High resolution melting real-time PCR detect and identify filarial parasites in domestic cats
2019-01-05DarawanNonsaithongSupitYotmekSomsriYotmekHathaiNochoteSirichitWongkamchaiSittirukRoytrakulUsaLekUthai
Darawan Nonsaithong, Supit Yotmek, Somsri Yotmek, Hathai Nochote, Sirichit Wongkamchai, Sittiruk Roytrakul, Usa Lek-Uthai✉
1Master of Science (Public Health) Program in Infectious Diseases and Epidemiology, Faculty of Graduate Studies, Mahidol University, Bangkok, Thailand
2Department of Parasitology and Entomology, Faculty of Public Health, Mahidol University, Bangkok, Thailand
3Center for Disease Control, Vector Borne Disease Control Center 11.3 Surat Thani Province, Thailand
4Department of Parasitology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
5National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency (NSTDA), Thailand Science Park, Pathum Thani, Thailand
Keywords:High resolution melting analysis Dried blood spot Brugia malayi Brugia pahangi Dirofilaria immitis
ABSTRACT Objective: To detect and identify filarial parasites in dried blood spots (DBS) collected from domestic cats using high resolution melting real-time PCR (HRM RT-PCR). Methods: A total of 208 DBS were collected from domestic cats in a brugian filariasis endemic areas in Surat Thani Province, southern Thailand. Microfilariae were found in 9 blood slides using Giemsa-stained thick blood film. The extracted DNA from blood spot volumes of 10 and 20µL DBS with positive filarial parasites in cats were performed using HRM RT-PCR method.The primers were designed based on the partial mitochondrial 12S rRNA gene for identifying Brugia malayi, Brugia pahangi, Dirofilaria immitis. All purified samples were then detected.Results: Using different volumes of 10 µL and 20 µL DBS could easily distinguish filarial parasites and showed similar results. PCR amplicons of Brugia malayi, Brugia pahangi and Dirofilaria immitis were determined at melting peak (temperature) of 75.70 ℃, 77.46 ℃, and 73.56 ℃, respectively. All 9 positive DBS samples showed positive Brugia pahangi and similar nucleotide sequences. Conclusions: This HRM RT-PCR method is able to diagnose, identify and discriminate filarial parasites collected from DBS, which is simple and inexpensive compared with other probe-based genotyping methods. Furthermore, this method is useful to survey, prevent and control filariasis.
1. Introduction
The World Health Organization (WHO) established the Global Programme to Eliminate Lymphatic Filariasis aiming to eliminate the disease by the year 2020. Brugian filariasis is zoonotic and has been reported as being diurnally subperiodic in Southeast Asia including Malaysia and Surat Thani Province, in southern Thailand[1,2]. Domestic cats are important reservoir hosts, and can be infected with Brugia malayi (B. malayi), Brugia pahangi (B.pahangi), Dirofilaria immitis (D. immitis) and Dirofilaria repens[3,4].From 2010-2013, Brugia spp. infection rates of 10%, 6%, 9% and 25% were reported in surveyed domestic cats[5]. The microscopic examination to discriminate B. malayi from B. pahangi in cats is difficult because of their similar microfilaria morphology[6,7].High resolution melting real-time PCR (HRM RT-PCR) has been applied to detect Brugia DNA in human blood samples[8]. Recently,effective HRM RT-PCR has been used to rapidly detect and identify microfilaria, providing a simple, fast and less expensive method than other conventional DNA-based assays. The polymorphic sequences in a single-tube assay based on the melting of double-stranded amplified DNA has been reported to differentiate filarial species,such as Brugia spp.[9,10], Wuchereria bancrofti, D. immitis[11] and Dirofilaria spp.[12]. Additionally, this method is able to detect filarial parasites in various species in a single PCR tube[13]. Collecting blood using dried blood spots (DBS) on filter paper is simple, inexpensive and less time consuming than whole blood samples. The DBS method could collect large numbers of field specimens economically,store blood at room temperature and properly transfer specimens for DNA extraction in a laboratory located in rural areas. The use of whole blood collection by anticoagulants for which anticoagulation is achieved through antithrombin-mediated inhibition of the coagulation factors[14] is limited owing to its complexation with ions. As we know, hemoglobin molecule contains four heme groups,which contain iron, therefore can inhibit PCR ability by the release of iron ions and disturb DNA polymerase activity, which has been implicated as the cause of DNA amplification[15]. Centrifugation and separation sera from blood clots were needed but the collected blood tube may be reduced by the infected materials[16]. Thus, this study has applied a method to extract DNA from DBS of domestic cat blood samples to identify DNA of the B. malayi, B. pahangi and D.immitis in a single reaction tube using the partial mitochondrial 12S rRNA gene by HRM RT-PCR.
2. Materials and methods
2.1. Source of positive and negative controls
Reference strains for the HRM method were B. malayi, B. pahangi,and D. immitis which were provided from TRS Labs (Athens,Georgia) and stored at the Department of Parasitology, Faculty of Medicine, Siriraj Hospital, Mahidol University. Blood samples collected from cats without filarial parasites infection were used as negative control.
2.2. Blood sample collection
A total of 208 domestic cat blood samples were collected including 100 samples from Thachana District and 108 samples from Phrasaeng District, Surat Thani Province, Thailand. Blood samples were collected from 10:30 am to 2:30 pm. For each sample, a 60 µL of thick blood film slide was prepared for microscopic examination(smooth oval shape in the center of the slide; 3 cm in length and 2 cm in width), and microfilariae was found in 9 blood slides. Those 9 microfilariae positive samples were marked as No. 1 to No. 9 (cat No.1=C1 to cat No.9=C9), for which 10 µL and 20 µL of blood were spotted onto Whatman® 3MM filter paper (Sigma-Aldrich, Merck,U.S.A.) for further use of DNA extraction.
2.3. Animal ethics approval
The Animal Ethics document was approved by the Faculty of Tropical Medicine, Animal Care and Use Committee at Mahidol University, Bangkok, Thailand (FTM-ACUC 012/2017).
2.4. Giemsa staining
All thick blood film slides were screened by Giemsa staining method according to standard WHO procedure. Each blood film was analyzed for microfilaria under a microscopy (40×).
2.5. DNA extraction from DBS /blood spot volume and DNA extraction method
Each collected DBS was punched out then soaked in 200 µL TE buffer and incubated at 37 ℃ for 1 h and overnight. The High Pure PCR Template Preparation Kit (Roche Diagnostics, Penzbery,Germany) was used to extract DNA according to manufacturer instructions.
2.6. High resolution melt analysis
Based on alignment of partial mitochondrial 12S rRNA genes of B. malayi, B. pahangi, and D. immitis (GenBank accession number AJ544843.1, AM779849.1, and FN391554.1, respectively), the primers were designed as previously described by Wongkamchai et al. to identify B. malayi, B. pahangi and D. immitis[13]. The sequences of these primers were as follows: forward primer, 5´-TTTAACCGAAAAAATATTGACTGAC-3´ and reverse primer, 5´-AAAAACTAAACAATCA TACATGTGCC-3´.
HRM RT-PCR was performed in 10 µL of reaction volume with separate tubes. Briefly, extracted DNA (3 µL) was added to PCR mixture consisting of ResoLight Dye (Roche Diagnostics), 3 mM MgCl2in 5 µL, 0.25 pmol of each primer with PCR grade water being adjusted to a final volume. Then 3 µL of DNA template of positive control and nuclease-free water of negative control were replaced.
Thermocycling was conducted in a thermal LightCycler® 480 instrument (Roche, Germany) as follows: an activation step at 95 ℃for 5 min; followed by 40 cycles of 95 ℃ for 10 s, 58 ℃ for 10 s, and 72 ℃ for 10 s. The HRM RT-PCR products were heated to 95.8 ℃for 1 min and then cooled to 40 ℃ for 1 min for which the HRM RT-PCR machine could increase at 1 ℃/s with 25 acquisitions per degree from 65 ℃ to 95 ℃. The final step was 40 ℃ cooled for 10 s. The different melting curves were plotted and analyzed using LightCycler® 480 gene scanning software (Roche Applied Science,Germany).
2.7. DNA sequencing
A High Pure PCR Product Purification Kit (Roche Diagnostics,Penzbery, Germany) was used to purify recombinant plasmids containing PCR products from B. malayi, B. pahangi and D. immitis.The CLUSTALW2 multiple alignment comprised the sequences analyzing program which could identify the polymorphisms of the partial mitochondrial 12S rRNA nucleotide sequence.
3. Results
3.1. Giemsa staining to detect microfilariae in blood samples
The identification process, using microscopic examination of Giemsa’s stain blood smear in 208 blood samples, showed 9 (4.3%)were positive for Brugia spp. microfilaria, which showed clear sheathed microfilaria and 2 terminal nuclei at the tail tip. In addition, some of the samples showed exsheated microfilariae (Figure 1).
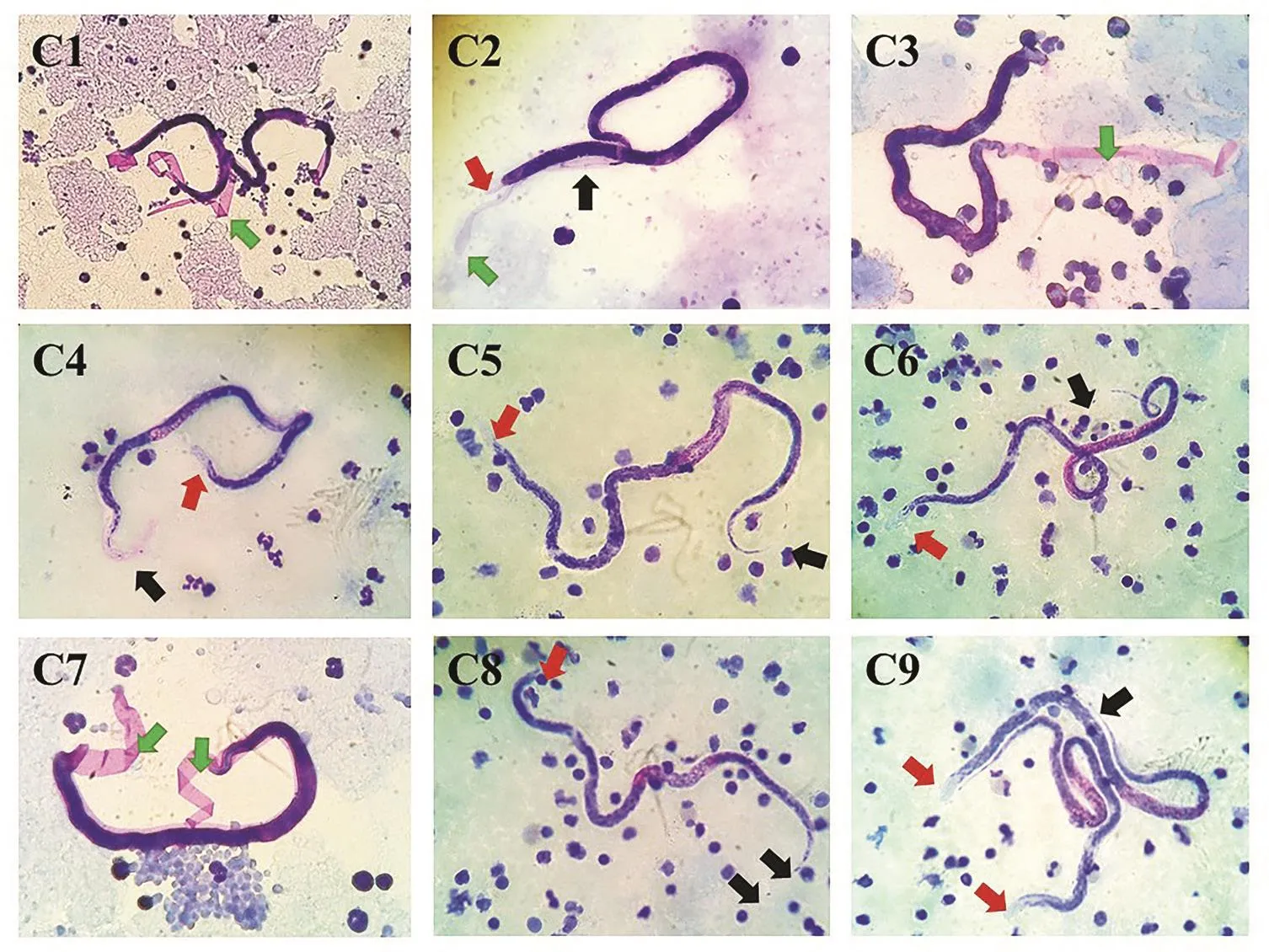
Figure 1. Microscopic examination of Brugia spp. microfilaria in domestic cats (cat No. 1-cat No. 9; C1-C9) by Giemsa staining method (40×).
3.2. Blood spot volume and DNA extraction method
The HRM RT-PCR assay was performed using universal primers that targeted a partial sequence of the partial mitochondrial 12S rRNA genes and were designed to detect B. malayi, B. pahangi, and D. immitis. This assay allowed to show the positive control samples clearly according to different melting temperature of the amplicons peaked at 75.70 ℃ (B. malayi), 77.76 ℃ (B. pahangi) and 73.56 ℃(D. immitis) as shown in Figure 2.
DNA were extracted from 10 µL and 20 µL volumes of B.malayi positive control DBS which were stored at the Department of Parasitology, Faculty of Medicine, Siriraj Hospital, Mahidol University. The amplicons of B. malayi from 10 µL DBS, soaked in 200 µL TE buffer for 1 h and 20 µL DBS, soaked in 200 µL TE buffer,overnight revealed a similar variation in melting peak at 74.84 ℃.And 20 µL DBS, soaked in 200 µL TE buffer, 1 h showed a peak at 75.81 ℃. However, B. malayi from 10 µL DBS, soaked in 200 µL TE buffer, overnight showed no melting peak. These results implied that the DBS volume that was spotted onto Whatman® 3MM filter paper should ideally be 20 µL. A negative cat blood sample was not amplified.

Figure 2. Temperature profiles of the derivative melt curves of the 12S rRNA gene for B. malayi (Bm), B. pahangi (Bp), and D. immitis (Di).
Since the pre-test results showed 10 µL DBS, 1 h and 20 µL DBS,overnight showed similar temperature curves, only the 20 µL DBS soaked with TE buffer for 1 h samples which took shorter time were selected for further analysis.The amplicons ofB. malayifrom 10 µL DBS, soaked in 200 µL TE buffer for 1 h and 20 µL DBS, soaked in 200 µL TE buffer, overnight revealed a similar variation in melting peak at 74.84 ℃ (10 µL DBS, 1 h and 20 µL DBS,overnight) and 20 µL DBS, soaked in 200 µL TE buffer, 1 h showed a peak at 75.81 ℃ (20 µL DBS, 1 h). However,B. malayifrom 10 µL DBS, soaked in 200 µL TE buffer, overnight showed no melting peak.
3.3. High resolution melt analysis to differentiate PCR products of blood samples
The universal primers that were designed to detect B. malayi, B.pahangi and D. immitis was used to detect the partial mitochondrial 12S rRNA gene. The sample clusters were classified by different melting plotted curves within groups.
Amplicons of positive control samples of B. malayi, B. pahangi,and D. immitis showed peak melting temperatures of 75.70 ℃, 77.76℃ and 73.56 ℃, respectively. PCR amplicons of the nine blood samples (20 µL soaked in TE buffer for 1 h) exhibited recognized melting peaks according to the temperature-shifted fluorescence differences of melting plots (Figure 3).
The extracted DNA from the DBS sample cat No. 1 (C1) showed a single melting peak at 76.88 ℃ (Figure 3A). The other (20 µL,1 h) curve was generated from a tested laboratory DBS sample of B. malayi, it was presented for melting temperature comparison.Amplicons from DBS samples of cat No. 2-cat No. 4 (C2-C4)(Figure 3B) cat No. 5-cat No. 9 (C5-C9) (Figure 3C) showed single peaks between 76.88 ℃ to 78.08 ℃ which had a mean of 77.45℃ confirming they were B. pahangi (C5=77.43 ℃, C6=77.25 ℃,C7=77.37 ℃, C8=77.88 ℃, C9=77.33 ℃).This method could easily distinguish filarial parasites that showed similar results. A negative cat blood sample was not amplified.

Figure 3. Temperature profiles of the derivative melt curves of the 12S rRNA gene for filarial parasites.
4. Discussion
In southern Thailand, infection by filarial parasites in reservoir hosts, especially cats, is an endemic disease that could represent an emerging infectious disease. In nature, domestic cats infected with B.malayi, B. pahangi and D. immitis is usually diagnosed by detecting the microfilariae morphology using Giemsa’s stain in blood smear slide. Normally, Giemsa-stained shows the pink-staining sheath of microfilaria, and sometime shows exsheated microfilariae that may appear similar to D. immitis[17]. In this research, Giemsa-stained TBF illustrated clear sheathed and terminal nuclei at the tail tip, while some samples exhibited Brugia spp. exsheated microfilaria.
Currently, based on HRM RT-PCR analysis, molecular techniques have been successfully applied for nematode differentiation and D. immitis differentiation[18], human hookworms[19], the quarantine nematode Bursaphelenchus spp.[20] and Brugia spp.[13]. Albonico et al.[12]applied HRM analysis to differentiate D. immitis and Dirofilaria repens in canine peripheral blood. The limit of detection using 2-fold serial dilutions of the genomic DNA of B. malayi and D. immitis was (19.4×10-12) g and (16.4×10-12) g, respectively[6]. It is also reported that the detection limit using 10-fold serial dilution for B.malayi was 2 copies, while for B. pahangi the limit was 4 copies,and for D. immitis the limit was 10 copies of DNA[11]. In addition it could detect and identify the species in one single step PCR by differentiation of the specific melting temperature peak[21]. Thus,HRM analysis could be used to differentiate the melting peaks according to the GC content, length and sequence composition of the PCR products.
This study applied the DNA extraction method and HRM RT-PCR method to extract DNA from DBS of domestic cat blood samples in order to identify DNA of the B. malayi, B. pahangi and D. immitis in a single reaction tube. The partial mitochondrial 12S rRNA gene of B. malayi, B. pahangi and D. immitis are highly conserved and contains genus-specific and species-specific sequence variations[13].Generally, extracted DNA from whole blood has been used for molecular assay. However, collection of whole blood samples involve imitations, and dry ice transportation to the laboratory site is needed. Alternatively, DBS on filter paper collection is more convenient. Collected DBS samples in study areas could be used to detect filarial parasites using extraction and amplification DNA kits[22]. DBS would greatly facilitate epidemiological surveys of samples collection, which can preserve specimens until proper monitoring for molecular assay is conducted.
Long term storage of DBS has diminished sensitivity[23] and DBS samples stored for 2 or more years at ambient temperature showed loss of positive results[24]. But stored DBS samples at room temperature, in a dark room for 3 to 9 months and 3.5 years at -20 ℃before DNA extraction have shown positive results[25,26]. Therefore,the use of silica gel is recommended to reduce humidity in the containers where the DBS are stored. Humidity and temperature are likely factors to influence the preserved DNA in the DBS[24,27].
In this study, different volumes of DBS (10 µL and 20 µL) and DNA extraction methods (DBS was soaked in 200 µL TE buffer for 1 h and overnight) were compared. DBS soaked in 200 µL TE buffer for 1 h and overnight demonstrated a similar variation in melting peaks of the B. malayi positive control. However, Brugia spp. from 10 µL DBS, soaked in 200 µL TE buffer overnight showed no melting peak. Different volumes of 50, 200, and 1 mL DBS, showed similar positive results while 5 µL DBS exhibited less sensitivity[28].On the other hand, DBS spotted with 20 µL was extracted, and amplified to analyze isolates of Plasmodium falciparum. All malaria cases were successfully amplified and characterized in plasmodial DNA from filter paper. Similar results performed in 20 µL of DBS showed 100% sensitivity for detecting DNA in blood samples[24]. In this study, we used 20 µL DBS soaked in 200 µL TE buffer for 1 h to identify filarial parasites using HRM RT-PCR. George and Moat[29]recommended that blood spots containing 20 µL might create an increased risk of having false-negative results.
TE buffer is commonly used in DNA extraction to dissolve DNA that has already been extracted and precipitated or used in DNA extraction protocols to maintain the stability of that DNA during extraction. Bereczky et al.[30] described the TE buffer-based DNA extraction method, and showed superior results, compared with two standard methods for extracting blood samples on filter paper stored for 15 and 29 months. This was similar to the 12 to 15 months stored blood samples of this study using Whatman®3MM filter paper(Sigma-Aldrich, Merck, U.S.A.) to collect blood samples.
In conclusion, filarial nematode species identification is recommended for zoonotic surveys especially in domestic cats using HRM RT-PCR analysis. A completely “closed tube” using a single pair of primers could be achieved, without requiring probe and electrophoresis. Therefore, this method was able to diagnose,identify and discriminate B. malayi, B. pahangi and D. immitis in DBS collected samples. Furthermore, this method was useful in survey, prevention and control of filariasis. Additionally, detecting filarial DNA using HRM RT-PCR from DBS is simpler and cheaper than other molecular methods, such as collecting, and transporting samples and using molecular diagnostic methods in filarial infection endemic areas.
Conflict of interest statement
The authors declare they have no conflicts of interest.
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- Salacca zalacca: A short review of the palm botany, pharmacological uses and phytochemistry
- Immune enhancement effect of an herb complex extract through the activation of natural killer cells and the regulation of cytokine levels in a cyclophosphamide-induced immunosuppression rat model
- Phytocompounds of Anonna muricata leaves extract and cytotoxic effects on breast cancer cells
- Screening of antiproliferative activity mediated through apoptosis pathway in human non-small lung cancer A-549 cells by active compounds present in medicinal plants
- Ultrasound-assisted extraction of antioxidant polyphenolic compounds from Nephelium lappaceum L. (Mexican variety) husk
- Can miRNA712_3p be a promising biomarker for early diagnosis of toxoplasmosis?
