Weighted Correlation Network Analysis (WGCNA) of Japanese Flounder (Paralichthys olivaceus) Embryo Transcriptome Provides Crucial Gene Sets for Understanding Haploid Syndrome and Rescue by Diploidization
2018-12-20ZHAOHaitao1DUXinxin1ZHANGKai1LIUYuezhong1WANGYujue1LIUJinxiang1HEYan1WANGXubo1andZHANGQuanqi1
ZHAO Haitao1, DU Xinxin1, ZHANG Kai1, LIU Yuezhong1, WANG Yujue1, LIU Jinxiang1, HE Yan1, WANG Xubo1, and ZHANG Quanqi1,
Weighted Correlation Network Analysis (WGCNA) of Japanese Flounder () Embryo Transcriptome Provides Crucial Gene Sets for Understanding Haploid Syndrome and Rescue by Diploidization
ZHAO Haitao1), 2), DU Xinxin1), 2), ZHANG Kai1), 2), LIU Yuezhong1), 2), WANG Yujue1), 2), LIU Jinxiang1), 2), HE Yan1), 2), WANG Xubo1), 2), and ZHANG Quanqi1), 2),3), *
1),,266003,2),,266003,3),,266237,
Artificial gynogenesis is of great research value in fish genetics and breeding technology. However, existing studies did not explain the mechanism of some interesting phenomena. Severe developmental defects in gynogenetic haploids can lead to death during hatching. After diploidization of chromosomes, gynogenetic diploids may dispense from the remarkable malformation and restore the viability, although the development time is longer and the survival rate is lower compared with normal diploids. The aim of this study was to reveal key mechanism in haploid syndrome of Japanese flounder, a commercially important marine teleost in East Asia. We measured genome-scale gene expression of flounder haploid, gynogenetic diploid and normal diploid embryos using RNA-Seq, constructed a module-centric co-expression network based on weighted correlation network analysis (WGCNA) and analyzed the biological functions of correlated modules. Module gene content analysis revealed that the formation of gynogenetic haploids was closely related to the abnormality of plasma proteins, and the up-regulation of p53 signaling pathway might rescue gynogenetic embryos from haploid syndromeregulating cell cycle arrest, apoptosis and DNA repair. Moreover, normal diploid has more robust nervous system. This work provides novel insights into molecular mechanisms in haploid syndrome and the rescue process by gynogenetic diploidization.
Japanese flounder; RNA-Seq; gynogenesis; haploid syndrome; weighted correlation network analysis
1 Introduction
Gynogenesis is a reproductive mode in which progeny is formed exclusively from maternal genetic information (Neaves and Baumann, 2011). In artificial gynogenesis, the haploid egg is activated by genetically inactivated spermatozoa and then develops into diploid offspring after chromosome diploidization (Schön., 2009; Bi and Bogart, 2010). In 1932, Amazon molly was found as the first gynogenetic fish species, and from then on stud- ies on artificial gynogenesis in fish have been developed rapidly (Hubbs and Hubbs, 1932; Makino and Ozima, 1943; Purdom, 1969; Chourrout and Quillet, 1982; Ihssen., 1990). Gynogenesis is of great value in aquaculture, including production of inbred lines, breeding of mono- sex fish stocks, identification of sex-determination me- chanisms, development of genetic maps, and even detec- tion of alternative gene splicing (Yamamoto, 1999; Arai, 2001; Fopp-Bayat, 2010; Wang., 2014).
Artificial gynogenesis includes two main procedures: 1) induction of gynogenetic haploid embryos by activation of embryogenesis using genetically inactivated sperma- tozoa; and 2) retrieval to diploid through suppressing the second meiosis of oocyte (meiogynogenesis) or the first cleavage of zygote (mitogynogenesis) (Felip., 2001; Komen and Thorgaard, 2007; Nichols, 2009). Over the decades, several methods, such as cold or heat shock, hydrostatic shock and chemical shock, have been devel- oped to restore diploids (Felip., 2001).
Gynogenetic haploids show characteristics of haploid syndrome, such as bending short thick body, the expan- sion and edema of cavum pericardial, distemperedness of blood circulation, and disorders of head and tail. Thus, haploid embryos are nonviable, and the vast majority of them die during hatching stage (Arai, 2001; Li., 2009). In contrast, gynogenetic diploids restore viability after chromosome diploidization, which has drawn the attention of researchers to explore the mechanisms. In Atlantic salmon, isozyme analysis for 24 biochemical loci confirmed that genes were fully activated in haploids as well as in diploids, and the defect of epiboly and involu- tion during gastrulation might be caused by extensive cell death and disorganization in blastoderm (Stanley, 1983;Araki., 2001). In goldfish, diploid-dependent regula- tion caused abnormal development of eyes in haploids (Luo and Li, 2003). A recent transcriptome analysis of flounder embryos showed the down-regulation of notch and Wnt signaling pathways in haploids (Fan., 2016). However, the regulation of gene co-expression network during formation of haploids is poorly understood. Addi- tionally, it remains unknown why the development of gynogenetic diploids are retarded than normal diploids.
Japanese flounder () is a com- mercially important marine teleost in East Asia. Recent developments in transcriptome sequencing techniques provide high-throughput approaches toward understand- ing the genome-wide gene expression and co-expression network in flounder. Based on thetranscriptome sequencing of a double haploid flounder, thousands of alternative splices and single nucleotide polymorphisms were discovered (Wang., 2014). The comparative transcriptome analysis of gonads revealed the molecular regulatory mechanism of gonadal development and ga- metogenesis (Fan., 2014; Zhang., 2016). Co- expression network analysis of sterile doubled haploid flounder provided insights into reproductive dysfunction (Zhang., 2015). In this study we measured genome- scale gene expression of haploid, gynogenetic diploid and normal diploid embryos using RNA-Seq, constructed mo- dule-centric co-expression network based on weighted gene co-expression network analysis (WGCNA) and ana- lyzed the biological functions of correlated modules. The findings will help to understand the mechanism responsi- ble for haploid syndrome and the rescue by gynogenetic diploidization.
2 Materials and Methods
2.1 Induction of Gynogenetic Embryos and Sample Collection
In May 2014, the induction of gynogenetic Japanese flounder was performed at Haiyang Yellow Sea Aquatic Product Co., Ltd. with a previously reported method (Yamamoto, 1999). The eggs from a female and the se- men from a male were divided equally into three groups. One portion of the semen was diluted 100-fold with Ca2+and Mg2+free Ringer’s solution (NaCl, 13.5gL−1; KCl, 0.6gL−1; NaHCO3, 0.02gL−1) and immediately irradiated with a dose of 112mJcm−2ultraviolet (UV) light. Then the semen was inseminated to one portion of eggs. At 2min after fertilization, eggs were used to induce meiogy- nogenesis by hydrostatic pressure treatment of 60MPa for 6min and this group was named gynogenesis group (g2n). The haploid group (n) was gained as g2n, but without hydrostatic pressure treatment. The control group (c2n) was produced by normal fertilization. The rearing temperature was maintained at 16±0.5℃. The embryonic developments of haploids, gynogenetic diploids and nor- mal diploids were confirmed according to a previous study (Liu., 2008). Approximately 50 embryos from each group at each developmental stage were collected in triplicate and preserved in RNAwait (Solarbio, Beijing, China) for RNA extraction. The developmental stages include thirty-two cells, high blastula, early gastrula, mid gastrula, late gastrula, neurula and hatching, namely, stages 1 to 7, respectively.
2.2 RNA Extraction, Library Construction and RNA-Seq
Total RNA was extracted from embryos of the seven stages of all three groups using Trizol reagent (Invitrogen, Carlsbad, CA , USA) for library construction and quanti- tative real-time PCR (qRT-PCR) analysis. Genomic DNA was removed using the DNase I Kit (TaKaRa, Dalian, China) and RNA was purified using an RNA purification Kit (Biomed, Beijing, China). RNA integrity and concen- tration were detected using gel electrophoresis, Thermo NanoDrop Spectrophotometer (Thermo Scientific, Carls- bad, CA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Library construction and RNA-Seq were completed by Novogene Sequencing Company (Beijing, China). Total RNA with the best quality in three replicates was used for library construction and RNA-Seq sequencing. A cDNA library was prepared using 3μg of total RNA per sample, according to the protocol for the NEBNext Ultra Direc- tional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA). Sequencing was performed using the Illumina HiSeq 2000 platform with 125bp paired- end reads. The raw reads were filtered to obtain clean reads, including removal of the adapter sequences and more than 10% sequences of unknown nucleotides and low-quality reads with Q value<13 (Cox., 2010). The RNA-Seq data has been uploaded to NCBI SRA (Se- quence Read Archive) database and the accession number is PRJNA382751.
2.3 Read Alignment, Expression Quantification and Differential Expression Analysis
The TopHat-Cufflinks-Cuffmerge-Cuffdiff pipeline was applied to process the clean data with default parameters. Firstly, the reads were mapped to flounder genome which was deposited at NCBI SRA (accession number PRJN- A352095) with TopHat v2.0.13 (Trapnell., 2009). The results were used to construct, identify and estimate the abundance by Cufflinks v2.2.0 (Trapnell., 2012). Fragments per kilobase of transcript per million frag- ments mapped (FPKM) was adopted to quantify the abundance of assembled transcripts. Then all the Cuff- links results were merged to assemblies by Cuffmerge v1.0.0 and the abundance of transcripts was re-estimated by Cuffdiff v2.2.0. Differential expression analysis was also performed by Cuffdiff between every two groups at each developmental stage. Genes with more than two fold change (log2FC>1 or <−1) in expression level and false discovery rate (FDR)<0.05 were considered as differ- entially expressed genes (DEGs).
2.4 GO and KEGG Pathway Enrichment Analysis
The enrichment analysis of GO terms was performed using Database for Annotation, Visualization, and Inte- grated Discovery (DAVID, v6.8, http://david-d.ncifcrf. gov) (Huang., 2009). The enrichment analysis of KEGG pathways was performed using the online Omic- Share tools (http://www.omicshare.com/tools). All ex- pressed genes were used as the background.
2.5 Gene Co-Expression Network Construction
Gene co-expression network was constructed using the R package WGCNA 1.51 (Langfelder and Horvath, 2008; Zhang and Horvath, 2005). A total of 19200 genes from 21 samples were used for module detection in one block with default settings. In order to obtain an appropriate scale-free topology, we chose a power of eight according to the scale free topology model (model fit index2=0.75). The intramodular connectivity (K) was calculated to measure the connection strength of a given gene to other genes in the particular module.
2.6 Identification of Gynogenesis-Responsive Modules and Visualization
The eigengene is the first principal component of the module and can represent the gene expression profile of the module. To identify the gynogenesis-responsive mod- ules, the eigengene value for each module was calculated and the association with each sample was used to identify responsive modules. Top 15% genes inKranking were counted as hub genes in a given module. The networks were visualized using Cytoscape v3.3.0 (Shannon., 2003).
2.7 qRT-PCR Validation
Twelve genes were chosen for qRT-PCR validation. Total RNAs of the three replicates were reverse-tran- scribed by M-MLV reverse transcriptase (TaKaRa). Pri- mers were designed using Primer Premier 5.0 and the sequence were listed in Table 1. qRT-PCR was performed on a LightCycler 480 real-time PCR system (Roche Ap- plied Science, Mannheim, Germany) with SYBR Green PCR Master Mix (TaKaRa) following the procedures de- scribed previously (Wang., 2015). All PCRs were performed in triplicate.was used as the reference gene (Zhong., 2008). The relative expression level was quantified by the 2−ΔΔCtcomparative Ct method.
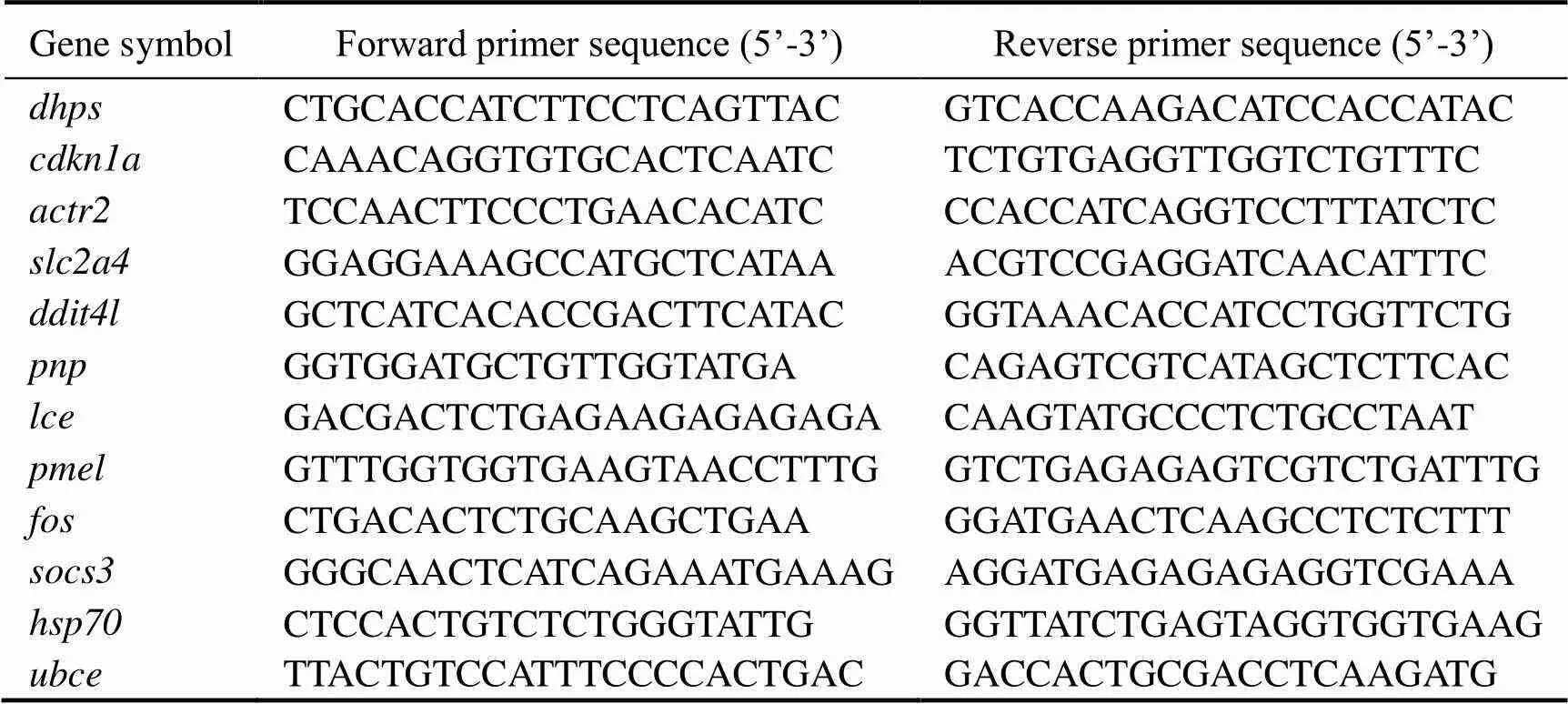
Table 1 List of primers used in quantitative RT-PCR
3 Results
3.1 Global Analysis of RNA-Seq Data
RNA-Seq data generated 596538812 clean reads from 21 samples at different developmental stages (Table 2). Among them, 458972511 were successfully mapped to Japanese flounder genome. The mapping rates ranged from 68.6% to 81.0% with an average mapping rate of 76.9%. Among the 24313 genes predicted in the genome, 21818 were detected as expressed.
To evaluate the global expression pattern, principal component analysis (PCA) and hierarchical cluster analy- sis were conducted for all 21 samples. PCA results showed that all the samples were roughly divided into seven clus- ters according to the developmental stages (Fig.1). Sam- ples from stages 1 and 2 were arranged closely while those from stages 3 to 7 were more dispersed. Cluster dendrogram showed the relatedness of all samples (Fig.2). Three groups at each stage showed high correlation ex- cept that St5_n was more similar to the groups at stage 4. Stage 1 and stage 2 were clustered into one branch while the others into another branch. Correlation between stages 1 and 2 was higher and these two stages were far different from the other stages. This phenomenon might be resulted from the employment of maternal mRNA to express zy- gotic genes.
3.2 Differentially Expressed Genes (DEGs)
In the differential expression analysis, there was no DEG in stage 1, while 297, 156 and 132 DEGs were de- tected in c2n. g2n, c2n. n, and n. g2n of the rest stages, respectively. In the result of GO enrichment analysis of c2n. g2n, lipid transport and lipid localiza- tion were the most significant terms. GO enrichment terms of DEGs in c2n. n included extracellular matrix structural constituent, structural molecule activity, cellular amino acid biosynthetic process and alpha-amino acid biosynthetic process. Several GO terms about peptidase inhibitor activity were enriched in n. g2n DEGs: ser- ine-type endopeptidase inhibitor activity, enzyme inhibi- tor activity, endopeptidase inhibitor activity, endopepti- dase regulator activity, peptidase inhibitor activity and peptidase regulator activity.
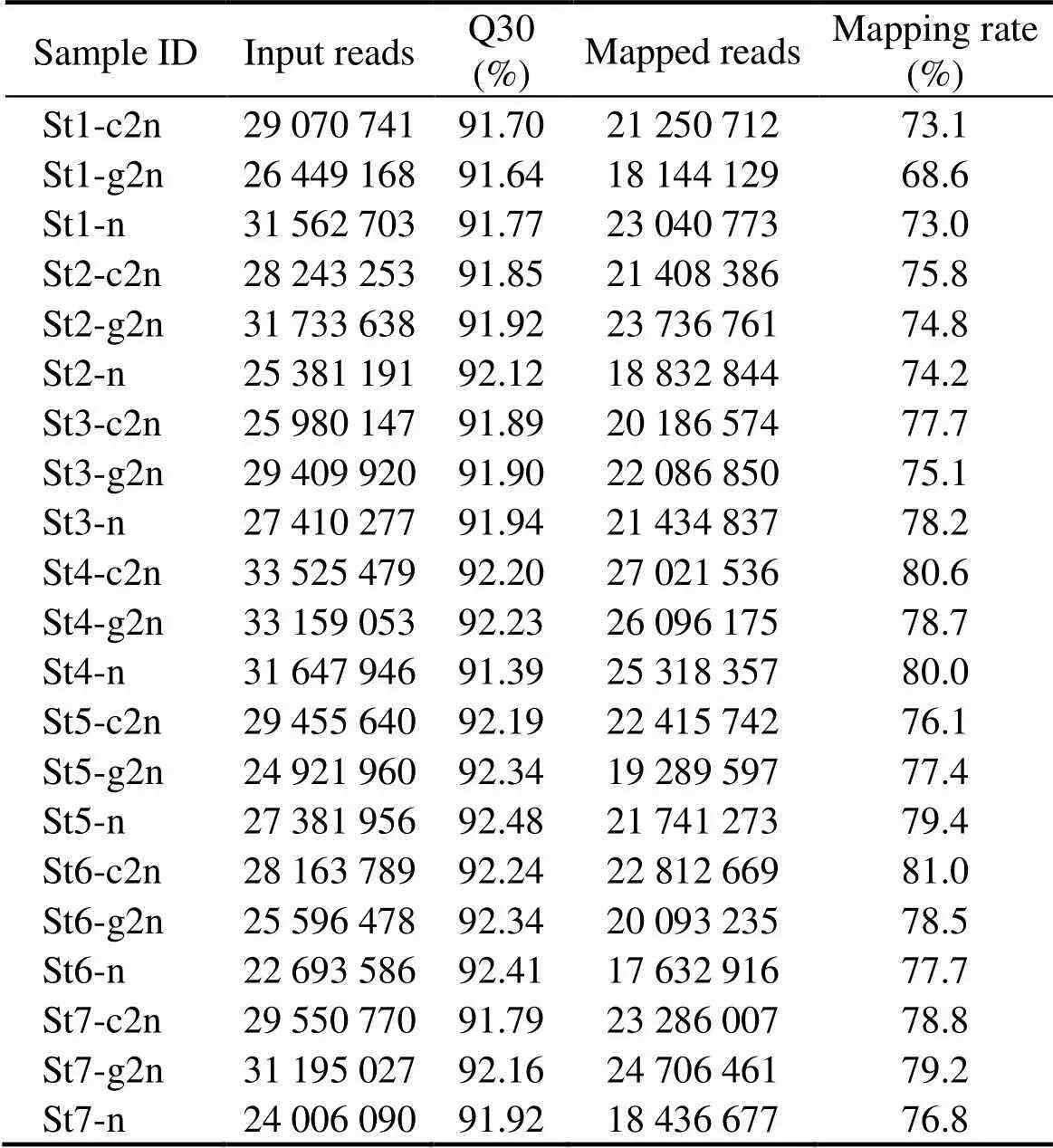
Table 2 Statistic summary of Japanese flounder embryo RNA-Seq data†
Notes:†, St1 to St7 indicate the seven stages of embryonic development: thirty-two cells, high blastula, early gastrula, mid gastrula, late gastrula, neurula and hatching. g2n, gynogenesis diploids; n, haploids; c2n, control diploids.
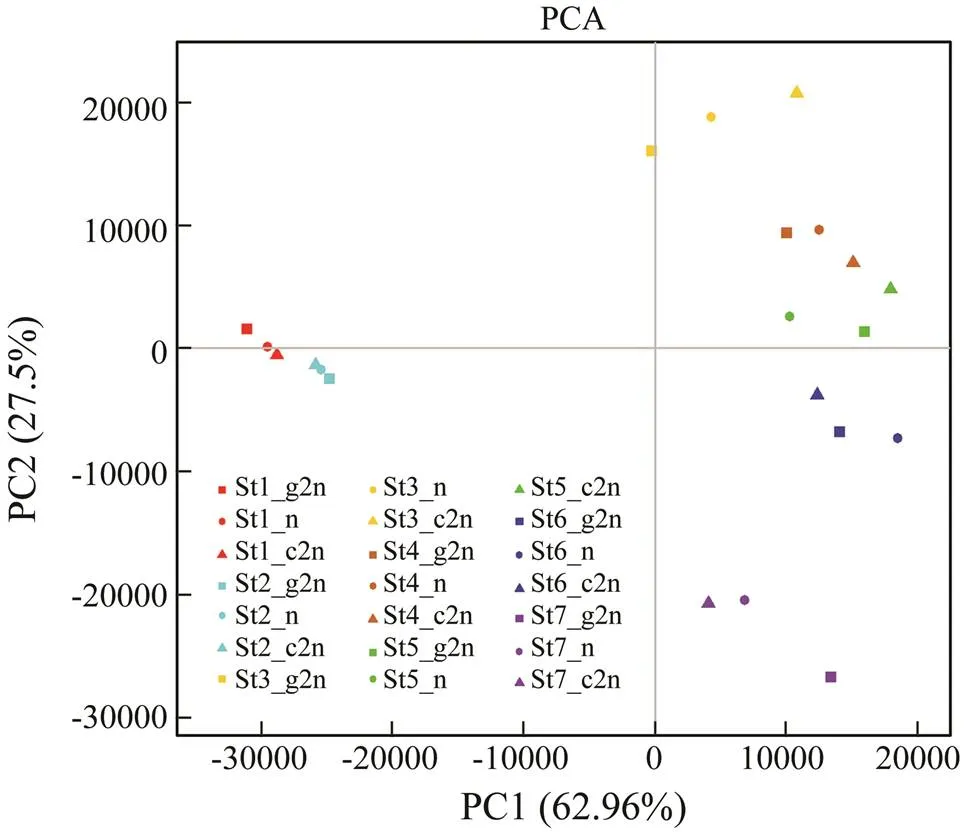
Fig.1 Principle component analysis (PCA) of the samples used for RNA-Seq analysis. PC1, the variation among sam- ples from the same stage. PC2, the variation among differ- ent stages. St1 to St7 indicate the seven stages of embryonic development: thirty-two cells, high blastula, early gastrula, mid gastrula, late gastrula, neurula and hatching. g2n, gy- nogenetic diploids. n, haploids. c2n, control diploids.
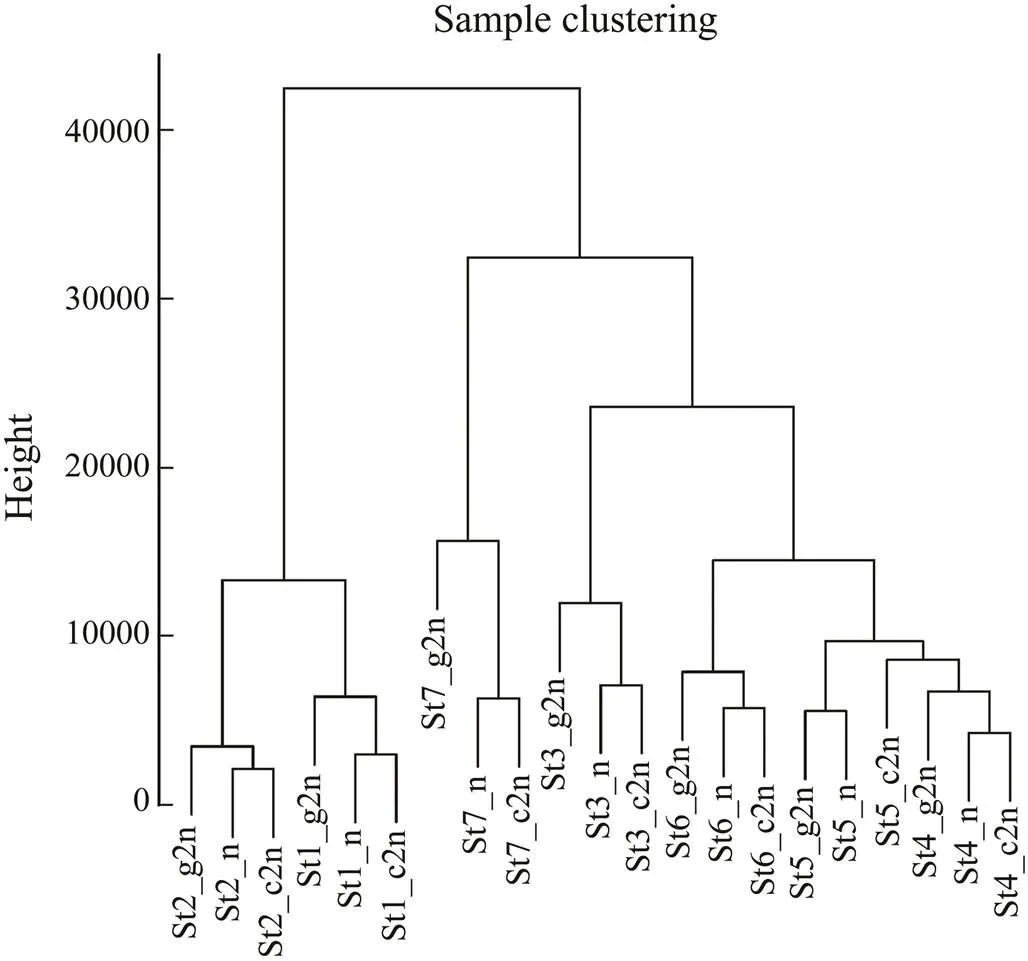
Fig.2 Cluster dendrogram showing global relationship of samples. The y axis shows the degree of variance. Gy- nogenesis diploids (g2n), haploids (n) and control diploids (c2n) were sampled from stages during Japanese flounder embryonic development, including thirty-two cells (St1), high blastula (St2), early gastrula (St3), mid gastrula (St4), late gastrula (St5), neurula (St6) and hatching (St7).
3.3 Gene Co-Expression Network
Gene co-expression network constructed by WGCNA provided a systems biology approach to understand the gene networks instead of individual genes.Thus WGCNA was adopted in this study to identify modules represent- ing functional categories. According to the scale-free to- pology model fit and mean connectivity, soft threshold powerwas set to 8 in our study, which produced an ap- proximate scale-free network with appropriate mean connectivity. After screening, a total of 21 samples and 19200 genes were used for co-expression network con- struction, and 16 modules with module size ranging from 31 to 5702 were acquired (Fig.3).
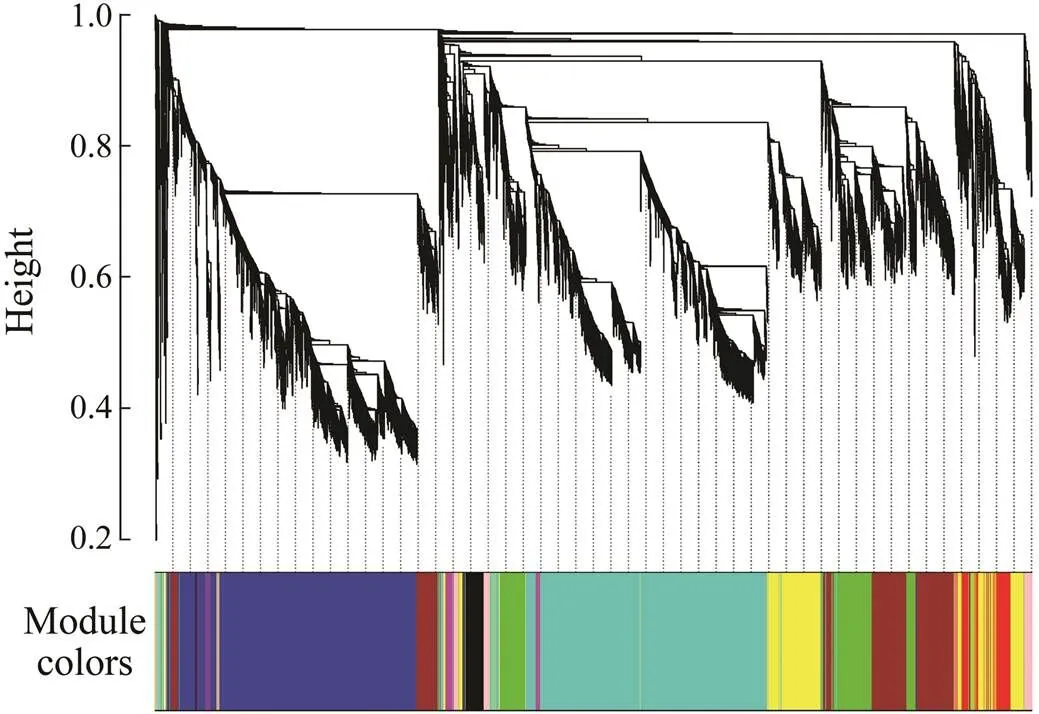
Fig.3 Hierarchical cluster tree showing co-expression mo- dules identified by weighted correlation network analysis (WGCNA). Each leaf of the tree indicates a gene. Hori- zontal color bars represent 16 modules labeled in different colors.
Notably, 6 of the 16 modules were identified as signi- ficantly highly expressed in one particular sample (>0.9,<0.01, Fig.4). Therefore, each of the 6 modules was considered to represent a specific sample (Fig.5). Of these six modules, genes were up-regulated in the correlated samples. Modules 16, 10, 14, 12, 11 and 15 corresponded to St5-c2n, St7-c2n, St6-n, St7-n, St3-g2n and St7-g2n, respectively. The networks of these modules (Fig.5) showed that each group (c2n, n and g2n) was correlated with two modules. According to the result of GO enrich- ment analysis, no GO term was significantly enriched in module 16. Among 31 genes of this module, only 15 geneswere annotated, including serine/threonine-protein kinase, tetranectin and intestinal-type alkaline phosphatase. Mo- dule 10 correlated with St7-c2n was significantly enrich- ed in neuron part, somatodendritic compartment, neuron projection, neurological system process, synapse and neu- ronal cell body, which were all involved in neurological system. Integral component of plasma membrane and in- trinsic component of plasma membrane were enriched in module 14. Module 12 corresponded to St7-n, which was enriched in potassium ion transmembrane transport, cel- lular potassium ion transport, G-protein coupled receptor signaling pathway, potassium ion transport, potassium channel activity, integral component of plasma membrane and intrinsic component of plasma membrane. Module 11 was significantly enriched in cellular response to DNA damage stimulus, protein kinase inhibitor activity and kinase inhibitor activity. Besides, p53 signaling pathway was enriched in this module. Module 15 was correlated with St7-g2n and enriched in cardiac muscle contraction.
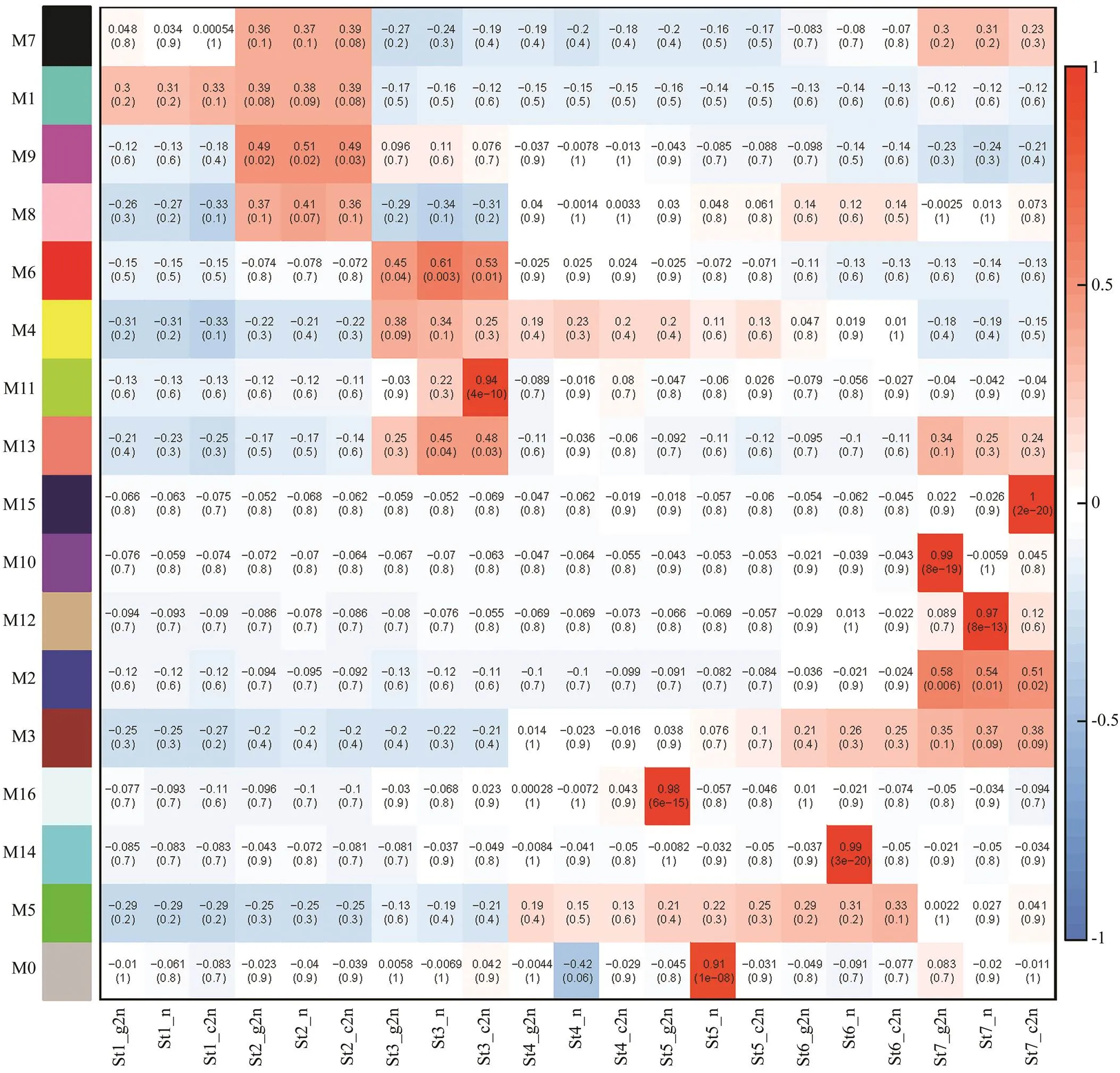
Fig.4 Correlation heatmap of module-sample association. Each row corresponds to a module. Each column corresponds to a sample. The color of row-column intersection indicates the correlation coefficient between the module and sample. The dark red means high correlation between module and sample. St1 to St7 indicate the seven stages of embryonic development: thirty-two cells, high blastula, early gastrula, mid gastrula, late gastrula, neurula and hatching. g2n, gynogenesis diploids. n, haploids. c2n, control diploids.
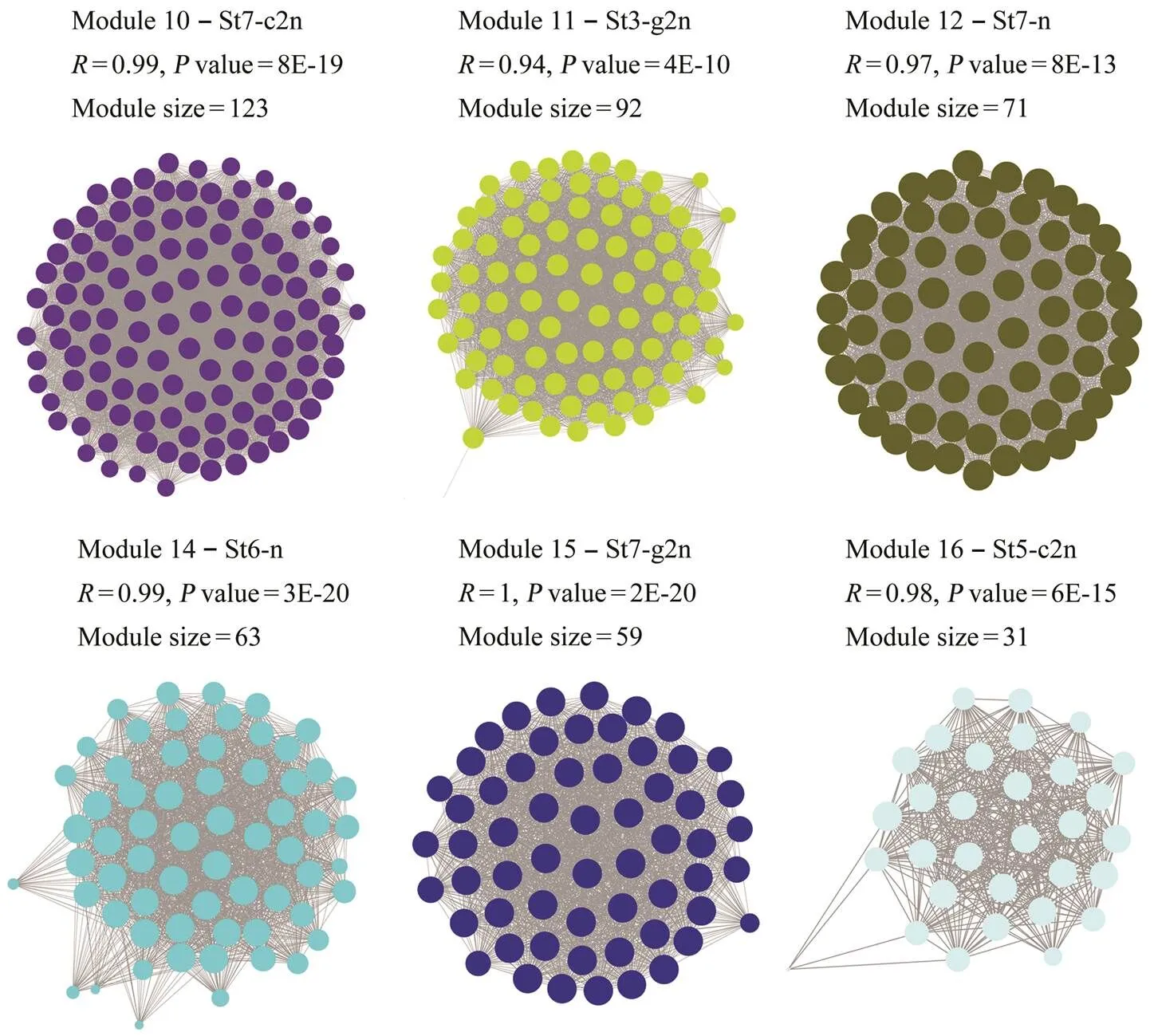
Fig.5 Correlation networks of six modules. Samples correlate to specific modules, and their correlation coefficient R, P value and module size are shown.
3.4 Validation of DEGs
The quantitative expressions of 15 DEGs from differ- ential analyses were tested by qRT-PCR, which found that 12 genes were down-regulated and three were up-regu- lated. Between c2n and g2n, four DEGs at stage 3, one at stage 4 and one at stage 7 were validated. Between c2n and n, two DEGs at stage 3, four at stage 4 and three at stage 7 were validated. All the validated DEGs showed the same changing patterns in RNA-Seq and qRT-PCR. The correlation (2=0.9628, Fig.6) suggested that the RNA-Seq data were reliable to evaluate the gene expres- sion levels.
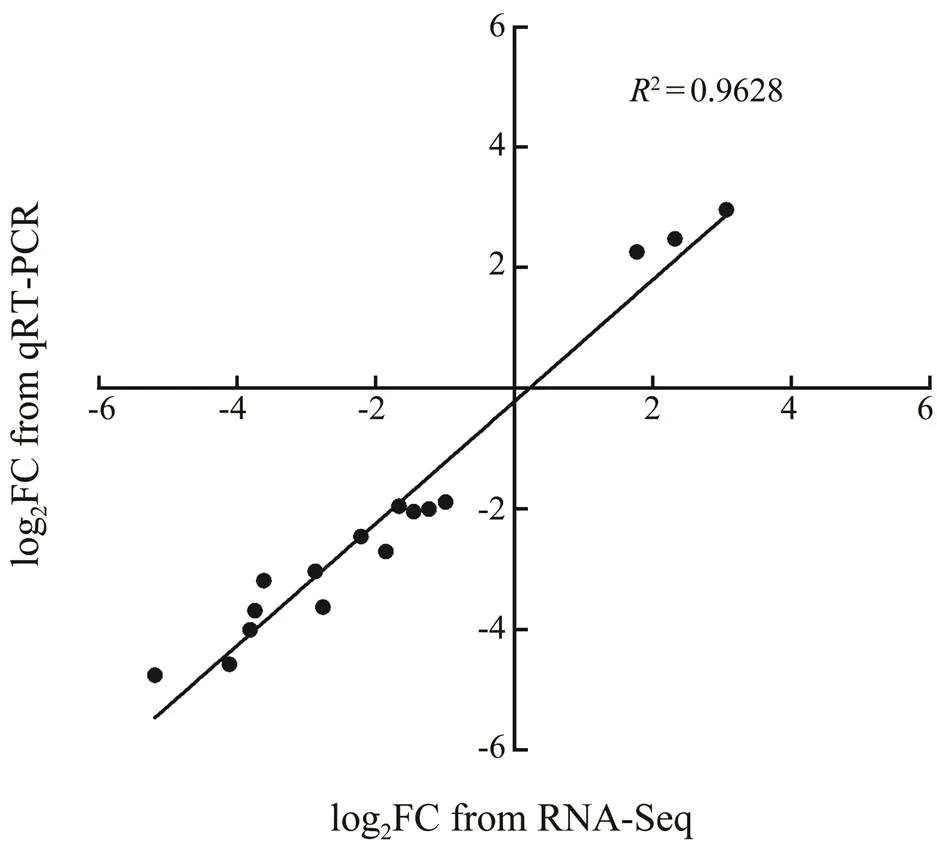
Fig.6 Consistency between quantitative real-time PCR (qRT-PCR, y axis) and RNA-Seq data (x axis). FC, fold change.
4 Discussion
Gynogenetic haploids and diploids exhibit several dis- tinctive characters during the process of embryonic de- velopment. Haploid embryos suffer a series of severe developmental disorders, which constitute haploid syn- drome ultimately resulting in failure to survive. Dramati- cally, after the chromosome doubling treatment, gynoge- netic diploid embryos restore viability. Additionally, compared with normal diploids, the development of gy- nogenetic diploid embryos take longer time and gynoge- netic diploid larvae show a remarkably low survival rate. However, the mechanism of these phenomena was still unclear. In this study, based on RNA-Seq data of Japanese flounder, WGCNA was used to reveal several novel in- sights into the expression characteristics of haploid, gy- nogenetic diploid and normal diploid embryos.
4.1 Gene Expression Profile Implicated Dosage Compensation in Haploids
In mammals, the changes of gene dosage often bring great deleterious effects to the organisms (Antonarakis., 2004). Gene dosage compensation was the war- ranty to control the appropriate product. In lower verte- brates, like fish, individuals with different ploidy were able to grow normally, which implied that fish may pos- sess an effective compensation control mechanism (Leg- gatt and Iwama, 2003). A study in allotriploid ofshowed that one of the three alleles was si- lenced and it was not a whole set of chromosomes that was inactivated (Pala., 2008). Researchers detected 24 biochemical loci in Atlantic salmon haploid and dip- loid embryos and found all of them were fully activated in both haploids and diploids (Stanley, 1983). The global expression profiles in our study showed that the differ- ences between haploid and diploid embryos were not as significant as one set of active chromosomes. In other words, haploids exhibited only limited alterations in the expression level of most genes, which was in accordance with the result in mouse embryos (Latham., 2002). The transcription levels might be increased to the degree as diploids through complex compensation mechanism.
4.2 Normal Diploids Might Own a More Robust Nervous System
Modules 16 and 10 correspond to control diploids at late gastrula and hatching stage respectively. Based on the GO enrichment analysis, module 10 was enriched for genes in neuron part such as astrotactin (), neurturin () and sodium- and chloride-dependent GABA trans- porter 2 (or). In vertebrates, central nervous system (CNS) histogenesis counts on the neuronal lami- nar formed by the glia-guided migration of postmitomic neurons (Chen., 1996b). Astn provides a receptor system and plays a role as a neuronal cell surface antigen in CNS neuronal migration (Edmondson., 1988; Fishell and Hatten, 1991). Low expression ofmRNA leads to slower neuronal migration, which might happen in haploids and gynogenetic diploids, restricting the gen- eration of laminar structure and the development of syn- aptic partner system (Pearlman., 1998; Adams., 2002). Nrtn is a neuronal survival factor, which can sup- port the survival of embryonic dopaminergic neurons and protect them from cell death induced by 6-OHDA (Horger., 1998; Heuckeroth., 1999). Moreover, Gat-2 is an important transporter of peripheral GABAergic me- chanisms (Schlessinger., 2012; Zhou., 2012; Paul., 2014). Therefore, we speculate that the devel- opment of nervous system especially CNS of haploids and gynogenetic diploids was defective or incomplete at hatching stage, while control diploids owned a robust nervous system, as reflected in module 10. Module 16 correspond to diploids specifically at late gastrula stage. Due to its small size (31 genes), few GO terms were en- riched. Among the annotated genes, several upstream genes like serine/threonine-protein kinase and tetranectin were found, which also implied that diploids might be superior to haploids and gynogenetic diploids in signal transduction in the nervous system (Scott and Soderling, 1992; Dahiya., 2017).
4.3 Haploid Syndrome Was Closely Related to Abnormality of Plasma Protein
Gynogenetic haploid embryos were induced by acti- vated egg division with UV-exposed spermatozoon, and all of them showed haploid syndrome. Modules 14 and 12 were identified to specifically correspond to stage 6 (neurula stage) and stage 7 (hatching stage) in the haploid group. Genes of module 14 were significantly enriched in integral component of plasma membrane and intrinsic component of plasma membrane. Most products of these genes were protein complexes embedded in the mem- brane, such as Claudin 1 (Cldn1), neuromedin K receptor (Nk3), protein ATP1B4 (Atp1b4) and Glypican 5 (Gpc5). Cldn1 protein has four transmembrane domains and con- stitutes a tight junction with occluding (Evans., 2007; Furuse., 1998). Nk3 has been reported as tachykinin neuropeptide embedded tightly in the plasma membrane (Masu., 1987). Atp1b4 is a subunit of Na+, K+-AT- Pase and is located in plasma membrane (Pestov., 2011). Gpc5 plays a role in cell growth and differentiation, controlling the cell surface heparin sulfate (Saunders., 1997). Up-regulation of these genes might change the structure and components of plasma membrane. These findings might help to explain the irregularity of the cel- lular contours in haploid embryos.
Module 12 was another haploid responsive module and was enriched in potassium ion transmembrane transport, cellular potassium ion transport, potassium ion transport, potassium channel activity and G-protein coupled recep- tor signaling pathway at the hatching stage. Furthermore, GO terms enriched at stage 6 were included. Notably, proteins encoded by these genes were also plasma mem- brane embedded ones. Among them, four potassium voltage-gated channel proteins (Kcng4, Kcne2, Kcnh7 and Kcna3) were found, all of which were from the same family of membrane signaling proteins (Yellen, 2002). Potassium channels were critically important in nervous system for normal functions in tissues like muscle and brain (Graves and Hanna, 2005). Specifically, mutations in voltage-gated potassium channels lead to dysfunction in central nervous system (Waters., 2006). Haploid embryos with abnormal expression of potassium volt- age-gate channels might suffer from severe neurodegen- erative diseases. Therefore, haploid syndrome might be closely related to abnormity of plasma proteins.
4.4 Up-Regulation of p53 Signaling Pathway Might Rescue Gynogenetic Embryos from Haploid Syndrome
Gynogenetic diploids undergo chromosome diploidiza- tion through inhibiting the second polar body release during the fertilized eggs. After chromosome diploidiza- tion, there is no haploid syndrome left. In this study, mo- dule 11 was detected to gynogenetic diploid responsive at early gastrula stage. According to the results of GO en- richment analysis, this module was involved significantly in the cellular response to DNA damage stimulus, protein kinase inhibitor activity and kinase inhibitor activity. The cellular response to DNA damage stimulus was consid- ered to be the reaction to the hydrostatic treatment. Meanwhile, in gynogenetic diploid at early gastrula stage, genes involved in the protein kinase inhibitor activity and kinase inhibitor activity were upregulated, including cy- clin-dependent kinase inhibitor 1 () and cyclin- dependent kinase 4 inhibitor B (). Cdks participate in extensive cellular processes, such as cell cycle regula- tion, neuronal physiological process, apoptosis, differen- tiation and transcription (Fischer., 2003). Their in- hibitors could induce cell growth arrest or apoptosis when the regulation of cell cycle was impaired (Malumbres and Barbacid, 2001). Hence, the up-regulation of these genes interfered in the normal cell cycle might result in delay of embryo development in gynogenetic diploids.
Module 11 was also enriched in p53 signaling pathway (Fig.7), which might indicate the characteristics of gyno- genetic diploid embryos from the upstream of Cdkn re- gulation. Transcription factor p53 is a critical node gath- ering pathways from comprehensive biological processes, such as cell cycle, apoptosis, DNA repair and damage prevention, angiogenesis and metastasis, to adapt to di- verse cellular insults (Harris and Levine, 2005). In mo- dule 11, five genes of p53 signaling pathway were de- tected. P21 is an inhibitor of G1 cyclin-dependent kinases in the upstream of cell cycle arrest (Xiong., 1993; Harper., 1993;Bunz., 1998). Pigs and Apaf-1 are essential components in Myc-induced apoptosis (Soengas., 1999). Sestrins play a role in DNA repair induction and damage prevention (Budanov., 2004). Mdm2 binds the transcriptional activation domain of p53 and the downstream genes are then suppressed, while p53 activates the expression of Mdm2 in an autoregulatory feedback loop (Chen., 1994; Chen., 1996a; Haupt., 1997). Based on the above information, we hypothesize that cellular insult of gynogenetic diploids might initiate cell cycle arrest and DNA repair, and sub- sequently, severely damaged cells step into apoptosis. The rescue mechanism of p53 signaling pathway is a defi- ciency of haploids, which is in accordance with our GO enrichment results.
Module 15 corresponded to gynogenetic diploids at hatching stage. Genes in this module were enriched in cardiac muscle contraction. This implies the impaired heart development at hatching stage in gynogenetic dip- loids, which might help to explain the low survival rate in gynogenetic diploids.
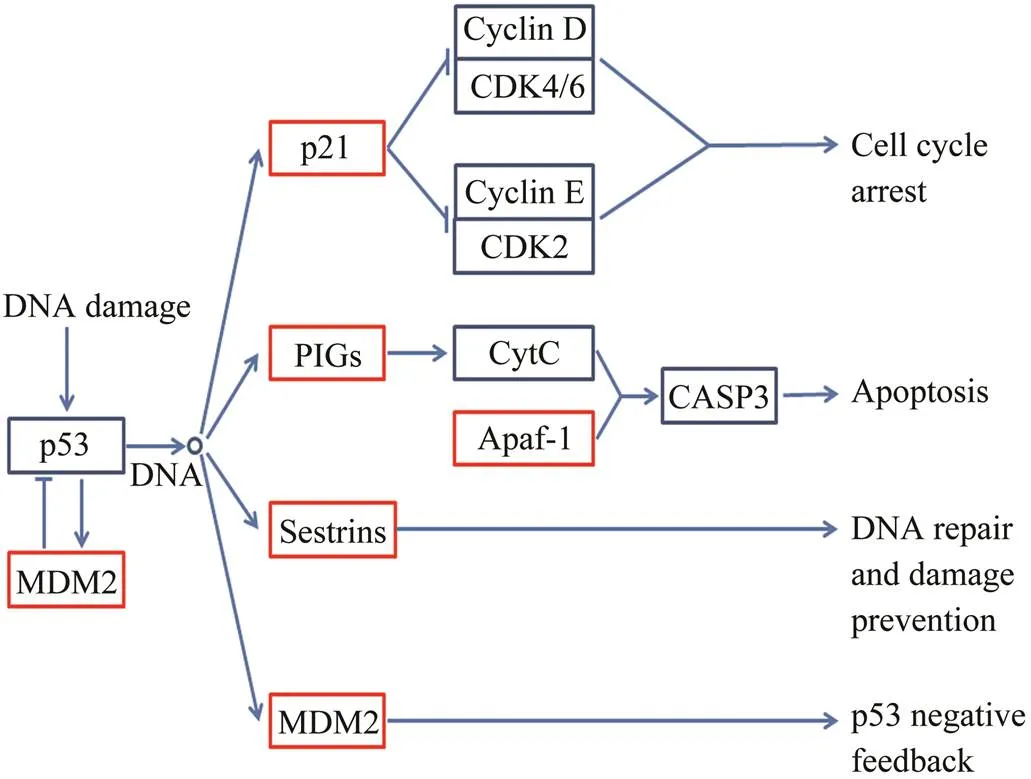
Fig.7 p53 signaling pathway enriched from module 11 correlated to gynogenetic diploids. Genes in module 11 are circled by red frame.
Acknowledgements
This work was supported by the Scientific and Tech- nological Innovation Project of Qingdao National Labo- ratory for Marine Science and Technology (No. 2015A SKJ02) and the National Natural Science Foundation of China (No. 31540063).
Adams, N. C., Tomoda, T., Cooper, M., Dietz, G., and Hatten, M. E., 2002. Mice that lack astrotactin have slowed neuronal migration., 129 (4): 965-972.
Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A., and Deutsch, S., 2004. Chromosome 21 and down syndrome: From genomics to pathophysiology., 5 (10): 725-738.
Arai, K., 2001. Genetic improvement of aquaculture finfish species by chromosome manipulation techniques in Japan., 197 (1): 205-228.
Araki, K., Okamoto, H., Graveson, A. C., Nakayama, I., and Nagoya, H., 2001. Analysis of haploid development based on expression patterns of developmental genes in the medaka., 43 (5): 591-599.
Bi, K., and Bogart, J. P., 2010. Time and time again: Unisexual salamanders (genus) are the oldest unisexual vertebrates., 10 (1): 238-251.
Budanov, A. V., Sablina, A. A., Feinstein, E., Koonin, E. V., and Chumakov, P. M., 2004. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD., 304 (5670): 596-600.
Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W., and Vogelstein, B., 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage., 282 (5393): 1497-1501.
Chen, C. Y., Oliner, J. D., Zhan, Q., Fornace, A. J., Vogelstein, B., and Kastan, M. B., 1994. Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway., 91 (7): 2684-2688.
Chen, J., Wu, X., Lin, J., and Levine, A. J., 1996a. Mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein., 16 (5): 2445-2452.
Chen, Z., Heintz, N., and Hatten, M. E., 1996b. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers., 272 (5260): 417-419.
Chourrout, D., and Quillet, E., 1982. Induced gynogenesis in the rainbow trout: Sex and survival of progenies production of all-triploid populations., 63 (3): 201-205.
Cox, M. P., Peterson, D. A., and Biggs, P. J., 2010. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data., 11: 485.
Dahiya, E. S., Mehndiratta, M. M., and Pillai, K. K., 2017. Plas- ma tetranectin as a potential clinical biomarker for epilepsy and correlation with clinical and social characteristics.,4 (1): 2-5.
Edmondson, J. C., Liem, R. K., Kuster, J. E., and Hatten, M. E., 1988. Astrotactin: A novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar micro- cultures., 106 (2): 505-517.
Evans, M. J., von Hahn, T., Tscherne, D. M., Syder, A. J., Panis, M., Wölk, B., Hatziioannou, T., McKeating, J. A., Bieniasz, P. D., and Rice, C. M., 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry., 446 (7137): 801-805.
Fan, Z., You, F., Wang, L., Weng, S., Wu, Z., Hu, J., Zou, Y., Tan, X., and Zhang, P., 2014. Gonadal transcriptome analysis of male and female olive flounder ()., 2014 (2014): 1-10.
Fan, Z., Wu, Z., Wang, L., Zou, Y., Zhang, P., and You, F., 2016. Characterization of embryo transcriptome of gynogenetic olive flounder., 18 (5): 545-553.
Felip, A., Zanuy, S., Carrillo, M., and Piferrer, F., 2001. In- duction of triploidy and gynogenesis in teleost fish with emphasis on marine species., 111 (1-3): 175-195.
Fischer, P. M., Endicott, J., and Meijer, L., 2003. Cyclin-dependent kinase inhibitors., 5: 235-248.
Fishell, G. O. R. D., and Hatten, M. E., 1991. Astrotactin pro- vides a receptor system for CNS neuronal migration., 113 (3): 755-765.
Fopp-Bayat, D., 2010. Meiotic gynogenesis revealed not homogametic female sex determination system in Siberian sturgeon (Brandt)., 305 (1): 174-177.
Furuse, M., Fujita, K., Hiiragi, T., Fujimoto, K., and Tsukita, S., 1998. Claudin-1 and-2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin., 141 (7): 1539-1550.
Graves, T. D., and Hanna, M. G., 2005. Neurological channel- opathies., 81 (951): 20-32.
Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K., and Elledge, S. J., 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases., 75 (4): 805-816.
Harris, S. L., and Levine, A. J., 2005. The p53 pathway: Positive and negative feedback loops., 24 (17): 2899-2908.
Haupt, Y., Maya, R., Kazaz, A., and Oren, M., 1997. Mdm2 promotes the rapid degradation of p53., 387 (6630): 296-299.
Heuckeroth, R. O., Enomoto, H., Grider, J. R., Golden, J. P., Hanke, J. A., Jackman, A., Molliver, D. C., Bardgett, M. E., Snider, W. D., Johnson, E. M., and Milbrandt, J., 1999. Gene targeting reveals a critical role for neurturin in the deve- lopment and maintenance of enteric, sensory, and parasym- pathetic neurons., 22 (2): 253-263.
Horger, B. A., Nishimura, M. C., Armanini, M. P., Wang, L. C., Poulsen, K. T., Rosenblad, C., Kirik, D., Moffat, B., Simmons, L., Johnson, E., Milbrandt, J., Rosenthal, A., Bjorklund, A., Vandlen, R. A., Hynes, M. A., and Phillips, H. S., 1998. Neur- turin exerts potent actions on survival and function of mid- brain dopaminergic neurons., 18 (13): 4929-4937.
Huang, D. W., Sherman, B. T., and Lempicki, R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources., 4 (1): 44-57.
Hubbs, C. L., and Hubbs, L. C., 1932. Apparent parthenogenesis in nature, in a form of fish of hybrid origin., 76 (1983): 628-630.
Ihssen, P. E., McKay, L. R., McMillan, I., and Phillips, R. B., 1990. Ploidy manipulation and gynogenesis in fishes: Cyto- genetic and fisheries applications., 119 (4): 698-717.
Komen, H., and Thorgaard, G.H., 2007. Androgenesis, gyno- genesis and the production of clones in fishes: A review., 269 (1): 150-173.
Langfelder, P., and Horvath, S., 2008. WGCNA: An R package for weighted correlation network analysis., 9 (1): 559-571.
Latham, K. E., Akutsu, H., Patel, B., and Yanagimachi, R., 2002. Comparison of gene expression during preimplantation development between diploid and haploid mouse embryos., 67 (2): 386-392.
Leggatt, R. A., and Iwama, G. K., 2003. Occurrence of poly- ploidy in the fishes.,13 (3): 237-246.
Liu, H. J., Wang, C. A., Zhu, X. C., Liu, Y. X., Zhang, X. Y., Hou, J. L., and Tang, N., 2008. Embryonic development of gynogenetic diploid and triploid Japanese flounder., 23 (3): 161-167.
Li, Z. H., Liu, H. J., Zhang, S. K., Jiang, X. F., and Wang, Y. F., 2009. Comparison of embryonic development in gynogenetic and diploid barfin flounder ()., 28 (12): 752-756.
Luo, C., and Li, B., 2003. Diploid-dependent regulation of gene expression: A genetic cause of abnormal development in fish haploid embryos., 90 (5): 405-409.
Makino, S., and Ozima, Y., 1943. Formation of the diploid egg nucleus due to suppression of the second maturation division, induced by refrigeration of fertilized eggs of the carp,., 13 (1): 55-60.
Malumbres, M., and Barbacid, M., 2001. Milestones in cell division: To cycle or not to cycle: A critical decision in cancer., 1 (3): 222-231.
Masu, Y., Nakayama, K., Tamaki, H., Harada, Y., Kuno, M., and Nakanishi, S., 1987. cDNA eloping of bovine substance-K receptor through oocyte expression system., 329 (6142): 836-838.
Neaves, W. B., and Baumann, P., 2011. Unisexual reproduction among vertebrates., 27 (3): 81-88.
Nichols, K. M., 2009.Clonal lines and chromosome manipulation for aquaculture research and production. In:. Overturf, K., ed., Wiley-Blackwell, Oxford, 408pp.
Pala, I., Coelho, M. M., and Schartl, M., 2008. Dosage com- pensation by gene-copy silencing in a triploid hybrid fish., 18 (17): 1344-1348.
Paul, A. M., Branton, W. G., Walsh, J. G., Polyak, M. J., Lu, J.Q., Baker, G. B., and Power, C., 2014. GABA transport and neuroinflammation are coupled in multiple sclerosis: Regu- lation of the GABA transporter-2 by ganaxolone., 273: 24-38.
Pearlman, A. L., Faust, P. L., Hatten, M. E., and Brunstrom, J. E., 1998. New directions for neuronal migration., 8 (1): 45-54.
Pestov, N. B., Zhao, H., Basrur, V., and Modyanov, N. N., 2011. Isolation and characterization of BetaM protein encoded by ATP1B4–A unique member of the Na, K-ATPase β-subunit gene family., 412 (4): 543-548.
Purdom, C. E., 1969. Radiation-induced gynogenesis and an- drogenesis in fish., 24: 431-444.
Saunders, S., Paine-Saunders, S., and Lander, A. D., 1997. Expression of the cell surface proteoglycan glypican-5 is de- velopmentally regulated in kidney, limb, and brain., 190 (1): 78-93.
Schlessinger, A., Wittwer, M. B., Dahlin, A., Khuri, N., Bonomi, M., Fan, H., Giacomini, K. M., and Sali, A., 2012. High selectivity of the γ-aminobutyric acid transporter 2 (GAT-2, SLC6A13) revealed by structure-based approach., 287 (45): 37745-37756.
Schön, I., Martens, K., and van Dijk, P., 2009.. Springer,Dordrecht,615pp.
Scott, J. D., and Soderling, T. R., 1992. Serine/threonine protein kinases., 2 (3): 289-295.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T., 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks., 13 (11): 2498-2504.
Soengas, M. S., Alarcon, R. M., Yoshida, H., Hakem, R., Mak, T. W., and Lowe, S. W., 1999. Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition., 284 (5411): 156-159.
Stanley, J. G., 1983. Gene expression in haploid embryos of Atlantic salmon., 74 (1): 19-22.
Trapnell, C., Pachter, L., and Salzberg, S. L., 2009. TopHat: Discovering splice junctions with RNA-Seq., 25 (9): 1105-1111.
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., Pimentel, H., Salzberg, S. L., Rinn, J. L., and Pachter, L., 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks., 7 (3): 562-578.
Wang, W., Wang, J., You, F., Ma, L., Yang, X., Gao, J., He, Y., Qi, J., Yu, H., Wang, Z., Wang, X., Wu, Z., and Zhang, Q., 2014. Detection of alternative splice and gene duplication by RNA sequencing in Japanese flounder,., 4 (12): 2419-2424.
Wang, Z., Liu, W., Song, H., Wang, H., Liu, J., Zhao, H., Du, X., and Zhang, Q., 2015. Comparative evolution of duplicated ddx3 genes in teleosts: Insights from Japanese flounder,., 5 (8): 1765-1773.
Waters, M. F., Minassian, N. A., Stevanin, G., Figueroa, K. P., Bannister, J. P., Nolte, D., Mock, A. F., Evidente, V. G. H., Fee, D. B., Müller, U., Dürr, A., Brice, A., Papazian, D. M., and Pulst, S. M., 2006. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes., 38 (4): 447-451.
Xiong, Y., Hannon, G. J., Zhang, H., Casso, D., Kobayashi, R., and Beach, D., 1993. p21 is a universal inhibitor of cyclin kinases., 366: 701-704.
Yamamoto, E., 1999. Studies on sex-manipulation and produc- tion of cloned populations in hirame,(Temminck et Schlegel)., 173 (1): 235-246.
Yellen, G., 2002. The voltage-gated potassium channels and their relatives., 419 (6902): 35-42.
Zhang, B., and Horvath, S., 2005. A general framework for weighted gene co-expression network analysis., 4 (1): 1-45.
Zhang, W., Liu, Y., Yu, H., Du, X., Zhang, Q., Wang, X., and He, Y., 2016. Transcriptome analysis of the gonads of olive flounder ()., 42 (6): 1581-1594.
Zhang, X., Hou, J., Wang, G., Jiang, H., Wang, Y., Sun, Z., Jiang, X., Yu, Q., and Liu, H., 2015. Gonadal transcriptome analysis in sterile double haploid Japanese flounder., 10 (11): e0143204.
Zhong, Q., Zhang, Q., Wang, Z., Qi, J., Chen, Y., Li, S., Sun, Y., Li, C., and Lan, X., 2008. Expression profiling and validation of potential reference genes duringembryogenesis., 10 (3): 310-318.
Zhou, Y., Holmseth, S., Guo, C., Hassel, B., Höfner, G., Huit- feldt, H. S., Wanner, K. T., and Danbolt, N. C., 2012. Deletion of the γ-aminobutyric acid transporter 2 (GAT2 and SLC6A13) gene in mice leads to changes in liver and brain taurine contents., 287 (42): 35733-35746.
September 13, 2017;
December 1, 2017;
September 13, 2018
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2018
. Tel: 0086-532-82031806 E-mail: qzhang@ouc.edu.cn
(Edited by Qiu Yantao)
杂志排行
Journal of Ocean University of China的其它文章
- Comparative Evaluation of Toleration to Heating and Hypoxia of Three Kinds of Salmonids
- Morphological Change in the Northern Red River Delta, Vietnam
- Internal Facies Architecture and Evolution History of Changxing Mouth-Bar Complex in the Changjiang (Yangtze) Delta, China
- The Holocene Environmental Evolution of the Inner Hangzhou Bay and Its Significance
- Provenance of Sediments Filling a Paleo-Channel that Formed on the Western Yellow Sea Continental Shelf During the Last Glacial Period
- Distribution and Characteristics of Hazardous Geological Features in the Marine Coastal and Offshore Areas of Zhejiang Province, East China Sea
