Effect of GmLEC1-A Expression on ABA Content at Germination Stage in Soybean (Glycine max)
2019-04-12JinYangmeiChenQingshanWangMinZhangFengBaiLeiWuXiaoxiaandSuAnyu
Jin Yang-mei, Chen Qing-shan, Wang Min, Zhang Feng, Bai Lei, Wu Xiao-xia*, and Su An-yu
1 College of Agriculture, Northeast Agricultural University, Harbin 150030, China
2 College of Resources and Environment, Northeast Agricultural University, Harbin 150030, China
Abstract: LEAFY COTYLEDON1 (LEC1) is a key regulator of seed maturation, which gives embryos the ability to withstand desiccation. In this study, a novel transcription factor that is homologous to LEC1 in soybean (Glycine max) was isolated from Dongnong 50 by homologous cloning and was named as GmLEC1-A (GenBank accession number: MF681785). Sequence analysis showed that GmLEC1-A contained conserved B regions, which were functional domains of H4 factor. The relative expression level of GmLEC1-A was the highest in seeds of the soybean cultivar Dongnong 50. To verify the function of GmLEC1-A, ectopic expression of Arabidopsis thaliana and over-expression of soybean plants were generated. In Arabidopsis, the expression of GmLEC1-A restored the tolerance of lec1 mutant to seed drying, indicating that GmLEC1-A was a functional homolog of LEC1,and it might regulate the maturation phase of seed development. In soybean, over-expression of GmLEC1-A caused slower growth and lower germination rates as compared to that of wild-type soybeans. Furthermore, over-expression of GmLEC1-A seemed to increase the level of endogenous abscisic acid (ABA) at the germination stage. These results suggested that GmLEC1-A had a function in regulating ABA content at the germination stage.
Key words: Glycine max, GmLEC1-A, Arabidopsis thaliana, seed development, abscisic acid (ABA)
Introduction
LEAFY COTYLEDON1 (LEC1) is a CCAAT-binding(CBF) transcription factor with a HAP3 subunit which occurs extensively in plants (Maity and Crombrugghe,1992; Mantovani 1999; Lee et al., 2003). LEC1 is a central regulator of seed development that can control many distinct aspects of embryogenesis in the early and late phases of embryonic development and identity (Meinke 1992; West et al., 1994; Lotan et al.,1998; Kwong et al., 2003). lec1 mutant embryos are intolerant to desiccation, and show defects in the expression of several maturation-specific genes (Parcy et al., 1997). Prominent characteristics of lec1 mutant include desiccation-intolerant embryos which can be rescued before the seeds desiccate, and rescued seeds do not display any obvious mutant phenotypes(Meinke, 1992; West et al., 1994). The expression of several genes encoding oil body proteins, storage proteins, and transcriptional regulators of the maturation stage is defective in lec1 mutants. Ectopic expression of LEC1 activates the genes involved in maturation and the accumulation of storage protein and lipid in vegetative organs (Harada, 2001; Kagaya et al., 2005; Mu et al., 2008; Junker et al., 2012).
Embryogenesis in higher plants has undergoes two distinct stages: early morphogenetic processes that give rise to embryonic cell types, tissues, and organ systems, and late maturation events that permit the fully developed embryo to go into dehydration and metabolic quiescence (West and Harada, 1993;Goldberg et al., 1994; Lau et al., 2012). Inflowering plants, the early stage of embryogenesis includes the formation of the shoot and root meristems, which can decide postembryonic development (Laux et al.,2004). The fates of single cells in the early embryo can be followed to determine their specific contributions to the seedling body (Haecker et al., 2004). The later stages of embryogenesis involve the establishment of the nutrient storage required for germination, and desiccation preparing the embryo for dormancy (Raz et al., 2001). In the plant life cycle, the transition between the early and late stages of embryogenesis is a critical threshold which is regulated by several plant growth regulators and key genes (Parcy et al., 1997,Ogas et al., 1997).
Seed germination is an important initial stage of growth and development that plays a crucial role in the survival of plants. The hormone abscisic acid(ABA) is a positive regulator of dormancy induction which can induce seed maturation, but is a negative regulator of seed germination and seedling growth(Kucera et al., 2005); Finch-Savage and Leubner-Metzger, 2006). These four master regulator genes LEAFY COTYLEDON 1 (LEC1), LEC2, ABSCISIC ACID INSENSITIVE 3 (ABI3), and FUSCA3 (FUS3)participate in ABA signaling regulation of gene expression during seed maturation (Finkelstein et al.,2002). However, only LEC1 affects ABA sensitivity of germination (Parcy et al., 1997).
Materials and Methods
Plant materials and culture condition
Soybean variety (Glycine max (L.) Merrill.): Dongnong 50 (DN50) seeds were obtained from the Key Laboratory of Soybean Biology of Chinese Education Ministry. DN50 seeds were grown in a plant growth chamber with 16 h light and 8 h dark at 25℃. Fifteen days post germination, when seedlings at thefirst-node stage (soybean growth phase V1) (Fehr et al., 1971)were used for experiments. The experiments were performed in the growth chamber with 16 h light at 26℃ and 8 h dark 18℃.
Arabidopsis thaliana ecotype Columbia-0 (Col-0)was used in this research. A. thaliana lec1 mutant T-DNA insertion line (Salk_118236C) was obtained from The Arabidopsis Information Resource (TAIR).
Isolation of GmLEC1-A and sequence analysis
According to the CDS sequence of Arabidopsis thaliana LEC1 gene (Genbank accession number:NC_003070.9) from TAIR website, the sequence of GmLEC1-A was obtained by blasting on the phytozome website (https://phytozome.jgi.doe.gov/pz/portal.html). The total RNA was isolated from seeds of DN50 using TRIzol reagent (Invitrogen,Shanghai, China) and reverse-transcribed into cDNA by ReverTra Ace1 qPCR RT Kit (TOYOBO, Dalian,China). Using the resulting cDNA as a template,GmLEC1-A gene was amplified by specific primers(GmLEC1-F: 5'-ATGGAAACTGGAGGCTTTCAT G-3' GmLEC1-R: 5′-GCTATGGAGCGAGCATTT GG-3') in a PCR reaction with following parameters:94℃ denaturation for 4.5 min, 35 thermal cycles of 94℃ for 30 s, 60℃ for 30 s, and 72℃ for 1 min,and a final extension at 72℃ for 10 min. The PCR product was ligated into Gateway-T (pGWC) (TaKaRa,Japan), and then transformed into E. coli DH5α cells(TIANGEN, Beijing, China) and sequenced (BioMed,Beijing, China).
The sequencing results of GmLEC1-A gene were aligned through the online BLAST program (http://www.ncbi.nlm.nih.gov/BLAST). The isoelectric point and molecular weight of GmLEC1-A were analyzed using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The nucleotide and amino acid sequences of GmLEC1-A were analyzed using the sequence alignment software DNAMAN (http://www.lynnon.com/). The protein structure and potential domains of GmLEC1-A were predicted by online InterPro program (http://www.ebi.ac.uk/interpro/).Homologous protein sequence similarity analysis was performed using the algorithm Blastp (proteinprotein BLAST: http://www.ncbi.nlm.nih.gov/blast).Three-dimensional (3D) structure of GmLEC1-A was predicted by the online Phyre 2 program (http://www.sbg.bio.ic.ac.uk/phyre2). The RasMol software 2.7.2.1.1 (http://www.OpenRasMol.org/Copyright.html) was used to generate a graphical representation of the protein structure. The phylogenetic relationships of GmLEC1-A homologs in plants were constructed using the neighbor-joining method with the program MEGA 5.2 (http://www.megasoftware.net).
Quantitative-RT-PCR analysis of GmLEC1-A expression
The expression of GmLEC1-A was determined through quantitative-RT-PCR (qRT-PCR) on an ABI 7500 Real-Time PCR Detection System (ABI,Shanghai, China) using the SYBR 1 Premix Ex Taq™ⅡKit (Tli RNaseH Plus, TaKaRa) according to the manufacturers' instructions. The total RNA was isolated from the root, stem, leaf, flower, and seed of DN50 soybean plants using the TRIzol Reagent(Invitrogen) following the manufacturers' protocol.About 1 μg of the total RNAs was used for thefirststrand cDNA synthesis using the M-MLV reverse transcriptase kit (TaKaRa). Each quantitative PCR reaction was performed with 1 μL of the resulting cDNA, 10 μL of 2×SYBR Green PCR Master Mix,and 1.5 μL of each primer in a total volume of 20 μL.The PCR cycling conditions were as the followings:95℃ denaturation for 30 s, 40 cycles of 95℃ for 5 s,60℃ for 20 s, and 72℃ for 20 s, and afinal extention of 60℃ for 1 min. The GmLEC1-A expression was determined using the gene-specific primers (RTGmLEC1-A-F: 5'-CAAGAGTGCGTGTCGGAGTA-3';RTGmLEC1-A-R: 5'-TTGGTGGCGTTTCCCT-3').The soybean GmACTIN4 (GenBank accession no. AF049106) and the Arabidopsis AtACTIN8(A. thaliana 18S rRNA gene GenBank accession no. X16077) were used as reference genes with the primer pairs GmActin4-F (5'-GTGTCAGCCATAC TGTCCCCATTT-3';) and GmActin4-R (5'-GTTTC AAGCTCTTGCTCGTAATCA-3'), and AtActin8-F(5'-TCCTCTAAATGACCAAGTTTG-3') AtActin8-R(5'-GGAAGGGRTGTATTTATTAG-3'), respectively.The relative expression levels of GmLEC1 in different tissues were determined using 2-ΔΔCtmethod with three technical replicates.
ldentification of transgenic lec1 plants
Plants homozygous for the T-DNA insert were identified via PCR by the left genomic primer (LP),right genomic primer (RP), and the left T-DNA border primer (LB) designed for lec1 (LP: 5'-ATGCGGTTTGCAAAACTAATG-3'; RP: 5'-AAAGATATGGAACGTGGAGGC-3'; LB: 5'-ATTTTGCCGATTTCGGAAC-3'). After confirmation of homozygous T-DNA insertion, gene knock-out was confirmed by RT-PCR with a gene-specific primer.
Arabidopsis thaliana transformation and germination rate analysis
Arabidopsis thaliana seeds were kept in dark condition at 4℃ for vernalization for 2 days. The seeds were sterilized in 10% sodium hypochlorite for 10 min. The waste liquid was removed and the remaining seeds were washed six times by distilled sterile water. The seeds were then planted on Murashige and Skoog (MS)solid medium, and transplanted to a sterilized soil and vermiculite mixture (sterilized soil: vermiculite was 1 : 1), when the plant grew 3-4 rosette leaves.Transformation proceeded during plant flowering by the infiltration method (Clough and Bent, 1998). A single colony of Agrobacterium was suspended into a 15 mL yeast extract peptone liquid culture medium containing selection antibiotics, and incubated at 28℃,with shaking at 185 r · min-1, until the absorbance at 600 nm was approximately 0.5. The Agrobacterium was collected by centrifugation for 15 min at 5 000×g.The flowers were immersed in the Agrobacterium liquid for 30 s and then moved into a thermostatic chamber to culture without light after being covered with fresh-keeping films. After 24 h, the plants were returned to the upright position and supplied with normal illumination. The lec1 mutant, transgenic lines, and wild-type Arabidopsis were grown in the same growth chamber, maintained at 22℃ with a 16 h light/8 h dark cycle (light intensity of 350 mol · m-2· s-1).At four-day post-germination, the rate of germination of the wild-type, lec1 mutant, and ectopically expressed Arabidopsis with 0, 1, 2, and 3 µmol · L-1ABA were compared.
Soybean transformation and phenotypic analysis of transgenic soybean
The Agrobacterium mediated soybean transformation was performed using the cotyledonary nodes of DN50 soybeans as explants (Paz et al., 2004).After being cultured in dark conditions, shoot regenerative proliferation, shoot elongation induction,root differentiation multiplication, and plantlet regeneration, the regeneration plants were transferred into pots and grown in the greenhouse.
DN50 soybean seeds were grown in a growth chamber with a 16 h photoperiod (light intensity of 350 mol · m-2· s-1) and 8 h dark cycle. Fifteen days after planting, seedlings at the first-node stage (soybean growth phase V1) (Fehr et al., 1971) were used for various treatments.
ABA measurements
For ABA measurements, cotyledons were frozen in liquid nitrogen and lyophilized. ABA was measured using an ELISA procedure (Kusaba et al., 1998).
Results
Cloning and sequence analysis of GmLEC1-A gene in soybean
The full-length GmLEC1-A (GenBank accession number: MF681785) mRNA in DN50 was 672 bp in length, containing an open reading frame of 669 bp encoding a protein of 223 amino acids. The protein for LEC1-A was predicted to have a molecular mass of 25.19426 ku with a theoretical iso-electric point of 6.30. Based on soybean genome in phytozome databases, there are homologs of LEC1 in the soybean genome, one on chromosome 7 and the other on chromosome 17, respectively. Phylogenetic analysis of GmLEC1-A and other LEC1 homologous from other plant species including crops, fruits, and vegetables showed that GmLEC1-A grouped with plant LEC1 family members (Fig. 1A). The conserved H4 DNA-binding domain resides between residues 59-158 amino acids (99 amino acids) in GmLEC1-A (Fig. 1B),which comprised of five α-helix structures, and five turn structures (Fig. 1C). The predicted structure of GmLEC1-A contained conserved B regions (Fig. 1D),which were functional domains of H4 factor.
Tissue specificity analysis of GmLEC1-A in soybean
Tissue specific expression of GmLEC1-A was analyzed by quantitative PCR in the root, stem, leaf,flower, and seed of DN50. Results showed that GmLEC1-A was expressed the highest in immature seeds, followed by flowers, indicating that GmLEC1-A played a role in regeneration of embryos and infloral morphogenesis.
Identification and analysis in ectopically expressed GmLECI-A Arabidopsis thaliana plants
Through the "double primer" genomic PCR method(http://signal.salk.edu/tdnaprimers.2.html), homozygous T-DNA insertion lec1 mutants were identifi ed and used for the following experiments (Fig. 3A).Transgenic Arabidopsis lines expressing GmLEC1-A were also produced. qRT-PCR showed high transcript abundance in the three transgenic lines, suggesting GmLEC1-A was transformed successfully in these three transgenic lines (Fig. 3B). The germination rate between desiccated seeds of wild-type, lec1, and lec1/GmLEC1-A Arabidopsis on the solid MS medium were then compared. None of lec1 mutant seeds germinated at four-day post germination (Fig. 3C).However, both wild-type and transgenic Arabidopsis seeds were germinated at the same time (Fig. 3C),although the germination rate of lec1/GmLEC1-A seeds was slightly reduced as compared to that of wild-type (Fig. 3C). The germination rates with external application of ABA were further compared.The mutant seeds which were rescued displayed hyposensitivity toward ABA in the germination stage, whereas lec1/GmLEC1-A seeds displayed more sensitivity to ABA than wild type in the germination stage (Fig. 3D).

Fig. 1 Characterization of GmLEC1-A
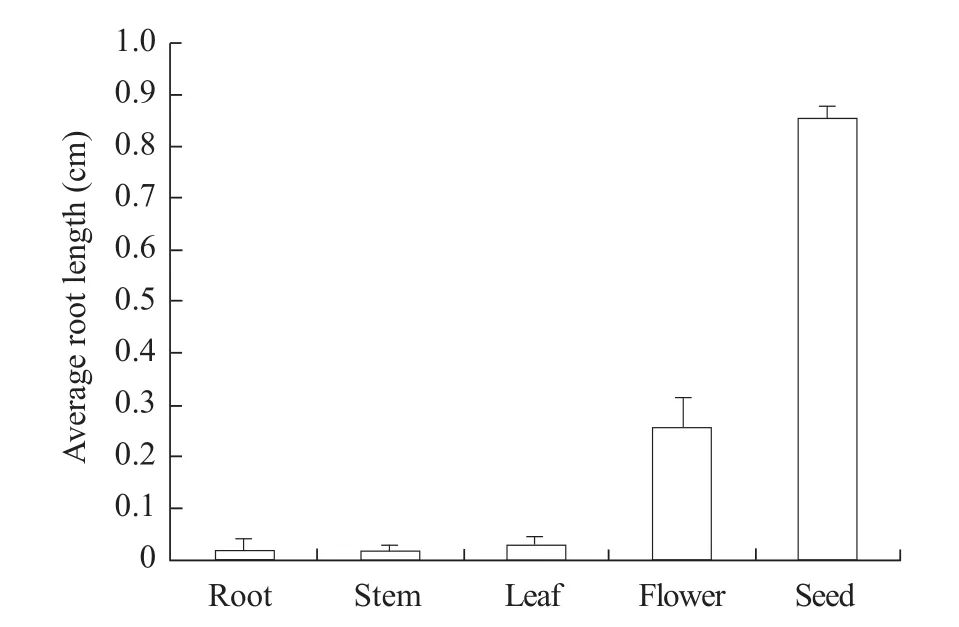
Fig. 2 Tissue expression of soybean GmLEC1-A gene
Identification and phenotype analysis of transgenic soybean plants over-expressing GmLEC1-A
Stable soybean transformants were obtained through the cotyledons mediated transformation. T1seeds grew in soil, and the first trifoliolate leaf was used to identify transformants at V1developmental stage.qRT-PCR showed that GmLEC1-A expression was significantly higher in the four soybean transgenic lines than in control soybean plants (Fig. 4A).To explore the relationship between GmLEC1-A and ABA, ABA content was measured after seed imbibition 0, 3, 6, 9, 12, and 24 h in over-expression and the control soybean seed. The ABA content of over-expression soybean seeds was significantly higher than that of the control seeds (Fig. 4B). The average stem length of four transgenic plants was measured on 1.5 days after germination. The result showed that the hypocotyl length of transgenic soybean lines was much shorter (Fig. 4 C, D). After 3 days, hypocotyl length between transgenic and the control had more difference(Fig. 4E, F). Thus, phenotypic analysis showed that the germination of control soybeans occurred more quickly and successfully than that of the transgenic soybean lines, and the hypocotyl length of the control soybeans was longer than that of over-expression soybeans. These results seemed to related to ABA content.

Fig. 3 Identification and analysis of lec1 mutant and Arabidopsis seedlings

Fig. 4 Analysis of GmLEC1-A transgenic soybean plants
Discussion
In this study, GmLEC1-A, which belongs to the LEC1 type in HAP3, a key regulator of seed maturation, was identified, and ectopic expression can cause dwarfism and low generation rate in plants.
LEC1 played a key regulatory role throughout the embryonic development of plants (Lotan et al.,1998). LEC1 was necessary for the maintenance of embryonic cell fate in the early embryonic stage. In the late embryonic stage, LEC1 participated in many steps of seed maturation, including the acquisition of drying tolerance and the accumulation of stored products. Previous studies showed that embryos of LEC1 deficient mutant lec1 were not tolerant to desiccation and that dried seeds were not active; lec1 homozygous seeds could be rescued to survival before desiccation. The rescued lec1 mutant homozygous seeds were fertile, and showed no mutant phenotype in the vegetative organs and floral organs of plants(West et al., 1994). In this study, the expression of GmLEC1-A restored the desiccation tolerance of lec1 mutants, indicating that GmLEC1-A was a functional homologue of LEC1, and might play a key role in the regulation of soybean embryo development.
The transformed plants in this study confirmed the integration and expression of GmLEC1-A gene by PCR and fluorescent protein detection. GmLEC1-A was initially active in developing T2seeds, and could be complementary to lec1 mutant, but was inactivated after germination, which was consistent with the ectopic expression of LEC1 in Arabidopsis thaliana(Stam et al., 1997) Lotan et al. (1998) and West et al. (1994) found embryonic-specific RNA in oleosins and in two 2S storage proteins in ectopic expressed 35S/LEC1 seedlings. These results indicated that the ectopic expression of LEC1 gene might result in the expression of specific genes required for the development of late embryo seeds in vegetative and reproductive tissues.
In this study, it was speculated that the expression of GmLEC1-A might have a relationship with ABA through the phenotype of transgenic plants, so the content of ABA was measured. The results showed that there was a great difference in ABA content between transgenic and the control soybean. GmLEC1-A transgenic seedlings grew short hypocotyls, suggesting that seed-specific genes might be prematurely expressed, resulting in abnormal morphogenesis of transgenic plants. In addition, lec1 mutant rose the possibility that LEC genes affected ABA responses.Ectopic expression of LEC1 activated FUS3, and FUS3 caused an increase in ABA levels. In this study,lec1 mutant was insensitive to ABA. Furthermore,over-expression seemed to increase the level of endogenous ABA at the germination stage. Therefore,the next study should examine the mechanisms behind the shortening of hypocotyls and increased ABA content of GmLEC1-A transgenic plants, in the context of the specific gene expressions required for seed development. Subsequent researches in our lab will study the associated changes in phenotype,which in combination with the current study, will provide valuable insight into the understanding of seed development and embryonic development in soybean,to the benefit of thefield and agriculture.
Conclusions
The function of the soybean GmLEC1 gene as well as relationship among the function of GmLEC1 and seed germination, and hypocotyl elongation were analyzed. In soybean, GmLEC1 over-expression led to lower hypocotyl elongation and higher ABA contents.These results indicated that GmLEC1 might play an important role in seed germination and elongation of soybean.
杂志排行
Journal of Northeast Agricultural University(English Edition)的其它文章
- Proteomic Studies of Petal-specific Proteins in Soybean [Glycine Max (L.)Merr.] Florets
- Compatibility Screening of Plant Extracts Synergistic with Osthole
- Bioinformatics and Expression Pattern Analysis of Tomato nsLTP 2-like cDNA full-length Gene Clone
- Molecular Differentiation of Different Pathogenic Phenotypes of Infectious Bursal Disease Viruses by RT-PCR Combined with Restriction Fragment Length Polymorphism (RFLP) Assay
- Effect of Xylazine Anesthesia on Glu and GABA Amino Acid Neurotransmitters in Rat
- Chlorophyll Content Retrieval of Rice Canopy with Multi-spectral Inversion Based on LS-SVR Algorithm
