Colorectal cancer:The epigenetic role of microbiome
2019-04-22HusseinSabitEmreCevikHuseyinTombuloglu
Hussein Sabit, Emre Cevik, Huseyin Tombuloglu
Hussein Sabit, Emre Cevik, Huseyin Tombuloglu, Department of Genetics, Institute for Medical Research and Consultations, Imam Abdulrahman Bin Faisal University, Dammam 31441,Saudi Arabia
Abstract
Key words: Colorectal; Cancer; Colorectal cancer; Epigenetics; Gut; Microbiota
INTRODUCTION
Colorectal cancer (CRC) is one of the primary causes of cancer-related deaths globally[1].It occurs as a result of complicated sequences involving mutation accumulation that is either genetic or epigenetic[2].The process of CRC carcinogenesis is a quite slow, starting with minor inflammation followed by adenomatous polyps in the epithelium, and finally adenocarcinoma[3].Given the crucial role of epigenetic changes in developing CRC, 95% of cases are sporadici.e.appear in patients with no family history for the disease.Meanwhile, minor ratio (3%) is attributed to hereditary nonpolyposis CRC, and 2% of cases are caused by other hereditary disorders such as MYH-associated polyposis and familial adenomatous polyposis[2,4].
Microorganisms occupy almost every part of the human body, armed with a huge number of genes, where it could interact, modulate, or disrupt a wide array of human genes especially in colonic cells[5].Interestingly, the human microbiome encodes for approximately 100-fold more proteins than the human genome itself.This microbiota comprise 1000 to 1500 bacterial species, and the composition of the microbiome is significantly diverse among individuals[6].These species belong to just a few phyla:
Firmicutes,Bacteroidetes(most prevalent),Proteobacteria,Verrucomicrobia,Actinobacteria,Fusobacteria, andCyanobacteria[7].Although distribution of the microbiota in terms of types and number is common in healthy individuals, it differs significantly in diseased persons.In addition to bacteria that compose the gut microbiome, eukaryotic fungal species have recently been found to co-exist with bacterial species, the major component of the microbiome[8].
It is well established that gut microbiota play critical roles in the progression of CRC eitherviatheir metabolites or interaction with their host intestinal epithelial cells[9].Imbalance of this microbiota has been associated with several disorders including inflammatory bowel disease and CRC[10].Nonetheless, several studies have related the changes in microbiota to a cause of disease, while others have indicated that these changes are merely a result; however, this issue demands further investigation[11].In this review, we highlight the recent studies that addresses the causal link between gut microbiota and CRC onset and progression.Meanwhile, the epigenetic changes underlying CRC and its microbial root will also be described.
CRC
CRC is one of the most prevalent malignant tumors and the third most common cause of cancer-related death worldwide[12].It is the third most common malignancy in males and the second in females, with a lifetime risk of about 6%[13].Being welldeveloped, CRC can metastasize -even after operation- to distant organs such as the liver and lungs, forming secondary CRC[14].The common risk factors underlying CRC involve genetics[15], environmental pollution[16], diet[17], age[18], alcohol consumption[19],smoking[20], obesity[21], and physical inactivity[22], with gut microbiota standing alone as a potent risk factor[23](Figure 1).It is well established that CRC arises due to accumulation of genetic mutations.Large studies showed that approximately 13000 mutations in 67 genes correlate with CRC.Among them, only 12 genes were found to be closely related to CRC[24].Different types of genomic instability predispose patients to CRC including microsatellite instability (MSI), in which frequent insertions and deletions are prevalent, and chromosomal instability (CIN), in which gain or loss of chromosomes prevail[25].CIN is responsible for about 85% of CRC cases, where loss of chromosomal segment/arm includes 15q11-q21, 17p12-13, and18q12-21 and gain of chromosomal segment/arm includes 1q32, 7p, 7q, 8q, 13q, 20p, and 20q[26,27].
Several genes are directly correlated with CRC.Examples includeAPCin which inactivation leads to triggering the Wnt signaling pathway to initiate colon polyps which can be benign (e.g., hyperplastic polyp), pre-malignant (e.g., tubular adenoma)or malignant (e.g., colorectal adenocarcinoma)[28].Furthermore, transforming growth factor receptor 2TGFBR2is involved in almost 30% of CRC cases.The downstream effector of this genes,i.e.KRASwas found to be activated in 55%-60% of CRC cases.Mismatch repair genes such asMLH1,MSH6,MSH2, andPMS2causing frameshift mutations were found to induceMSI, triggering the development of CRC[29](Figure 2).
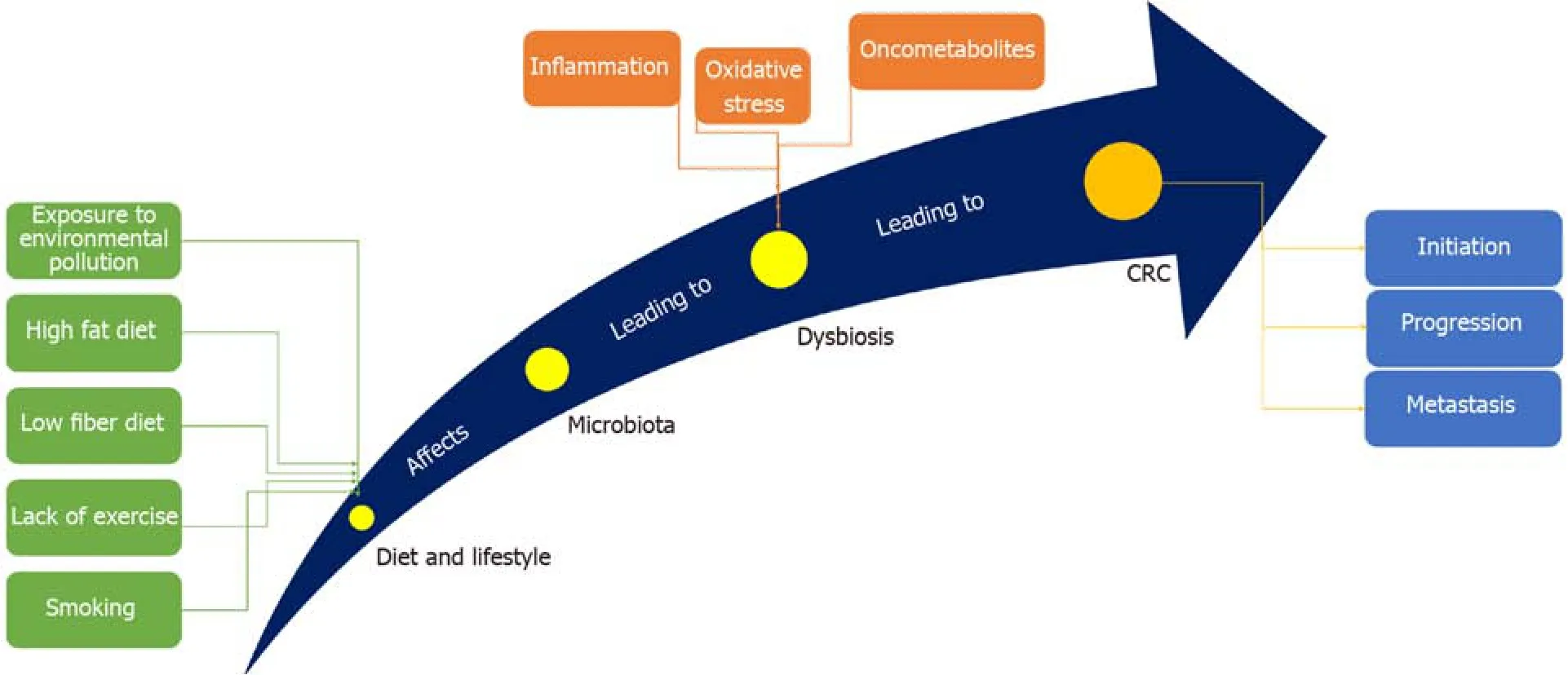
Figure 1 The way gut microbiota induces CRC.
Epigenetic regulation of gene expression analysis is a validated tool to correlate gene expression changes with cancer development[2,30].Through the last three decades,solid common knowledge has been established to indicate that the perturbation of epigenetic mechanisms leads to cancer initiation and progression[29,31].Identification of CRC epigenetic changes has revealed that almost all CRCs have abnormally methylated genes.Although rare data have been provided to highlight the pattern of specific histone modifications in CRC, certain histone modifications (such as acetylation, methylation, and phosphorylation) have been found to work in harmony with DNA methylation to regulate CRC-related gene expression that is involved in cancer initiation and progression[1,32].Therefore, a deep understanding of epigenetic changes related to CRC pathogenesis might help develop epigenetic-based biomarkers for CRC diagnosis and prognosis, and hence, epigenetic-based therapy[29].
GUT MICROBIOME
Function
In the normal adult person, the gut microbiota comprise approximately 1014bacterial cells that live in commensalism with the host, where they substantially facilitate various aspects in the host health and disease[33].The normal gut microbiota are rich in anaerobic bacteria, which are 100- to 1000-times more than aerobic and facultative anaerobic bacteria, respectively[34].The colon has a reductive environment devoid of oxygen, which allowsBacteroidetesandFirmicutesto be the dominant phyla followed byActinobacteriaandVerrucomicrobia[35].For bacteria, the colon represents a suitable niche as it provides them with elevated pH, nutrients, and low concentration of bile salts and pancreatic secretions.These conditions, indeed, are favored by bacteria to flourish and proliferate[10].Commensal bacteria start colonization of the host during birth and continue to variate in number and type along with the host development[36].It is well established that our gut microbiome is responding to any dietary shift,where switching from polysaccharides-rich diets to that high in animal fat eventually leads to a hasty shift in the gut microbiome[37].Commonly in the gut, the prevailed microbial product is lipopolysaccharide (LPS), produced by Gram-negative bacteria,function to stimulate the innate immunity, thus, protecting against inflammation that leads to cancer[38](Table 1).
Protective role
Gut microbiota is crucial for numerous characteristics of host biology[10,39].They enable the host to digest and metabolize indigestible polysaccharides[40].The gut microbiota plays an important role in maintaining gut homeostasis[41].Furthermore, gut microbial community also participates effectively in the normal cellular proliferation.To keep its habitat for millions of years, several gut microbiota essentially protect the host against CRC[42].Reports have indicated that enterotoxigenicBacteroides fragilisis capable of induce apoptosis in CRC cells[42].Generally, diet is metabolized by microbiota into potent oncometabolites and tumor-suppressive metabolites.Whereas,the same microbiota can digest fiber into short-chain fatty acids (SCFAs)[43].It is well known that SCFAs have anti-inflammatory properties and can increase the level of colonic regulatory T cells (Tregs) and protect the host against colitis[43,44].The most common SCFAs produced in the gut are acetate, propionate, and butyrate[45].Butyrate is one of the important sources of energy, where it provides the cells with 5%-15% of its caloric requirements.Faecalibacterium prausnitziiandEubacterium rectaleare the main gut bacterial species that produce butyrate[44,46].Butyrate controls cell proliferation, differentiation, and apoptosis among other functions in colon cells[23,47].It exerts also a preventive role where it ameliorates the harmful effects of N-nitroso compounds, a product that accumulates in the colon upon intake of heat-treated and processed meat[48](Figure 3).It has been indicated thatClostridiumspecies enhances Treg cell abundance by increasing the production of potent anti-inflammatory molecules such as cytokine interleukin 10 (IL-10)[49].Lactic acid bacteria have also shown protective properties against CRCviadifferent mechanisms that include strengthening the mucosal barrier and altering luminal secretions, resulting in underpinning of the host immune system.Ursodeoxycholic acid (UDCA, ursodiol) is a metabolic byproduct of intestinal bacteria, with a chemical structure that resembles deoxycholic acid (DCA)[50].While DCA promotes the initiation of CRC, UDCA function to prevent the disease.It was reported that UDCA inhibits the expression of cyclooxygenase-2 (COX-2) by Ras-dependent and RAS-independent mechanisms in CRC cells[51].UDCA prevents colon cells from the harmful effect of DCAviainhibiting the DCA-induced extracellular signal-regulated kinase and Raf-1 kinase activity and the activation of epidermal growth factor receptor (EGFR)[52].
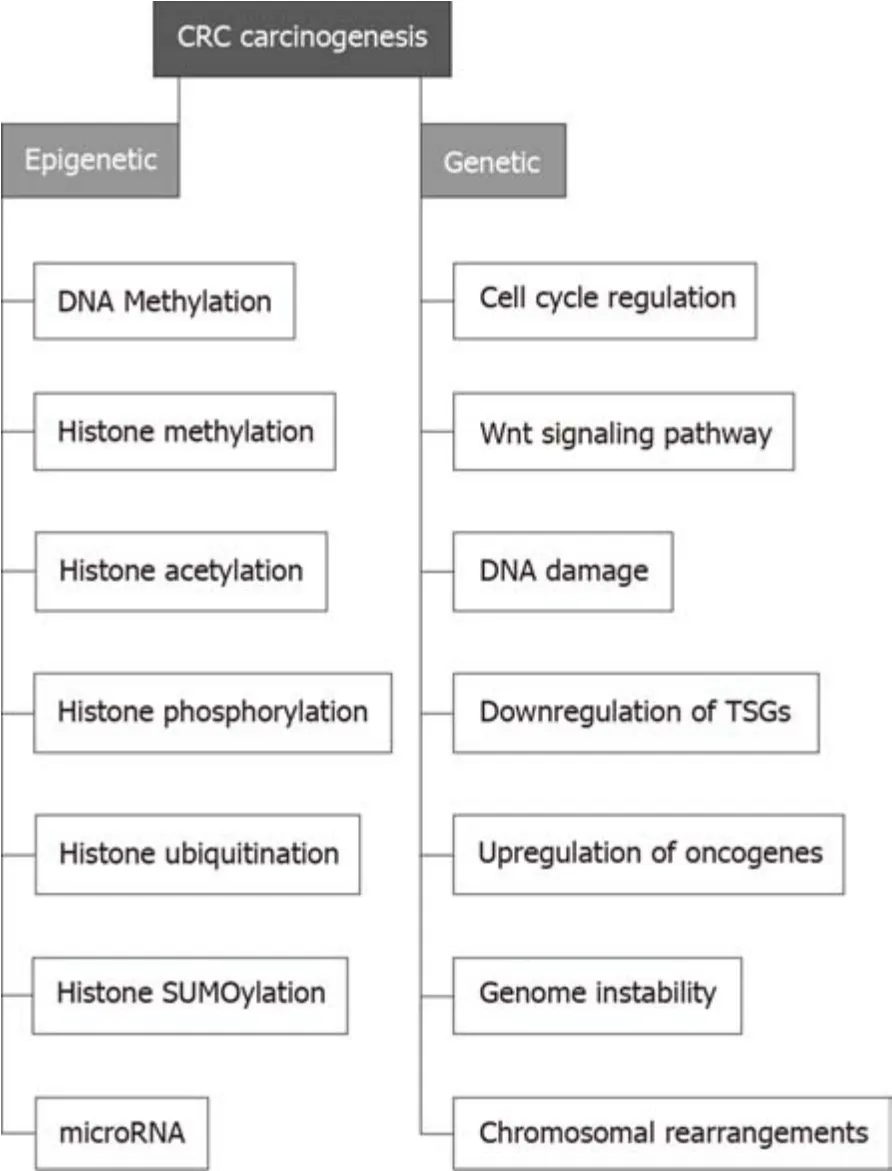
Figure 2 Different pathways through which CRC develops.
Pro-carcinogenic role
Microbiota-mediated carcinogenesis is a complex process that takes place through changing host cell proliferation, influencing the host cell immune system, and metabolizing dietary factors[53].A plethora of research has suggested that an imbalance in normal intestinal microbiota can trigger inflammatory conditions by producing carcinogenic metabolites that lead to cancer formation, and about 16% of human cancers are triggered by bacteria[36,53].Gut bacteria can attack intestinal epithelial cells to cause inflammation, that in turn, increase the risk of developing CRC[54].For CRC to occur, the microbiota-host interaction must be dysregulated,resulting in disruption of cellular homoeostasis[55].The major component of the gut immune system, Peyer’s patch, is robustly influenced by gut microbiota[56].The host diet can trigger gut microbiota to be involved in the early stages of CRC carcinogenesis[57].Upon metabolism of saturated fatty acid- and sugar-rich diets, gut bacteria produce harmful procarcinogenic products including polyamines hydrogen sulfide, secondary bile acids such as DCA and lithocholic acid (LCA), and reactive oxygen species (ROS), which induce chronic inflammation, and hence elevate the susceptibility of cells to develop CRC[58].DCA is a metabolite of the gut microbiota that induces cancer stemness by regulating the muscarinic 3 receptor/Wnt intracellular signaling pathway[59].It can also trigger the production of Nur77 protein,which is positively correlated with CRC when upregulated[60].Meanwhile, DCA induces CRCviadownregulation of miR-199a-5p that degradesCAC1, the tumor suppressor gene plays a role in CRC[61].LCA (aka 3α-hydroxy-5β-cholan-24-oic acid), a secondary bile acid synthesized by gut microbiota, is verified to a promoter of CRC[62].Gut bacteria produce LCA by utilizing DCA[63].Both LCA and DCA can enhance cancer stemness[64].Furthermore, LCA and DCA activate the EGFR signaling pathway,inducing DNA damage, and causing oxidative stress, apoptosis, mutation, and activation of the protein kinase C pathway[59].

Table 1 Gut microbiota are involved in CRC carcinogenesis
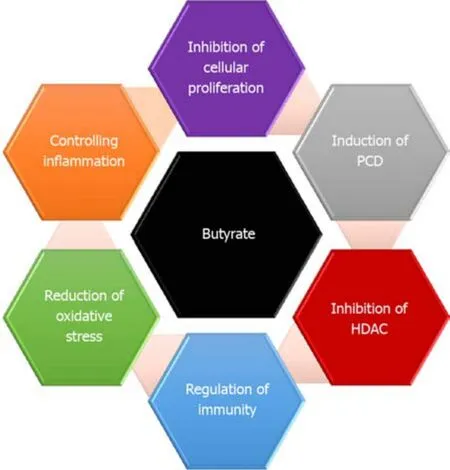
Figure 3 Different functions of butyrate in protecting against CRC.
Trimethylamine (TMA) is solely synthesized by gut microbiota (in humans) from various dietary nutrients including choline and carnitine (found in red meat)[65].It reacts with flavin monooxygenase to produce trimethylamine-N-oxide (TMAO), a microbial metabolite involved in CRC progression[66].A high incidence rate of CRC was noticed in omnivorous individuals, as they produce more TMAO compared to vegans and vegetarians who show low incidence rate[67].The genetic pathway by which TMAO triggers CRC remains unclear.
Furthermore, specific gut bacteria such asEnterococcus faecaliswas found to induce COX-2, that generates pro-proliferative signals through prostaglandin E(2) (PGE2)[68].Several Gram-negative bacteria produce LPS that activates TLR4, COX-2, and then PGE2leading to inhibition of programmed cell death and increase cell proliferation[69].Moreover, there is an increased resistance to macrophage killing and MAPK activation in those who have thepks(polyketide synthase) island inE.coliisolated from CRC patients[70].Activated TLR also enhances angiogenesis through MAPK and NF-κB signaling networks[71](Figure 4).
Other CRC-related bacterial metabolites were highlighted including fragilysin[72].This extracellular 20 kDa zinc-dependent metalloproteinase metabolite, produced byB.fragilis, could hydrolyze the extracellular domain of E-cadherin and activate the βcatenin nuclear signaling, leading to induction of CRC[73].Meanwhile fragilysin can damage the tight junction of the intestines, increases intestinal permeability[74].Colibactin is a bacterial-derived genotoxin first reported in 2006 by Nougayrede and colleagues.It is a hybrid polyketide-non ribosomal peptide produced through an intricate biosynthetic mechanism and encoded by thepkspathogenicity island[75].E.colistrains harboring thispksisland were found to be associated with CRC[76].Moreover, colibactin, a kind of bacterial toxin synthetized by thepksgenomic island can trigger chromosomic instability and DNA damage that might lead eventually to CRC[77].
GUT MICROBIOTA AND EPIGENETIC MODIFICATION

Figure 4 Different gut microbiota generate different oncometabolites.
Several epigenetic changes are common in CRC including DNA methylation, histone modification, and miRNA-mediated post-transcriptional regulation[28,78].Abnormal epigenetic modifications (AKA epimutations) occur in the promoter regions of tumorsuppressor genes and proto-oncogenes.These epimutations were reported in several malignancies including CRC, where many genes such asMLH1,LKB1,APC,p16INK4a, andGATA4represent common targets[2,40,79].Microbial community in our guts are armed with an arsenal of genes that produces millions of proteins, let alone their outpouring of metabolites[67,80].This microbiota produces low-molecular-weight substances that interact within the human cells with different targets to trigger genomic and epigenomic changes[81].Research teams everywhere highlight the association between gut microbiota and human diseases; thus, all these diseases should be revisited once again to elucidate the actual role played by microbial community.Being very stable, DNA might not be affected by microbial metabolites,and this is pointing to a more fragile component in our cells; epigenome.
DNA methylation
Linking diseases to epigenetic changes was first addressed in 1983[82].Based on that first note, numerous researches indicated that cancer cells undergo global hypomethylation along with site-specific hypermethylation in the promotors of cancer-related genes[83].A bunch of reports have indicated that the microbial metabolites can modulate epigenetic landscape of the host gut’s cellsviamodifying the methylation pattern of cancer-related genes, as they represent a validated source of microbial-induced epigenetic change.Thus, the deep understanding of how epigenetic modifications influenced by the gut microbiota take place could offer possible therapeutic targets to prevent and treat CRC[10,80].
In DNA methylation, DNMTs add methyl group (CH3) to the fifth carbon atom in the cytosine residue using the intracellular methyl substrateS-Adenosyl methionine(SAM) as a methyl donor to convert the normal cytosine into 5-methylcytosine (5-mC)[84].Meanwhile, ten-eleven translocation enzyme can reverse this processviathe oxygenation of 5-mC to produce 5-hydroxymethylcytosine[85].It is well known that folate is the main source of SAM, and this vitamin could be produced byBifidobacteriumandLactobacillus, common probiotic bacteria[86].Folate is required for DNA methylation (5-methyltetrahydrofolate) or DNA synthesis (5-formyltetrahydrofolate and 5, 10-methenyltetrahydrofolate)[87].A study indicated that volunteers administeredBifidobacteriumshowed a high concentration of folate in their feces, meaning that these probiotic bacteria were capable to generate it and hence,affect DNA methylation pattern[88].On the other hand, deficiency of folate synthesis might contribute to DNA hypomethylation, which is an established phenomenon in CRC[89].Meanwhile, pathogenic bacteria such asHelicobacter pylorithat infects the stomach and causes gastritis or gastric ulcers or in severe infection gastric cancer can induce several epigenetic changes[90].A comparison between gastric biopsies excised from patients with gastritis (upon infection withHelicobacter pylori) and healthy individuals showed that chronic gastritis was associated with promoter hypermethylation ofE-cadherin(CDH1),MGMT,WIF1, andMLH1[91].Although studies that address the relationship between the microbiome and epigenetic changes in CRC are rare, a population-based study reported thatFusobacterium nucleatumwas associated with genomic hypermutation independent ofCIMPandBRAFmutations[92].Other study indicated thatFusobacteriumwas correlated with theCIMPphenotype, wild-typeTP53,hMLH1methylation, genomic hypermutation, andCHD7/8mutation[93].These studies strongly suggest the contribution ofF.nucleatumto the epigenetic changes.
Histone modification
In addition to DNA methylation changes, histone modification patterns are also altered in human cancers[84,94].Some bacterial metabolites such as short chain fatty acids (butyrate and acetate) can induce epigenetic changes in colonic cells[23,45].Butyrate, a byproduct of the fermentation process of undigested dietary carbohydrates and proteins carried out byFirmicutes, has been shown to regulate over 4000 genes including many involved in apoptosis and cell cycle regulation[95].It is known also to inhibit histone deacetylases and induce hyperacetylation of histones,that lead to changes in the expression of critical cell cycle regulatory genes such asCCND3andCDKN1Ain intestinal cells.Butyrate triggers epithelial generation of ROS and function also to suppress NF-kB, the protein complex controls DNA transcription[44,96].Furthermore,Bacteroides thetaiotaomicronstimuli the inflammatory signaling by inhibiting NF-κB pathway through binding to IkB (inhibitor ofκB),inhibitory component of the NF-κB pathway[97].It was reported that infection withListeria monocytogenes(L.monocytogenes) can cause deacetylation of histone H3K18 in many genes in colonic cells such asSMAD1,IRF2,SMARCA2andCXCL12[98].L.monocytogenesexecute the deacetylation process by translocating NAD-dependent deacetylase sirtuin 2 to the host nucleus.By doing so,L.monocytogenesepigenetically regulates cell cycle-related genes and modulate the host immune response to enable its invasion[99].
MICRORNAS (MiRNAs) AND CRC
MiRNAs are a class of small single-stranded non-coding RNA molecules that are evolutionarily conserved and encoded by nearly 1% of the genome in most species[100].MiRNAs were found to be involved in initiation, progression, and metastasis of CRC,where it regulate of various cancer-related gene expression at the post-transcriptional level[101].Deregulated miRNAs identified in different types of cancers might put us a step forward towards understanding the tumor microenvironment, which necessitate deep investigation of their actual role in cancer progression and spreading[102].Numerous miRNAs were found to be associated with CRC, such as miR-21, Let-7,miR-145, miR-221, miR-17-19 cluster, and miR-143[103].Table 2 highlights some of miRNAs related to CRC development, progression, and metastasis.Studies addressed the expression levels of different miRNA in CRC, reported that miR-31, miR-20, miR-25, miR-223, miR-133b, miR-92, miR-93, miR-135a, miR-203, miR-183, and miR-17 were upregulated in CRC, while miR-26b, miR-192, miR-145, let-7a, miR-143, miR-215,miR-16, and miR-191 were downregulated in patients with CRC[13,104].Some miRNAs were suggested to serve as diagnostic markers for CRC, including miR-133a, miR-145,miR-484-5p, miR-139, miR-143, and miR-106a[105], while another study indicated different set of miRNAs that could be used as biomarkers, including miR-125b, miR-125a, miR-143miR-30a-3p, and miR-145[106].However, this variation in miRNA list might be attributed to the samples used in the identification process (cell line, tissue,blood or stool) and to the techniques employed.Reports also highlight the role of human diet in modulating the expression of miRNA[107].For example, butyrate was found to regulate the expression of Let-7, miR-18-106a, miR-25-106b, and miR-17-92a in CRC[96].The later miRNA cluster (miR-17-92a) was found to be associated with c-Myc to inhibit the activity of PTEN and promotes PI3K-Akt-mTOR axis raising the cell survival in early stage adenoma in CRC[108].
CONCLUSION
Gut microbiota is an enhancer to our second brain; the intestine.With millions of proteins expressed by the microbiota’s arsenal, human could make use of various kinds of dietary ingredients, that otherwise will be rubbish-in/rubbish-out.Although genetic factors and age play a role in the pathogenesis of CRC, still gut microbiota has the lion’s share in this complicated process.Armed with an enormous number of identified and yet-to-be-identified metabolites, this population of bacteria can modify the gut’s cells methylation pattern and histone structure causing inflammation, that lead eventually to cancer.It is quite important to keep these microorganisms under focus by deeply investigate their intricate communications with our cells.By doing so,we would be able to avoid at least life-threatening diseases such as CRC[1].
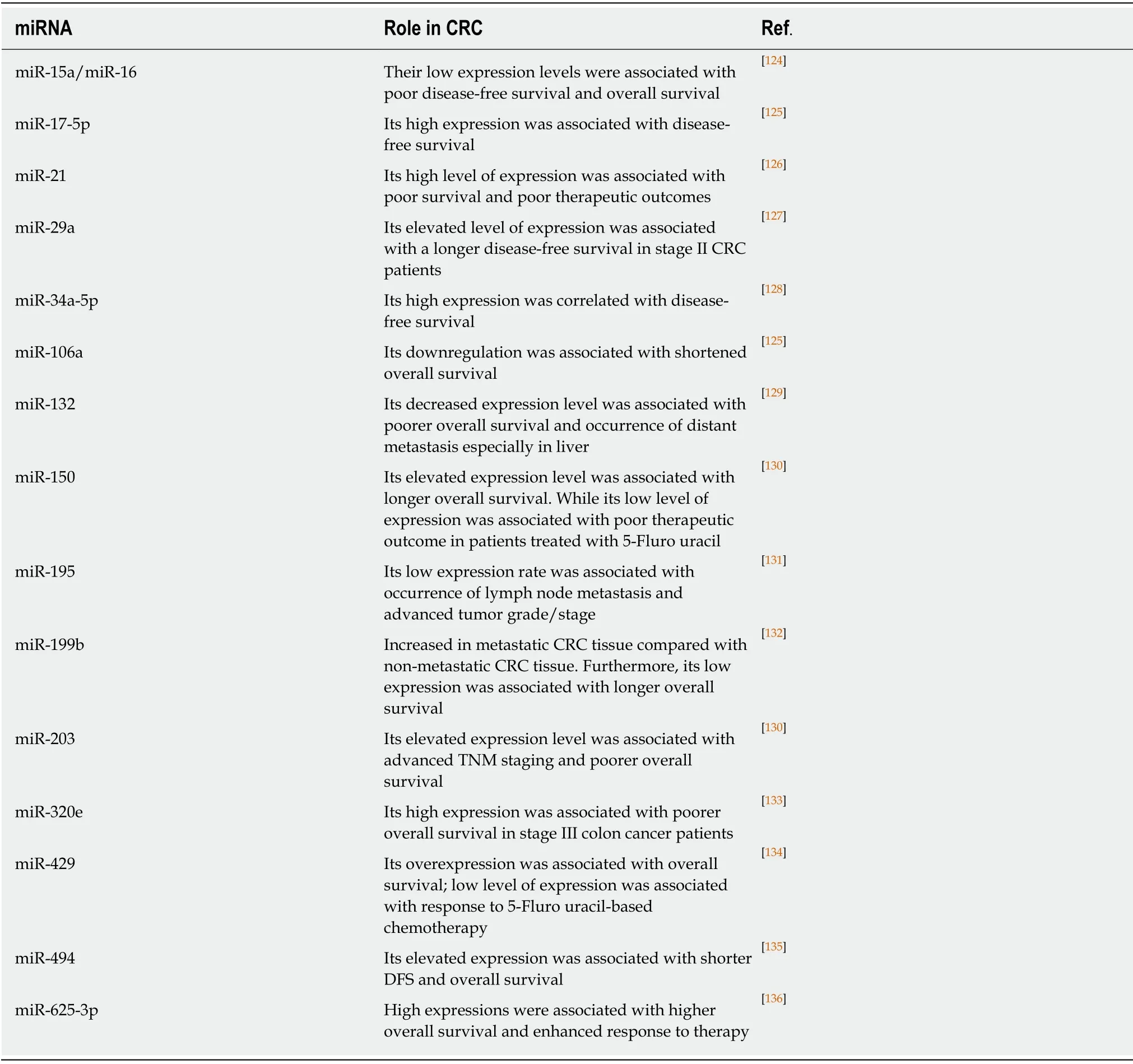
Table 2 A list of representative miRNAs identified in tumor tissues that are of prognostic value in CRC patients
杂志排行
World Journal of Clinical Cases的其它文章
- Human podocyte injury in the early course of hypertensive renal injury
- Relationship between acute hypercarbia and hyperkalaemia during surgery
- Surgical treatment ofpatients with severe non-flail chest rib fractures
- Super-selective arterial embolization in the control of acute lower gastrointestinal hemorrhage
- End-stage liver disease score and future liver remnant volume predict post-hepatectomy liver failure in hepatocellular carcinoma
- Treatment of hemorrhoids:A survey of surgical practice in Australia and New Zealand
