称各异,但是核心组分都含有PCGF6、E2F6、RING1B、L3MBTL2和HP1γ等因子(表1),本质上属于同一复合体或十分相近的同类复合体。为了叙述简洁,本文在后续介绍中将其统称为PRC1.6复合体。

表1 PRC1.6复合体的发现及组分
2.2 PRC1.6复合体各组分的分子功能
如上所述,PRC1.6复合体是一个多组分蛋白复合体,其组分大致可以分成与催化组蛋白修饰相关的因子和与复合体装配或靶向定位相关的因子。同其他PRC1亚型类似,PRC1.6复合体最主要的酶学活性是由RING1B提供的E3泛素连接酶活性,催化产生H2AK119ub1;而RYBP、PCGF6等因子对于RING1B的活性发挥起到重要的促进作用[14,21,23]。在该复合体中包含异染色质结合蛋白HP1γ,说明其可能介导PRC1.6与甲基化的H3K9之间存在功能互作;除此之外,Gao等[21]还在PRC1.6复合体中检测到了组蛋白去乙酰化酶HDAC1/2,说明该复合体在某些情况下可能与组蛋白去乙酰化酶协同作用抑制靶基因的转录。
PRC1.6蛋白复合体各组分的已知分子功能见表2。在ESCs中通过免疫共沉淀实验发现PCGF6与L3MBTL2、MAX、RING1B和RYBP具有相互作用,敲除时阻断了L3MBTL2与复合体中RING1B、RYBP和MAX等组分的互作,这说明整个复合体的完整性在一定程度上依赖于PCGF6[24]。另有研究表明PCGF6可通过招募RING1B到靶基因上催化H2AK119位点的泛素化并抑制其表达[22]。MGA对于PRC1.6复合体的装配和稳定性发挥着更加关键的作用,在HEK293T细胞中敲除发现不仅该复合体各特异性组分在靶基因上的富集程度均受到明显抑制,且其中几个核心特异组分的蛋白总量明显下降,如PCGF6、L3MBTL2和E2F6[25]。从L3MBTL2的RNA干扰实验可知,部分靶基因的上调与其基因上富集的H2K119ub1水平的明显下降相伴发生,说明该因子对于PRC1.6复合体在靶基因区域组蛋白H2AK119位点的泛素化是必需的。同时,体外实验还发现L3MBTL2能够诱发染色质凝缩[20]。
目前,关于PRC1.6复合体的靶向定位的研究结果显示该复合体的定位机制较为复杂,由多个组分协同执行,包括MAX/MGA和E2F6/DP-1/2这两对转录因子形成的异二聚体以及L3MBTL2[25](表2)。MAX和MGA通过各自的bHLHZip (basic Helix- Loop-Helix-Zipper)结构域结合形成异二聚体,并特异性识别E-boxes序列CACGTG;此外MGA的氮端还包含另一个DNA结合结构域,可识别T-box序列AGGC/TGC/TGA,该异二聚体在PRC1.6复合体的组装、稳定性及靶基因识别上都发挥着重要作用[25,26]。另一对转录因子是E2F6和DP-1或DP-2,二者结合共同识别E2F家族识别序列GCGGGA[27]。与E2F家族的其他转录激活因子不同,E2F6通常对其结合的序列调控的靶基因产生转录抑制效应。L3MBTL2包含4个MBT结构域,结合组蛋白H3和H4,但该结合是否依赖于组蛋白的甲基化状态目前还有争议[28,29]。

表2 PRC1.6复合体中各组分的主要生化及分子功能
3 PRC1.6复合体在胚胎发育中抑制生殖谱系相关基因的表达
近年来不断积累的针对PRC1.6组分的RNA干扰或基因敲除实验已经充分证实PRC1.6复合体在维持ESCs的自我更新及调控其分化能力方面,尤其在抑制生殖谱系相关基因表达中发挥重要作用。在小鼠中,该复合体组分的敲除往往造成胚层及/或胚外谱系不同程度的发育异常,多数情况下产生胚胎致死表型(表3),由此可见,PRC 1.6复合体在ESCs以及胚胎发育过程中发挥至关重要的调控功能。
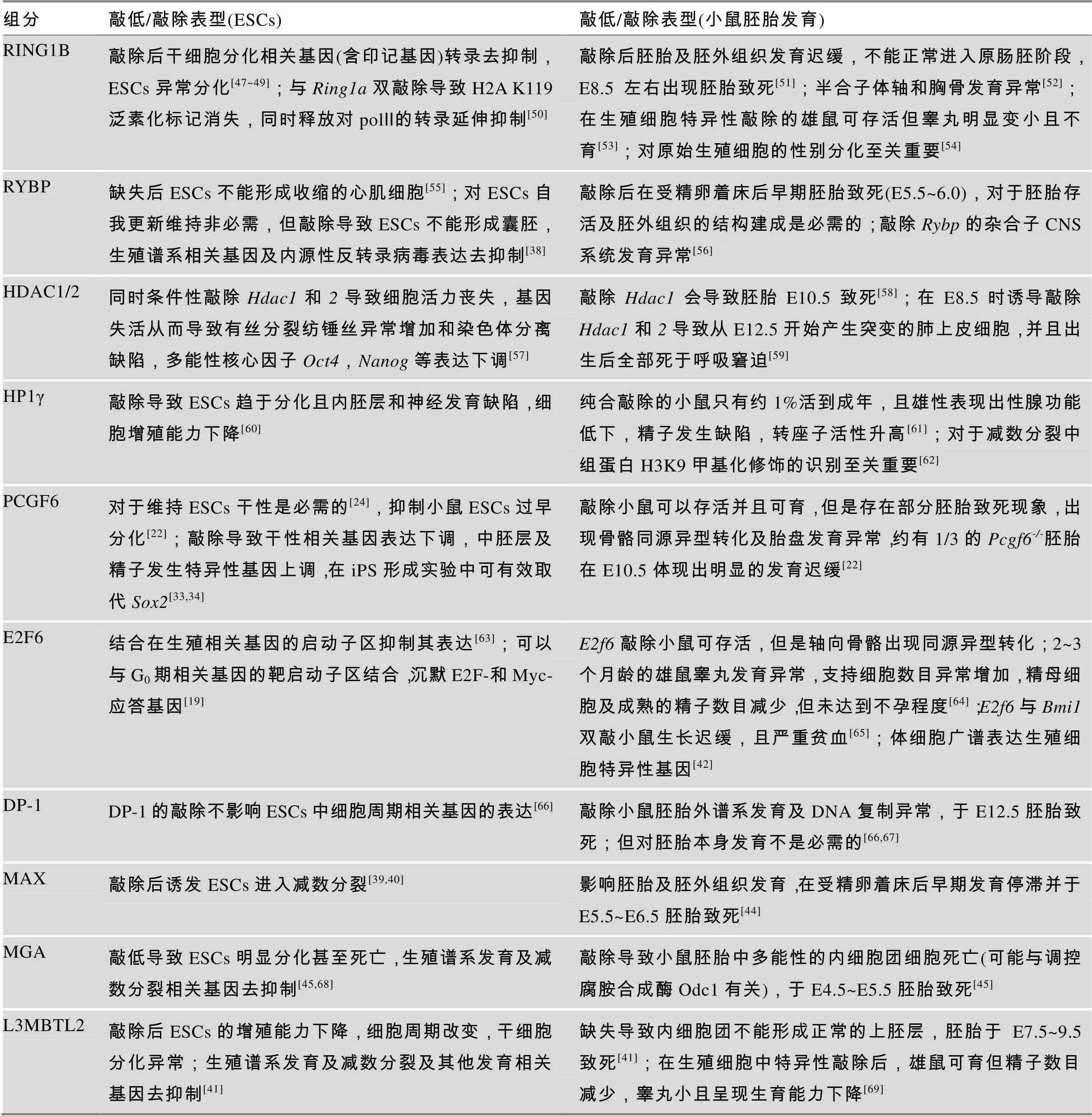
表3 PRC1.6组分下调对ESCs及小鼠胚胎发育或性腺发育的影响
3.1 PRC1.6复合体参与ESCs干性维持及调控ESCs正常分化
PRC1.6复合体的组分RING1B、MAX、MGA、HP1γ、PCGF6和L3MBTL2等都已被报道对于ESCs的干性维持是必需的,这些组分在被敲除后均会引起ESCs分化异常(表3)。另有一些因子如RYBP虽然对于ESCs的自我更新是非必需的,但对于ESCs的正常分化却是不可缺少的[38]。2013年,Maeda等[39]在小鼠ESCs中通过siRNA文库从864个候选基因中筛选能够抑制生殖细胞分化路径的重要基因,筛选到多个PRC1.6复合体组分,包括、和,其中敲低效应最为显著,可诱发ESCs进入类似减数分裂的状态。Suzuki等[40]在ESCs中诱导性敲除后也发现表达上调的基因主要包括减数分裂和精子发生过程中的相关基因,如、和等。通过细胞免疫荧光实验发现在Dox诱导敲除10天后,ESCs发生类似减数分裂细胞的形态学变化,出现减数分裂前期(细线期和偶线期)相关蛋白SYCP3的表达,因此PRC1.6复合体功能异常导致减数分裂相关基因的异常高表达,使ESCs越过原始生殖细胞(primordial germ cells, PGCs)直接形成类似减数分裂前期的细胞,说明MAX对于调控减数分裂的起始至关重要[40]。
在小鼠ESCs中PRC1.6复合体不同组分的ChIP-Seq数据显示,该复合体在全基因组中靶向上万个位点(由PCGF6、L3MBTL2和MGA共同靶向);当逐个敲除不同组分时,这些PRC1.6复合体靶基因中转录本水平发生上调的基因数目一般为数百个,如敲除导致882个基因上调[25,26];敲除导致421个[25]或167个[41]基因上调。这类对PRC1.6复合体调控最敏感的基因中包含不同类型的发育相关基因(如神经发育基因),但最显著的是特异性地包含生殖谱系及减数分裂关键基因[24,25,40,41],这是其区分于其他PRC1复合体亚型所独具的功能。在敲除的ESCs中表达显著上调(>10倍)的基因中有49个都是在精子发生过程中必不可少的基因[22]。在ESCs中敲除后发现与减数分裂相 关的基因、、和表达量上调[25]。同样,敲除的ESCs也呈现出生殖谱系相关基因特异性上调的现象[38]。另有研究表明,PRC1.6复合体同时抑制体细胞中生殖谱系相关基因的表达,如敲除基因的小鼠呈现出生殖谱系相关基因(和等)在体细胞中的异常表达[42,43]。这些实验证据都支持PRC1.6复合体在ESCs等其他非生殖细胞类型中发挥稳定抑制生殖谱系特异性基因表达的作用,维持这些细胞类型的身份,限制其向生殖细胞谱系异常分化。
3.2 PRC1.6复合体对于正常胚胎发育必不可少
PRC1.6复合体对于哺乳动物胚胎发育的正常进行是必需的,但每个组分被敲除后小鼠所呈现的胚胎发育异常状况却有所区别,这一定程度上反映出每个组分在该复合体功能发挥中的权重不同以及部分组分可能因为参与多种复合体而具备更为多样的发育调控功能(表3)。有研究表明,敲除小鼠在E5.5~6.5时表现出胚胎及胚外组织发育阻滞,且不具备明显的胚胎特征或胚胎与胚外组织分界不清晰[44]。的表达在小鼠E3.5时的内细胞团中以及E4.5~E6.5时具全能性的上胚层(epiblast)中均可检测到,当使用Mga/Mga纯合敲除,内细胞团在E4.5后随即发育停滞,细胞增殖无明显变化但细胞凋亡增加,上胚层不能正常形成[45]。由Mga衍生出来的Mga等位基因呈现出低表达表型,约一半Mga/Mga纯合小鼠在出生及哺乳期死亡,其余能存活并可育[45]。早期通过原位杂交实验发现在胚胎发育E9.5~10.5间广谱表达,在肢芽、腮弓及尾部区域高表达[26]。为进一步澄清其在E4.5之后的发育调控功能,近期研究发现表达低剂量的Mga/Mga杂合转基因小鼠在E7.5~E12.5之间死亡,主要由于E7.5之前上胚层全能性细胞发育停滞导致[46],所以Mga对于上胚层的正常分化是必不可少的。缺失会导致多能性ESCs增殖及分化异常,在E7.5左右胚胎因不能正常形成原肠胚而致死,致死原因尚不明确[41]。
4 PRC1.6复合体调控雄性性腺发育及精子生成过程中生殖谱系基因的顺次激活
PRC1.6复合体不仅可以影响胚胎发育中生殖谱系相关基因的表达,还可以影响精子生成和雄性性腺发育。从表3中可知,当分别敲除和时,小鼠都可存活,但会表现出不同程度的睾丸发育不良及精子生成障碍。特异性敲除生殖细胞中的小鼠呈现出睾丸发育异常和不育表型,GSCs数目明显下降并且异常分化[53]。最新研究表明,L3MBTL2在小鼠精原细胞减数分裂前期高表达,特异性敲除生殖细胞中的雄鼠虽然可育但表现出睾丸重量减轻,附睾中的精子数目减少及异常精子增多,并出现睾丸早衰现象[69]。进一步研究发现这些雄鼠的精原细胞在减数分裂前期出现染色体异常联会,且影响延长型精子中组蛋白乙酰化水平和精子成熟过程中组蛋白向鱼精蛋白的转变,说明参与精子生成过程进而影响性腺发育。这些研究结果证明了PRC1.6复合体确实在精子发生及性腺发育过程中具有重要的作用。
近期关于PRC1.6复合体对生殖细胞分化过程中生殖谱系基因的表达调控也有相应的报道。Maezawa等[53]构建了生殖细胞特异性敲除的小鼠,并从出生7天的雄鼠中分离出未分化的精原细胞(Thy1+标记)和已初步分化的精原细胞(c-Kit+标记)。RNA-Seq检测结果揭示,与野生型相比Ring1b小鼠的Thy1+细胞中分别有116个基因表达下调和69个基因表达上调,c-Kit+细胞中有1381基因表达下调和269个基因表达上调。GO分析发现大多数上调基因的功能与精子发生没有直接的关联,而下调基因中富含调控精子分化的重要功能基因。Thy1+和c-Kit+的细胞中因敲除表达下调的基因中有78个重叠,包含许多精子发生过程中必要的调控基因(如、和),其中下调最显著的是,后续ChIP实验证实是RING1B的直接靶基因。c-Kit+细胞中因敲除特异性下调的基因主要用于调节精原细胞分化(如、和),减数分裂的开启(如和)和piRNA的调控(如、和);在正常精子发生中这些基因会在Thy1+精原细胞向c-Kit+精原细胞分化的过程中被高度激活,且进一步研究证实RING1B与SALL4可以共同靶向并上调这群基因的表达。该研究表明RING1B可与SALL4协同调控生殖谱系基因的顺次激活,这对生殖谱系细胞的正常分化及进入减数分裂等过程至关重要。
5 PRC1.6复合体是精子发生表观遗传调控网络的重要组成部分
精子发生是一个受到多因素调控的复杂发育过程。表4简要概括了精子发生过程中已知的表观遗传调控机制的大致类型。参与甲基化的各类DNA甲基转移酶(DNMTs)的表达及活性在生殖细胞发育过程中呈现明显的动态变化,这些DNMTs的异常往往直接导致生殖细胞发育异常以及个体雄性不育[70]。胚胎发育及生殖细胞发育过程中呈现动态变化的组蛋白甲基化、乙酰化等修饰对于建立细胞谱系的特异性身份至关重要[71,72]。其他组蛋白修饰,如泛素化、磷酸化以及近几年新发现的组蛋白巴豆酰化等对精子发育也十分重要[73,74]。在精子发育过程中,组装核小体的经典组蛋白在特定发育阶段被不同睾丸特异性组蛋白变体所替换,逐渐减弱DNA和组蛋白之间的作用,为大多数组蛋白在单倍体精子成熟的过程中被鱼精蛋白所替代做准备[75,76]。非编码RNA如piRNA介导的基因沉默机制是精子发育及其他情况下抑制可转座元件活性的重要途径,对于精子的正常发育必不可少[77];同时生殖细胞特异性表达的lncRNAs或miRNAs等在精子发育过程中也都起到重要的调控作用[78~81]。另外,近几年发现RNA的修饰尤其是甲基化修饰可参与RNA的转录后加工、核输出、翻译及RNA稳定性调节等,在造血干细胞分化、精子发生、神经发育等发育过程中发挥重要作用[82,83]。

表4 精子发育过程中的表观遗传调控方式
如表4所示,生殖谱系基因的表达受到非常复杂精密的表观遗传调控网络的调控,PRC1.6复合体作为该表观遗传调控网络的一部分,与该调控网络的其他“版块”之间存在着紧密的交互作用。目前已知PRC1.6复合体与DNA甲基化、组蛋白H3K9me3和H3K27me3等重要的表观遗传修饰存在协同作用,且这些协同互作呈现出明显的动态变化(图1)。例如,在胚胎发育早期E2F6对于减数分裂相关基因的转录抑制主要通过与PRC2协同完成,但不依赖于DNA甲基转移酶DNMT3B[98];然而,在小鼠体细胞中,E2F6则更多地通过招募DNMT3B对靶基因启动子进行甲基化来实现对这些生殖谱系特异性基因表达的有效抑制[99]。有趣的是,虽然小鼠ESCs中E2F6与DNMT3B之间没有协同关系,但MAX与DNA甲基转移酶DNMT1,DNMT3A,DNMT3B之间存在明显的协同作用并抑制生殖谱系相关基因的转录[100]。除了PRC1.6复合体自身包含HP1γ,可实现对转录抑制标记H3K9me3的结合之外,最近有研究揭示MAX与H3K9甲基转移酶SETDB1 可协同介导生殖谱系相关基因的转录抑制,但该机制并不依赖于PRC1.6复合体[100]。另外,在PCGF6 敲除的小鼠ESCs中,piRNA互作蛋白PIWIL1和PIWIL2的表达量均明显上升,意味着PRC1.6复合体通过抑制piRNA调控途径中重要RNA结合蛋白的表达水平来控制其活性[24]。除此之外,PRC1.6复合体组分自身的表达水平受到严密调控,目前这方面的报道相对较少,已知RYBP[101]和E2F6[102]等均可以受到miRNA的调控。
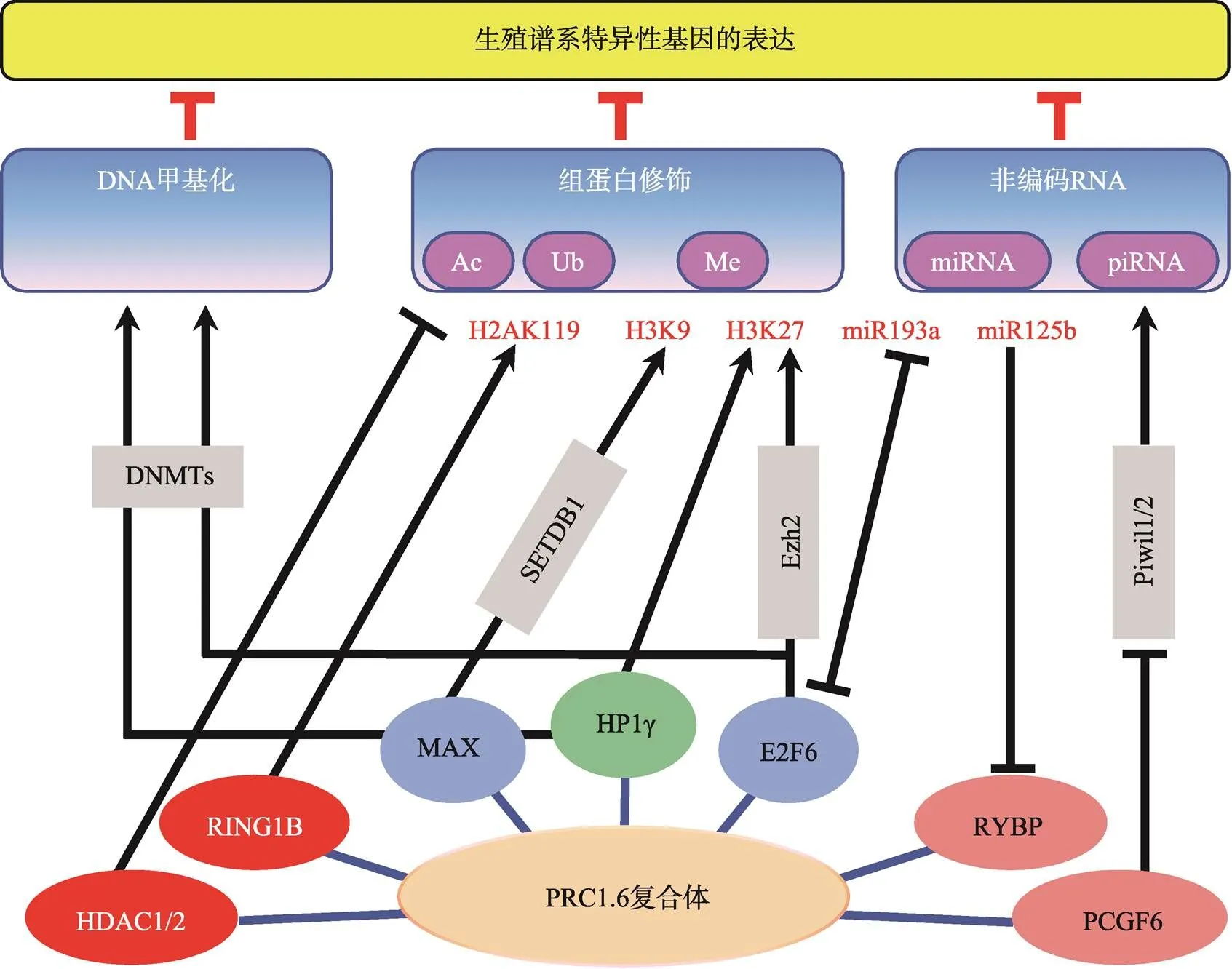
图1 PRC1.6复合体与其他表观遗传调控机制的相互作用
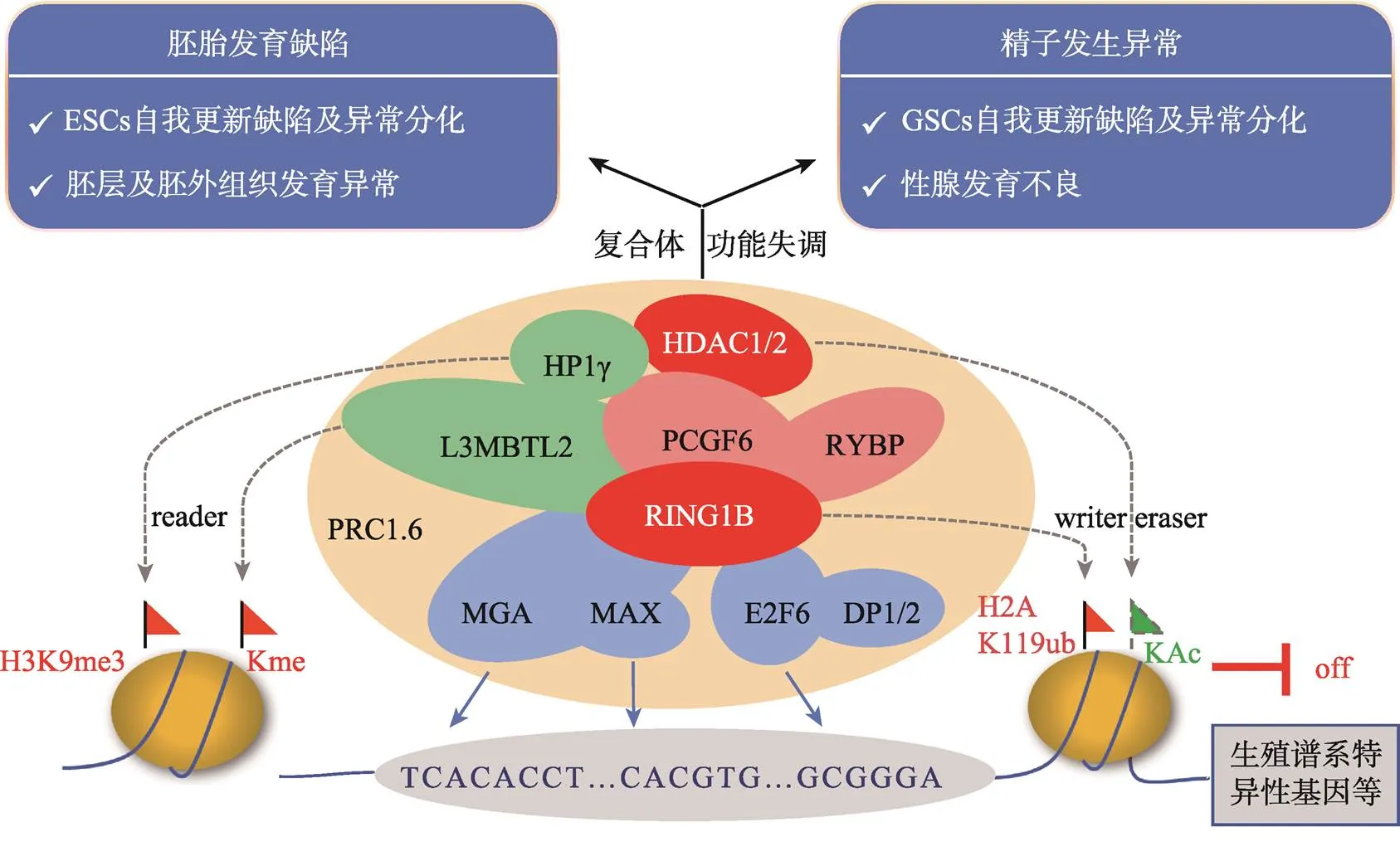
图2 PRC1.6复合体的组成、分子功能及活性异常产生的发育缺陷
标注红色的组分为具有酶活性或辅助酶活性的因子;标注蓝色的组分为具有DNA结合能力的转录因子;标注绿色的组分为具有染色质结合能力的因子。
6 结语与展望
本文总结了多梳蛋白PRC1.6复合体主要组分的分子功能及其在胚胎发育和精子发生中所扮演的重要角色(图2)。胚胎发育过程中PRC1.6复合体通过抑制生殖谱系相关基因及其他发育调控基因的表达确保了胚胎的正常分化。PRC1.6复合体也参与精子发生的表观遗传调控,顺次激活其不同阶段生殖谱系相关基因的表达,是正常精子发生及性腺发育的必要条件。尽管在过去的几年中有关PRC1.6复合体的组成及作用机制等方面取得了可喜的进展,但是对于该复合体具体的工作机制包括组分之间及整个复合体与其他表观调控途径之间的协调关系等的研究还有待深入。如PRC1.6各组分在调节减数分裂及精子形成过程中的具体分工还不够明确,以及该复合体如何实现对体细胞及不同分化阶段的生殖细胞中相关特异性基因的差异调控等。随着基因编辑技术以及现代分子生物学的进一步深入发展,PRC1.6各组分的具体功能以及它们之间的相互作用机制一定会研究得更加清晰,并将为与PRC1.6复合体组分突变或功能缺陷相关的男性不育的诊断与治疗带来新的思路。
[1] Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting., 2017, 171(1): 34–57.
[2] Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer., 2004, 3(4): 396–400.
[3] Lewis EB. A gene complex controlling segmentation in Drosophila., 1978, 276(5688): 565–570.
[4] Struhl G. A homoeotic mutation transforming leg to antenna in., 1981, 292(5824): 635– 638.
[5] Geisler SJ, Paro R. Trithorax and polycomb group- dependent regulation: a tale of opposing activities., 2015, 142(17): 2876–2887.
[6] Ingham PW. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes., 1983, 306(5943): 591–593.
[7] Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation., 2014, 10(12): e1004851.
[8] Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-polycomb complex activates gene expression in the CNS., 2014, 516(7531): 349–354.
[9] Morey L, Santanach A, Blanco E, Aloia L, Nora EP, Bruneau BG, Di Croce L. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms., 2015, 17(3): 300–315.
[10] Margueron R, Li GH, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms., 2008, 32(4): 503–518.
[11] Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks., 2009, 461(7265): 762–767.
[12] Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1B., 2006, 25(11): 2465–2474.
[13] Li ZZ, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi1-Ring1b polycomb group ubiquitin ligase complex., 2006, 281(29): 20643– 20649.
[14] de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation., 2004, 7(5): 663–676.
[15] Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment., 2014, 7(5): 1456–1470.
[16] Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins., 2014, 15(5): 412–428.
[17] Gil J, O'Loghlen A. PRC1 complex diversity: Where is it taking us?, 2014, 24(11): 632–641.
[18] Connelly KE, Dykhuizen EC. Compositional and functional diversity of canonical PRC1 complexes in mammals., 2016, 1860(2): 233– 245.
[19] Ogawa H, Ishiguro K, Gaubatz S , Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells., 2002, 296(5570): 1132–1136.
[20] Trojer P, Cao AR, Gao ZH, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, Farnham PJ, Reinberg D. L3mbtl2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure., 2011, 42(4): 438–450.
[21] Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes., 2012, 45(3): 344–356.
[22] Endoh M, Endo TA, Shinga J, Hayashi K, Farcas A, Ma KW, Ito S, Sharif J, Endoh T, Onaga N, Nakayama M, Ishikura T, Masui O, Kessler BM, Suda T, Ohara O, Okuda A, Klose R, Koseki H. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes., 2017, 6: e21064.
[23] Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. The polycomb- associated protein RYBP is a ubiquitin binding protein., 2006, 580(26): 6233–6241.
[24] Zhao W, Tong H, Huang Y, Yan Y, Teng H, Xia Y, Jiang Q, Qin J. Essential role for polycomb group protein PCGF6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity., 2017, 292(7): 2773–2784.
[25] Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6., 2018, 14(1): e1007193.
[26] Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif., 1999, 18(24): 7019–7028.
[27] Cartwright P, Müller H, Wagener C, Holm K, Helin K. E2f-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription., 1998, 17(5): 611–623.
[28] Stielow C, Stielow B, Finkernagel F, Scharfe M, Jarek M, Suske G. SUMOylation of the polycomb group protein L3MBTL2 facilitates repression of its target genes., 2013, 42(5): 3044–3058.
[29] Guo Y, Nady N, Qi C, Allali-Hassani A, Zhu H, Pan P, Adams-Cioaba MA, Amaya MF, Dong A, Vedadi M, Schapira M, Read RJ, Arrowsmith CH, Min J. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2., 2009, 37(7): 2204–2210.
[30] Rose NR, King HW, Blackledge NP, Fursova NA, Ember KJI, Fischer R, Kessler BM, Klose RJ. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes., 2016, 5: e18591.
[31] Zimberlin CD, Lancini C, Sno R, Rosekrans SL, McLean CM, Vlaming H, van den Brink GR, Bots M, Medema JP, Dannenberg JH. HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis., 2015, 29(5): 2070–2080.
[32] Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin., 2005, 19(3): 381–391.
[33] Yang CS, Chang KY, Dang J, Rana TM. Polycomb group protein PCGF6 acts as a master regulator to maintain embryonic stem cell identity., 2016, 6: 26899.
[34] Zdzieblo D, Li X, Lin Q, Zenke M, Illich DJ, Becker M, Müller AM. Pcgf6, a polycomb group protein, regulates mesodermal lineage differentiation in murine ESCs and functions in iPS reprogramming., 2014, 32(12): 3112–3125.
[35] Huang Y, Zhao W, Wang C, Zhu Y, Liu M, Tong H, Xia Y, Jiang Q, Qin J. Combinatorial control of recruitment of a variant PRC1.6 complex in embryonic stem cells., 2018, 22(11): 3032–3043.
[36] Bandara LR, Buck VM, Zamanian M, Johnston LH, La Thangue NB. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulating transcription factor DRTF1/E2F., 1993, 12(11): 4317–4324.
[37] Rowland BD, Bernards R. Re-evaluating cell-cycle regulation by E2Fs., 2006, 127(5): 871–874.
[38] Hisada K, Sánchez C, Endo TA, Endoh M, Román- Trufero M, Sharif J, Koseki H, Vidal M. Rybp represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells., 2012, 32(6): 1139–1149.
[39] Maeda I, Okamura D, Tokitake Y, Ikeda M, Kawaguchi H, Mise N, Abe K, Noce T, Okuda A, Matsui Y. Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells., 2013, 4: 1754.
[40] Suzuki A, Hirasaki M, Hishida T, Wu J, Okamura D, Ueda A, Nishimoto M, Nakachi Y, Mizuno Y, Okazaki Y, Matsui Y, Izpisua Belmonte JC, Okuda A. Loss of Max results in meiotic entry in mouse embryonic and germline stem cells., 2016, 7: 11056.
[41] Qin JZ, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, Hock H. The polycomb group protein L3mbtl2 assembles an atypical PRC1- family complex that is essential in pluripotent stem cells and early development., 2012, 11(3): 319–332.
[42] Pohlers M, Truss M, Frede U, Scholz A, Strehle M, Kuban RJ, Hoffmann B, Morkel M, Birchmeier C, Hagemeier C. A role for E2F6 in the restriction of male-germ-cell-specific gene expression., 2005, 15(11): 1051–1057.
[43] Storre J, Schäfer A, Reichert N, Barbero JL, Hauser S, Eilers M, Gaubatz S. Silencing of the meiotic genes Smc1beta and Stag3 in somatic cells by E2F6., 2005, 280(50): 41380–41386.
[44] Shen-Li H, O'Hagan RC, Hou H Jr, Horner JW, Lee HW, DePinho RA. Essential role for Max in early embryonic growth and development., 2000, 14(1): 17–22.
[45] Washkowitz AJ, Schall C, Zhang K, Wurst W, Floss T, Mager J, Papaioannou VE. Mga is essential for the survival of pluripotent cells during peri-implantation development., 2015, 142(1): 31–40.
[46] Burn SF, Washkowitz AJ, Gavrilov S, Papaioannou VE. Postimplantation Mga expression and embryonic lethality of two gene-trap alleles., 2018, 27: 31–35.
[47] Jørgensen HF, Giadrossi S, Casanova M, Endoh M, Koseki H, Brockdorff N, Fisher AG. Stem cells primed for action: polycomb repressive complexes restrain the expression of lineage-specific regulators in embryonic stem cells., 2006, 5(13): 1411–1414.
[48] Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins EZH2 and RNF2 direct genomic contraction and imprinted repression in early mouse embryos., 2008, 15(5): 668–679.
[49] Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, Koseki H. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity., 2008, 135(8): 1513–1524.
[50] Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase ii at bivalent genes in mouse ES cells., 2007, 9(12): 1428–1435.
[51] Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1B) deficiency causes gastrulation arrest and cell cycle inhibition., 2003, 100(5): 2468–2473.
[52] Suzuki M, Mizutani-Koseki Y, Fujimura Y, Miyagishima H, Kaneko T, Takada Y, Akasaka T, Tanzawa H, Takihara Y, Nakano M, Masumoto H, Vidal M, Isono K, Koseki H. Involvement of the polycomb- group gene Ring1B in the specification of the anterior- posterior axis in mice., 2002, 129(18): 4171–4183.
[53] Maezawa S, Hasegawa K, Yukawa M, Sakashita A, Alavattam KG, Andreassen PR, Vidal M, Koseki H, Barski A, Namekawa SH. Polycomb directs timely activation of germline genes in spermatogenesis., 2017, 31(16): 1693–1703.
[54] Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. PRC1 coordinates timing of sexual differentiation of female primordial germ cells., 2013, 495(7440): 236–240.
[55] Ujhelly O, Szabo V, Kovacs G, Vajda F, Mallok S, Prorok J, Acsai K, Hegedus Z, Krebs S, Dinnyes A, Pirity MK. Lack of RYBP in mouse embryonic stem cells impairs cardiac differentiation., 2015, 24(18): 2193–2205.
[56] Pirity MK, Locker J, Schreiber-Agus N. RYBP/DEDAF is required for early postimplantation and for central nervous system development., 2005, 25(16): 7193–7202.
[57] Jamaladdin S, Kelly RD, O'Regan L, Dovey OM, Hodson GE, Millard CJ, Portolano N, Fry AM, Schwabe JW, Cowley SM. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells., 2014, 111(27): 9840–9845.
[58] Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression., 2002, 21(11): 2672–2681.
[59] Wang Y, Tian Y, Morley MP, Lu MM, Demayo FJ, Olson EN, Morrisey EE. Development and regeneration of Sox2+ endoderm progenitors are regulated by a Hdac1/2-Bmp4/Rb1 regulatory pathway., 2013, 24(4): 345–358.
[60] Caillier M, Thénot S, Tribollet V, Birot AM, Samarut J, Mey A. Role of the epigenetic regulator HP1γ in the control of embryonic stem cell properties., 2010, 5(11): e15507.
[61] Brown JP, Bullwinkel J, Baron-Lühr B, Billur M, Schneider P, Winking H, Singh PB. Hp1gamma function is required for male germ cell survival and spermatogenesis., 2012, 5(1): 18.
[62] Takada Y, Naruse C, Costa Y, Shirakawa T, Tachibana M, Sharif J, Kezuka-Shiotani F, Kakiuchi D, Masumoto H, Shinkai Y, Ohbo K, Peters AHFM, Turner JMA, Asano M, Koseki H. Hp1γ links histone methylation marks to meiotic synapsis in mice., 2011, 138(19): 4207–4217.
[63] Leseva M, Santostefano KE, Rosenbluth AL, Hamazaki T, Terada N. E2f6-mediated repression of the meiotic Stag3 and Smc1β genes during early embryonic development requires Ezh2 and not the de novo methyltransferase Dnmt3b., 2013, 8(8): 873–884.
[64] Storre J, Elsässer HP, Fuchs M, Ullmann D, Livingston DM, Gaubatz S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6., 2002, 3(7): 695–700.
[65] Courel M, Friesenhahn L, Lees JA. E2f6 and Bmi1 cooperate in axial skeletal development., 2008, 237(5): 1232–1242.
[66] Kohn MJ, Leung SW, Criniti V, Agromayor M, Yamasaki L. Dp1 is largely dispensable for embryonic development., 2004, 24(16): 7197–7205.
[67] Kohn MJ, Bronson RT, Harlow E, Dyson NJ, Yamasaki L. Dp1 is required for extra-embryonic development., 2003, 130(7): 1295–1305.
[68] Hu G, Kim J, Xu QK, Leng YM, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal., 2009, 23(7): 837–848.
[69] Meng C, Liao J, Zhao D, Huang H, Qin J, Lee TL, Chen D, Chan WY, Xia Y. L3mbtl2 regulates chromatin remodeling during spermatogenesis., 2019. doi: 10.1038/s41418-019-0283-z. [Epub ahead of print]
[70] Uysal F, Akkoyunlu G, Ozturk S. DNA methyltransferases exhibit dynamic expression during spermatogenesis., 2016, 33(6): 690–702.
[71] Ge SQ, Lin SL, Zhao ZH, Sun QY. Epigenetic dynamics and interplay during spermatogenesis and embryogenesis: Implications for male fertility and offspring health., 2017, 8(32): 53804–53818.
[72] Xiong J, Wang H, Guo G, Wang S, He L, Chen H, Wu J. Male germ cell apoptosis and epigenetic histone modification induced by Tripterygium wilfordii Hook F., 2011, 6(6): e20751.
[73] Sheng K, Liang X, Huang S, Xu W. The role of histone ubiquitination during spermatogenesis., 2014, 2014: 870695.
[74] Liu S, Yu H, Liu Y, Liu X, Zhang Y, Bu C, Yuan S, Chen Z, Xie G, Li W, Xu B, Yang J, He L, Jin T, Xiong Y, Sun L, Liu X, Han C, Cheng Z, Liang J, Shang Y. Chromodomain protein CDYl acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis., 2017, 67(5): 853–866.e5.
[75] Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease., 2014, 15(4): 259–271.
[76] Bao J, Bedford MT. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis., 2016, 151(5): R55–70.
[77] Russell SJ, Stalker L, LaMarre J. Piwis, piRNAs and retrotransposons: complex battles during reprogramming in gametes and early embryos., 2017, 52(Suppl. 4): 28–38.
[78] Hong SH, Kwon JT, Kim J, Jeong J, Kim J, Lee S, Cho C. Profiling of testis-specific long noncoding RNAs in mice., 2018, 19(1): 539.
[79] Bie B, Wang Y, Li L, Fang H, Liu L, Sun J. Noncoding RNAs: potential players in the self-renewal of mammalian spermatogonial stem cells., 2018, 85(8–9): 720–728.
[80] Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis., 2017, 8: 35.
[81] Wichman L, Somasundaram S, Breindel C, Valerio DM, McCarrey JR, Hodges CA, Khalil AM. Dynamic expression of long noncoding RNAs reveals their potential roles in spermatogenesis and fertility., 2017, 97(2): 313–323.
[82] Yang Y, Chen YS, Sun BF, Yang YG. RNA methylation: regulations and mechanisms., 2018, 40(11): 964–976.杨莹, 陈宇晟, 孙宝发, 杨运桂. RNA甲基化修饰调控和规律. 遗传, 2018, 40(11): 964–976.
[83] Peer E, Rechavi G, Dominissini D. Epitranscriptomics: regulation of mRNA metabolism through modifications., 2017, 41: 93–98.
[84] Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis., 2006, 104(1): 228–233.
[85] Hata K, Kusumi M, Yokomine T, Li E, Sasaki H. Meiotic and epigenetic aberrations in Dnmt3l-deficient male germ cells., 2006, 73(1): 116-122.
[86] de Vries M, Ramos L, Housein Z, de Boer P. Chromatin remodelling initiation during human spermiogenesis., 2012, 1(5): 446–457.
[87] Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sèle B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases., 2000, 79(12): 950–960.
[88] Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin., 2005, 438(7071): 1176– 1180.
[89] Lu LY, Wu JX, Ye L, Gavrilina GB, Saunders TL, Yu XC. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis., 2010, 18(3): 371–384.
[90] Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen JJ, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids., 2012, 26(24): 2737–2748.
[91] Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, Okabe M, Nishimune Y. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility., 2005, 25(16): 7107–7119.
[92] Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind toMILI protein in mouse testes., 2006, 442(7099): 203–207.
[93] Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation., 2015, 12(8): 1234–1243.
[94] Moritoki Y, Hayashi Y, Mizuno K, Kamisawa H, Nishio H, Kurokawa S, Ugawa S, Kojima Y, Kohri K. Expression profiling of microRNA in cryptorchid testes: MiR-135a contributes to the maintenance of spermatogonial stem cells by regulating FoxO1., 2013, 191(4): 1174–1180.
[95] Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, Brinster RL. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells., 2011, 108(31): 12740–12745.
[96] Li L, Wang M, Wang M, Wu X, Geng L, Xue Y, Wei X, Jia Y, Wu X. A long non-coding rna interacts with gfra1 and maintains survival of mouse spermatogonial stem cells., 2016, 7: e2140.
[97] Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, Zhang Y, Song W, Jia G, Yang X, He C, Tong MH. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis., 2017, 27(10): 1216–1230.
[98] Leseva M, Santostefano KE, Rosenbluth AL, Hamazaki T, Terada N. E2f6-mediated repression of the meiotic stag3 and Smc1β genes during early embryonic development requires Ezh2 and not the de novo methyltransferase Dnmt3b., 2013, 8(8): 873–884.
[99] Velasco G, Hubé F, Rollin J, Neuillet D, Philippe C, Bouzinba-Segard H, Galvani A, Viegas-Péquignot E, Francastel C. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues., 2010, 107(20): 9281–9286.
[100] Tatsumi D, Hayashi Y, Endo M, Kobayashi H, Yoshioka T, Kiso K, Kanno S, Nakai Y, Maeda I, Mochizuki K, Tachibana M, Koseki H, Okuda A, Yasui A, Kono T, Matsui Y. DNMTs and SETDB1 function as co-repressors in Max-mediated repression of germ cell-related genes in mouse embryonic stem cells., 2018, 13(11): e0205969.
[101] Li HH, Lai P, Jia JP, Song YW, Xia Q, Huang KM, He N, Ping WF, Chen JY, Yang ZZ, Li J, Yao MZ, Dong XT, Zhao JC, Hou CH, Esteban MA, Gao SR, Pei DQ, Hutchins AP, Yao HJ. RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions., 2017, 20(4): 462–477.
[102] Cheng FHC, Lin HY, Hwang TW, Chen YC, Huang RL, Chang CB, Yang W, Lin RI, Lin CW, Chen GCW, Mai SY, Lin JMJ, Chuang YM, Chou JL, Kuo LW, Li C, Cheng ASL, Lai HC, Wu SF, Tsai JC, Chan MWY. E2f6 functions as a competing endogenous RNA, and transcriptional repressor, to promote ovarian cancer stemness., 2019, 110(3): 1085–1095.
Controlling the spatiotemporal expression of germ line specific genes by PRC1.6 complex
Xiaowei Sun1, Hongyang Li1, Jian Wang1, Bo Cheng1,2
Polycomb repressive complex 1 (PRC1) is a class of epigenetic regulatory complexes that normally represses gene expression by catalyzing and/or recognizing chromatin modifications. PRC1 mainly functions in stem cell maintenance, cell differentiation, cell cycle regulation and related processes. PRC1 also have aberrant functions which has been implicated in many types of developmental diseases and cancers. Mammalian PRC1 complexes are divided into six subtypes based on their composition and function; subtypes include PRC1.1 to PRC1.6. Each PRC1 subtype regulates a unique collection of target genes. The PRC1.6 complex subtype plays key roles in specifically repressing transcription of genes controlling germ cell development in embryonic stem cells and other somatic cell types. Recent research demonstrates that the PRC1.6 complex is also crucial for the timely activation of the germ line of specific genes during spermatogenesis, which is essential for proper gonad development. In this review, we summarize the identification of molecular functions of each core component of the PRC1.6 complex including how it recognizes and represses germ line specific genes. We also update the biological roles of this complex in regulating the spatiotemporal expression of germ line specific genes during embryonic development, gonad development, and spermatogenesis. Lastly, the crosstalk between the PRC1.6 complex and the other main epigenetic regulatory mechanisms involved in controlling spermatogenesis is discussed. Our discussion of the PRC1.6 complex in regulating germ line specific genes informs the studies of molecular processes of spermatogenesis and contributes to the understanding of the pathogenic mechanisms of male infertility.
polycomb repressive complex 1 (PRC1); PRC1.6; transcriptional repression; germ line specific genes; spatiotemporal expression
2019-01-27;
2019-03-14
国家自然科学基金项目(编号:31471233, 31771447),中央高校基本科研业务费(编号:lzujbky-2017-it51, lzujbky-2018-k05)和教育部细胞活动与逆境适应重点实验室开放基金项目(编号:lzujbky-2017-kb05, lzujbky-2018-kb05)资助[Supported by the National Natural Science Foundation of China (Nos. 31471233, 31771447), the Fundamental Research Funds for the Central Universities (Nos. lzujbky-2017-it51, lzujbky-2018-k05) and the Foundation of the Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations (Nos. lzujbky-2017-kb05, lzujbky-2018-kb05)]
孙晓伟,在读博士研究生,专业方向:细胞生物学。E-mail: sunxw18@lzu.edu.cn李宏阳,在读硕士研究生,专业方向:细胞生物学。E-mail: hyli17@lzu.edu.cn孙晓伟和李宏阳并列第一作者。
程博,博士,教授,研究方向:真核基因表达调控。E-mail: bocheng@lzu.edu.cn
10.16288/j.yczz.18-332
2019/4/1 9:34:53
URI: http://kns.cnki.net/kcms/detail/11.1913.R.20190401.0934.001.html
(责任编委: 苗龙)
