GCRV 096 VP6 protein and its impacts on GCRV replication with different genotypes in CIK cells
2018-10-17XiuyingYanLingfangXiongJieLiYaWangZaoheWuJichangJianYuDing
Xiuying Yan,Lingfang Xiong,Jie Li,Ya Wang,Zaohe Wu,Jichang Jian,Yu Ding
Guangdong Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals,Guangdong Ocean University,Zhanjiang,Guangdong,524088,China
Keywords:Grass carp reovirus(GCRV)Protein VP6 Replication Functional analysis
ABSTRACT Grass carp reovirus(GCRV)is the causative agent of grass carp hemorrhagic disease,which has detrimental effects on the grass carp aquaculture industry.There are four known genotypes of GCRV and strains of different GCRV genotypes differ greatly.In this study,the diversity of the protein VP6 from different GCRV stains and the effect of genotype GCRV 096 on replication was investigated in CIK cells.Our results showed that the VP6 protein of GCRV 096(genotype І)exhibited limited homology to that of GCRV GD108(genotypeШ),with few residues conserved in predicted protein-protein interaction domains.GCRV 096 VP6 protein was expressed and puri fied and an antiserum against it was characterized.Addition of puri fied VP6 protein or antiserum to culture media of CIK cells inhibited the replication of GCRV 096 in these cells.In contrast,replication of GCRV GD108(genotypeШ)was not affected in CIK cells under the same condition.Overall,our results indicated that the protein VP6 and VP6 antiserum did not provide cross-protection against GCRV strains and this can be attributed to differences among GCRV genotypes.It will be important to consider multiple GCRV genotypes in the development of effective GCRV vaccines and other therapies against grass carp hemorrhagic disease.In addition,bioinformatics analysis also suggested that the protein VP6 may have a role in the process of GCRV infection.This study lays the foundation for the prevention of grass carp hemorrhagic disease and further detailed studies on the pathogenesis of GCRV.
1.Introduction
Grass carp reovirus(GCRV)is the causative agent of grass carp hemorrhagic disease,which leads to serious economic losses in the grass carp aquaculture industry(Altenburq&Graham,1980;Subramanian,Hetrick,&Samal,1997).GCRV is a non-enveloped,double-stranded RNA virus with a double-layered icosahedral capsid.To date,more than twenty GCRV strains and four GCRV genotypes have been reported.The representative strains of GCRV genotypeⅠis GCRV 873,which infects C.idellus kidney(CIK)cells and induces obvious cytopathic effects(CPE).The representative strain of GCRV genotypeⅡis AGCRV,which was found in America.The representative strains of GCRV genotypeⅢis GCRV HZ08,which does not induce obvious CPE in infected CIK cells.The representative and onlystrain of GCRV genotypeⅣis GCRV 104(He et al.,2017;Wang et al.,2012;Yan,Wang,Xiong,Jian,&Wu,2014;Zong,Zhang,&Lv,2016).The identity of the nucleotide sequences among the different genotypes isolates is lower than 46%,but among the same genotype isolates is higher than 90%(Huang et al.,2016;Yan et al.,2014).GCRV genotype Ⅰ,Ⅲ and Ⅳ have been reported in China,and GCRV genotype І and GCRV genotype Ш are commonly found in China(Huang et al.,2016;Rao&Su,2015;Wang et al.,2012;Yan et al.,2014;Ye,Tian,Deng,Chi,&Jiang,2012).GCRV genotypeⅠis associated with low virulence and a long latent period before the induction of a mild host immune response,while GCRV genotypeⅡis associated with high virulence,a short latentcyand stimulates a strong and extensive host immune response(He et al.,2017).Substantial progress has been achieved through studies of GCRV,grass carp hemorrhagic disease,and antiviral immunity of grass carp(Fan et al.,2013;Pei,Ke,Chen,&Zhang,2014;Yan et al.,2015);however,the pathogenic mechanisms underlying GCRV infection and the functions of GCRV proteins have yet to be fully elucidated.The prevention and treatment of grass carp hemorrhagic disease also requires a further detailed investigation of different GCRV genotypes.
The protein VP6 is a component of both the GCRV core and its virion,and forms a clamp between the inner and outer capsids.VP6 is a homolog ofσ2(sigma-2)of mammalian orthoreovirus(MRV).The structural proteins(VP1-VP7)are conserved among all GCRV strains,to some extent(Cheng,Fang,Shah,Atanasov,&Zhou,2008).VP6 has been deemed an antigen with strong immunogenicity and the vp6 gene is a candidate for production of GCRV vaccines against some GCRV strains(Xiao et al.,2014;Zeng et al.,2016);however,there has been very little research into vaccine for all three GCRV genotypes.Investigation of cross immunity of GCRV strains and the function of the VP6 protein will assist in the development of GCRV vaccines,and in the prevention and treatment of grass carp hemorrhagic disease.
Grass carp(Ctenopharyngodon idella)kidney(CIK)cells are suitable for antiviral studies(Peng,Yang,&Su,2012),since they exhibit anti-GCRV immune responses when challenged with this virus(Chen et al.,2013;Li,Zhang,Sun,Wang,&Wang,2009;Peng et al.,2012;Rao et al.,2015).Transcription of the GCRV genome is divided into early and late phases.Early transcription begins 4h after CIK cells are infected with GCRV,and involves the transcription of viral dsRNA template by RNA transcriptase.Late transcription occurs 10h after CIK cells are infected with GCRV and involves generation of transcripts of late genes from progeny virus templates(Zhang&Ke,1993).Direct antiviral activity may suppress the proliferation of GCRV in CIK cells via inhibition of the early stage of the virus replication cycle(Li et al.,2009;Rao et al.,2015).
In this study,GCRV 096 and GCRV GD108 were included as representatives of GCRV genotypes І and Ш,respectively.GCRV 096 can induce signi ficant CPE in CIK cells,while GCRV GD108 can replicate in CIK cells without obvious CPE(Yan et al.,2014;Ye et al.,2012).The identity of GCRV 096 with GCRV 873(genotypeⅠ)based onthevp4 gene,thevp6 geneandthevp7 geneisrespectively 99.3%,99.4%and 92%.The identity of GCRV GD108 with GCRV HZ08(genotypeШ)based on the vp4 gene,the vp6 gene and the vp7 gene is respectively 96.3%,98.6%and 96.5%(Yan et al.,2014).The aim of the present study was to investigate the impact of the protein VP6 in GCRV 096 on GCRV replication in CIK cells by real-time quantitative PCR(qRT-PCR)and western blotting,and to analyze the function of the protein VP6 using bioinformatics.This study provides a foundation for the development of GCRV vaccines and elucidation of the pathogenic mechanisms underlying GCRV infection.
2.Materials and methods
2.1.Viruses,cells,vectors,bacteria,and animals
GCRV 096 and GCRV GD108 were isolated respectively,from diseased grass carp in Xiaogan,Hubei Province and Zhongshan,Guangdong Province by ultracentrifugation(Fang,Ke,&Cai,1989),and stored in our laboratory.The widely used GCRV sensitive cell line,CIK(Ma et al.,2011;Ye et al.,2012;Zhang et al.,2010),was purchased from ShenzhenInspectionandQuarantineBureau,China.
The vectors pMD18-T and pET-28a(+)were purchased from Takara Co.,Ltd.E.coli DH5αwas purchased from Takara(Dalian,China)and maintained in our laboratory.Adult New Zealand white rabbits(approximately 2.5kg)were purchased from the Experimental Animal Center of Guangdong Medical College(experimental animal quality certi fication:0001906)for immunization experiments.The rabbits were fed a standard laboratory chow in the animal facility of our laboratory.
2.2.Virus culture and puri fication
Cell culture,viral infection,and propagation were performed as previously described(Fang et al.,1989).GCRV 096 particles were extracted by differential centrifugation at 250-6000 x g,followed by ultracentrifugation of the supernatant at 35000 x g,4°C for 2.5h.The puri fied virus pellet was resuspended in phosphatebuffered saline(PBS,pH 7.4)and then stored at-70°C until required.
2.3.RT-PCR and sequence analysis
GCRV 096 genomic RNA was extracted from puri fied GCRV 096 virions using a viral RNA extraction kit(Takara,Dalian,China).cDNA was prepared from GCRV 096 genomic RNA using an RT-PCR kit(Takara,Dalian,China)with random primers and M-MLV reverse transcriptase.
Primers for ampli fication of the GCRV 096 vp6 gene were designed using homologous sequences in GenBank,as follows:5′-TGTGATGGCACAGCGTCAG-3′and 5′-GTTAGACGAACATCGCCTG C-3′.PCR cycling conditions consisted of an initial denaturation at 95°C for 3min,followed by30 cycles of 94°C for 30 s,59°C for 60 s,and 72°C for 70s,and a final extension step of 30 min at 72°C.The PCR reaction mix(50μl)included 33μl sterile water,3μl dNTPs(2.5 mmol/l each),2μl each primer(10pmol/l),5μl 10×buffer(containing Mg2+),100ng cDNA,and 0.25μl Ex Taq polymerase(5 U/μl,Takara,Dalian,China).The amplicon was puri fied from an agarose gelusing a Gel Extraction Kit(Takara,Dalian,China),ligated into the pMD18-T vector at 16°C with T4 DNA ligase,and then used to transform E.coli DH5α.The resulting recombinant plasmid was veri fied by sequencing in Sangon Biotech(Shanghai,China).
The open reading frame(ORF)of the vp6 gene was predicted using DNAstar software.The identity and similarity of the GCRV 096 vp6 gene with known vp6 genes,and their corresponding amino acid sequences was determined using NCBI BLAST and DNAstar software.Multiple alignments of the VP6 protein sequences were generated using DNAstar.
2.4.Expression of the protein VP6 in vitro
The primer pair,5′-CGCGGATCC ATGGCACAGCGTCAGTT-3'(underlined bases indicate a BamHI restriction site)and 5′-ACGCGTCGACTTAGACGAACATCACC-3'(underlined bases indicate a SaII restriction site),was designed to amplify the vp6 gene ORF.The PCR reaction mix and cycling conditions were the same as described above(RT-PCR),except that the annealing temperature was 56°C.
The ampli fied fragment was puri fied using a DNA gel extraction kit(Takara,Dalian,China).The puri fied PCR product was digested using the restriction enzymes BamH I and SaI I,and then ligated into the vector,pET-28a(+),using T4 DNA ligase(Takara,Dalian,China).The resulting recombinant expression plasmid(designated pET28a-vp6)was transformed into E.coli DH5αcompetent cells treated with CaCl2.Finally,pET28a-vp6 was transformed into BL21(DE3)competent E.coli for protein expression.All plasmids were checked by restriction enzyme digestion and PCR ampli fication and were also sequenced by Sangon Biotech(China)to con firm the presence of the correct insert.
Colonies of transformed BL21(DE3)cells positive for the pET28a-vp6,were cultured overnight in Luria-Bertani media supplementedwith50 mg/lkanamycin at37°Cwith shaking(200rpm).Then,bacteria were inoculated into fresh medium,grown to optical densities of 0.4-0.6 at 600nm,and induced with isopropyl-thiogalactoside(IPTG)(Takara,Dalian,China),at a final concentration of 1 mM,at 37°C for 1-5 h.Bacteria were then induced with 0.2-1.0 mM IPTG at 37°C for 4h,to determine the optimal concentration for induction of VP6 protein expression.In addition,to determine the optimum temperature for induction,bacteria were induced using 0.2 mM IPTG at 20°C,28°C,and 37°C for 4 h.Bacteria in each treatment group were collected and lysed,and both the supernatant and pellet of the cell lysates denatured by boiling for 5 min in SDS-PAGE loading buffer(Takara,Dalian,China).Samples were then loaded onto 12%SDS-PAGE gels for electrophoresis.
2.5.Puri fication of recombinant protein and western blotting
To purify the recombinant protein,bacterial cultures were harvested by centrifugation at 10,000 x g for 20 minat 4°C.Pellets were resuspended in a 1/10 volume of PBS(pH 7.4)and sonicated on ice for 30 min.The lysate was centrifuged at 12000 x g for 30 min at 4°C.The supernatant was discarded and the pellets resuspended in washing buffer(0.5%(v/v)Triton X-100,300mM NaCl,10mM EDTA,10 mM DTT,and 50 mM Tris-HCl,pH 8.0)and clari fied using 0.22-μm filter disks.
Since the recombinant VP6 protein carried an N-terminal Histag,it could be puri fied using HisTrap HP column charged with Ni2+ions(GE Healthcare,Sweden).Before eluting the target protein,non-speci fic proteins were washed from the column using binding buffer(20mM imidazole,500 mM NaCl,20 mM NaH2PO4).Target proteins were eluted using elution buffer(500 mM NaCl,20 mM NaH2PO4,and a linear gradient of 50-300mM imidazole).
Puri fied proteins were run on 12%SDS-PAGE and then transferred to nitrocellulose membranes.Blots were blocked in 5%nonfat milk,0.1%Tween 20 in PBS at room temperature for 1 h,and then incubated overnight at 4°C with mouse monoclonal anti-His antibody(1:1000;Invitrogen),followed by incubation for 2 h with horseradish peroxidase conjugated goat anti-mouse antibody(Takara,Dalian,China)(1:3000).Finally,the relevant substrate(Takara,Dalian,China)was added to visualize the proteins and the blot photographed.
2.6.Preparation of antiserum against the GCRV 096 VP6 protein
Normal serum samples were collected before immunization of rabbits and served as a negative control.Puri fied VP6 protein(500μg)and an equal amount of Freund's complete adjuvant(Sigma)were mixed until completelyemulsi fied and the emulsi fied mixture was administered to immunize rabbits using multi-point injections in the back.Two weeks after the first immunization,a booster immunization was performed using an emulsi fied mixture consistingof the same amounts of the puri fied VP6 protein(500μg)and of Freund's incomplete adjuvant(Sigma).Additional booster immunizations were performed every two weeks,until a total of four boosters had been administered.Eight days after the last booster immunization,rabbits were euthanized and blood samples collected.Antiserum waspuri fied using the octanoicacidammonium sulfate method(Grodzki&Berenstein,2010),then separated and stored at-20°C until used for ELISA and western blots,to determine its titer and speci ficity of antigen-binding.
2.7.Replication of GCRVs in CIK cells
Three types of experimental media(containing 2%FBS)were prepared:medium A,B,and C.Medium A was a mixture of 100μg puri fied VP6 proteinwith 5ml of M199,and media B and C were1:2 and 2:1(v:v)mixtures of antiserum with M199,respectively.Controls were incubated in M199(containing 2%FBS)alone.Experimental medium was sterile filtered and stored at 4°C.
Trypsinized CIK cells in the logarithmic phase of growth were divided equally between each well of six-well cell culture plates(2 ml per well)and then incubated at 25°C.After cells adhered to the wells,the original M199 medium(containing 2%FBS)was substituted for experimental medium(see above)or fresh M199(controls).After incubation for 12,24,and 48h,the experimental medium was removed and sterile GCRV 096 virus or GCRV GD108 sterile virus suspension(approximately 2130μg/ml)was added at 50μl per well.Virus adsorptionwas promoted byincubation for 1h,followed by the removal of the virus suspension,and addition of M199 medium(containing 2%FBS).Post-infection(4,8,and 13h)(Zou&Fang,2000),cells were collected.GCRV 096 or GCRV GD108 viral RNA was extracted from equal volume aliquots of cells from each treatment group and reverse transcription performed.Finally,levels of the viral vp6 gene in CIK cells were detected using qRTPCR.
2.8.Real-time quantitative PCR ampli fication,statistics and analysis,and western blotting
Primers were designed for qRT-PCR ampli fication based on GCRV 096 and GCRV GD108 vp6 gene sequences,as follows:096F(5′-GATCAGGGACCGATTTATCA-3′);096R (5′-AGGCTGTCTGTGCTA AGTG-3′);108F(5′-ACGGGTGGTAATACGCTTT-3′);and 108R(5′-TT ACATTCAACTTG CTTAT-3′).Primers based on the internal reference genes β-actin and GAPDH were: βF(5′-GGCTGT GCTGTCC CTGTA-3′); βR (5′-GGGCATAACCCTCGTAGAT-3′);GF(5′-GTGA ATCGGATACTC TCGG-3′);and GR (5′-GCAGACATAACGCAAG GCA-3′).Primers were synthesized by Sangon Biotech(China).
The components of the qRT-PCR reactions(25μl)were 9.5μl sterile water,0.5μl of each primer(10 pmol/l),12.5μl SYBR®Premix Ex Taq™,and 2μl cDNA.The PCR cycling conditions were an initial denaturation at 95°C for 3 min,followed by 40 cycles of 95°C for 10 s and 55°C for 30 s.qRT-PCR experiments were performed in triplicate.
Data generated by qRT-PCR were analyzed using the 2-△△CTmethod.Experimental data were analyzed using SPSS software and one-way Analysis of variance.P<0.05 was considered to indicate a statistically signi ficant difference.
The infected CIK cells and media were centrifuged at 35000 x g,4°C for 2.5h.Then,proteins were extracted from the precipitation for western blotting(using the same protocol indicated in “2.5.”above).Antibodies were mouse monoclonal anti-GAPDH antibody(1:1000;Invitrogen),mouse monoclonal anti-VP6 antibody(prepared in Vipotion,Guangzhou,China),and goat anti-mouse antibody(Takara,Dalian,China)(1:3000).
2.9.Bioinformatics analysis of the protein VP6 function
Interactions of the VP6 protein were predicted using bioinformatics.Peptide-binding domains,glycosylation sites,interaction sites,and the protein-protein interactions of GCRV 096 VP6 were predicted using DomPrep(http://lilab.uwo.ca/DomPep.html),NeOGlyc(http://www.cbs.dtu.dk/servi ces/NetOGlyc/),SPPIDER(http://sppider.cchmc.org),InterPro (www.ebi.ac.uk/interpro),Phobius(http://phobius.sbc.su.se/),and DIP(http://dip.doe-mbi.ucla.edu).
3.Results
3.1.Cloning of the GCRV 096 vp6 gene and sequence analysis
Viral RNA was extracted from puri fied GCRV 096 virus particles,reverse transcribed,and the vp6 gene ampli fied by RT-PCR(Fig.1A).The vp6 gene was ligated to the pMD18-T vector to construct the recombinant plasmid,pMD18-vp6.PCR replication and sequencing were used to con firm that the recombinant plasmid pMD18-vp6 was correct.The cloned vp6 gene(GenBank accession number,HQ452490)contained a 1239 bp ORF,encoding 412 amino acids.The predicted molecular weight and isoelectric point of the VP6 protein were 44.6 kDa and 8.5,respectively.
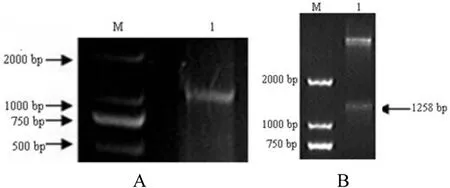
Fig.1.Ampli fication of the vp6 gene in GCRV 096(A)and double restriction enzyme digestion with BamH I and Sal I of the pET28a-vp6 plasmid(B).A.M:DL2000;1:PCR products of the vp6 gene.B.M:DL 2000;1:Double restriction enzyme digestion of the pET28a-vp6 plasmid.
Comparative analysis indicated amino acid sequence homology levels of the GCRV 096 VP6 protein(Wang et al.,2012;Yan et al.,2014)with that of GCRV of genotype Ⅰ,Ⅱ,Ⅲ,and Ⅳ were approximately 99%,58.7%,19.5%,and 19.2%.The similarity between the GCRV 096 VP6 amino acid sequence and that of GCRV GD108 was 19.4%.There were 99 conserved amino acid residues between GCRV 096 and GCRV GD108 VP6 proteins.BLAST analysis showed that GCRV 096 VP6 shared 18%amino acid sequence similarity to the MRVσ2(sigma-2)protein,which was considered to be a homolog.
3.2.Expression and puri fication of recombinant GCRV 096 VP6 protein
To construct the prokaryotic recombinant expression plasmid pET28a-vp6,the vp6 gene was ligated to the pET28a vector.The vp6 insert sequence in pET28a-vp6 was con firmed by PCR,double restriction enzyme digestion(Fig.1B)and sequencing.
The recombinant expression plasmid,pET28a-vp6,was transformed into E.coli BL21(DE3)and its expression induced.Initial experiments demonstrated that induction for 6 h with 0.4mM IPTG at 37°C was optimal for VP6 expression.SDS-PAGE demonstrated that a protein with a molecular weight consistent with the theoretical value was expressed(Fig.2A;the fusion tag is 2.77 kDa).The expressed recombinant VP6 protein was puri fied by af finity chromatography,eluted with different concentrations of imidazole,and all eluated fractions collected and subjected to electrophoresis.The results indicated that elution with 200 mM imidazole was optimal(Fig.2B).Western blotting using a His-Tag monoclonal antibody demonstrated that the GCRV 096 VP6 protein was successfully expressed and puri fied(Fig.2C).
3.3.Preparation of antiserum against the GCRV 096 VP6 protein
Three rabbits were immunized with the puri fied recombinant GCRV 096 VP6 protein.Antiserum(polyclonal antibody)was prepared after the fourth booster immunization.ELISAs were performed using plates coated with the expressed VP6 protein to test the reactivity of the antiserum.The results demonstrated that the titer of antiserum was 1:32000.Moreover,western blot analysis showed that the antiserum could speci fically recognize the GCRV 096 VP6 protein.
3.4.Replication of GCRVs in CIK cells
The levels of the vp6 geneandthe VP6 proteininCIK cell samples incubated with VP6 protein or the antiserum were analyzed by qRT-PCR and western blotting.The results demonstrated that levels of the vp6 gene and the VP6 protein altered over time(Fig.3)and were lower in experimental samples treated with VP6 protein or antiserum than in control samples.The level detected of the vp6 gene and the VP6 protein was lowest when CIK cells were incubated with the VP6 protein or antiserum for 48h;therefore,in subsequent experiments CIK cells were incubated with VP6 protein or antiserum for 48 h.
The levels of the vp6 gene and the VP6 protein respectively were analyzed by qRT-PCR and westernblotting in experimental samples treated with VP6 protein or antiserum.In samples infected with GCRV 096,levels of the vp6 gene and the VP6 protein changed over time post infection(Fig.4)and replication of GCRV 096 decreased signi ficantly 8 or 13 h post infection;however,in samples incubated with the VP6 protein or antiserum and then challenged with GCRV GD108,there was no signi ficant change in the level of the GCRV GD108 vp6 gene.
3.5.Prediction of GCRV 096 VP6 protein function
Bioinformatics analysis showed that the VP6 protein contains a PDZ domain(AVALAQVMFV,residues 403-412)and thirteen SH2 domains.Aglycosylationsitewasidenti fiedataminoacid400.The3D structure of the VP6 protein in the PDB database includes a D chain and an E chain;117 interaction sites were predicted in the D chain,comparedwith114suchsitesintheEchain.Moreover,aminoacid24 in the D chain and 21 in the E chain were predicted to be involved in protein-protein interactions and were conserved in both the GCRV 096 and GCRV GD108 VP6 proteins.Bioinformtics analysis indicated that GCRV 096 VP6 is non-cytoplasmic and exhibits homology with sigma1/sigma2 reoviral family proteins.The VP6 protein was predicted to potentially interact with ubiquitin carboxyl-terminal hydrolase 33,Rho GTPase-activating protein 4,slit homolog 2 protein,and SLIT-ROBO Rho GTPase-activating protein 2.
4.Discussion
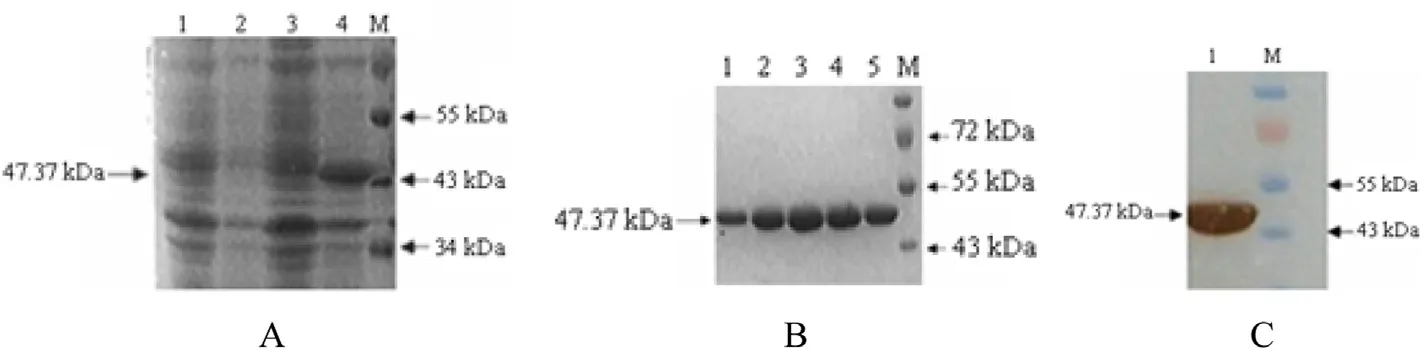
Fig.2.Expression of the recombinant protein VP6 in E.coli BL21(A),puri fication(B)and western blotting of the expressed recombinant protein VP6(C).A.M:Standard protein marker;1:pET-28a(extract of transformed pET-28a BL21 without IPTG induction);2:pET-28a(extract of transformed pET-28a BL21 with IPTG induction);3:pET28a-vp6(extract of transformed pET-28a BL21 without IPTG induction);4:pET28a-vp6(extract of transformed pET-28a BL21 with IPTG induction).B.M:Standard protein marker;1-5:The Puri fied recombinant protein VP6 with 90,150,200,250,300 mmol/l imidazole in the elution buffer,respectively(as arrows indicated).C.M:Standard protein marker;1:The expressed recombinant proteinVP6.

Fig.3.The levels of the vp6 gene(A)and the VP6 protein(B)in CIK cells with the challenge of GCRV 096.1:Control group;2:Medium B;3:Medium C;4:Medium A.Notes:h is the incubation time of CIK cells with the proteinVP6 or antiserum.a stands for signi ficant difference.Medium A:mixture of the puri fied protein VP6 100μg with 5 ml of M199;Medium B:mixture of antiserum with M199 in volume ratio 1:2;Medium C:mixture of antiserum with M199 in volume ratio 2:1.
New GCRV strains are frequently reported and there are major differences among those belonging to different GCRV genotypes(Rao&Su,2015).For example,homologyrates among the VP6 proteins in GCRVs of different genotypes range from 11.1%to 45.4%(Huang et al.,2016).There is no obvious pattern in the geographical distribution of GCRV genotypes(Yan et al.,2014).The diversity of GCRVs makes it dif ficult to prevent and treat grass carp hemorrhagic disease.Veri fication of GCRV protein function and the pathogenic mechanisms underlying infection with this virus will lay the foundation for the prevention and treatment of grass carp hemorrhagic disease.
In the current study,we raised highly effective antiserum against theGCRV 096 VP6 protein,indicating that theVP6 proteinis anideal immunogen for targeting of GCRV 096.However,we identi fied numerous differences between the VP6 proteins in GCRV 096 and GCRV GD108,with only 19.4%homology and very few conserved sites.In the 3D structure of the VP6 protein,conserved interaction sites were identi fied at amino acids 24 and 21 of the D and E chains,respectively.Questions remain regarding other differences between GCRV 096 and GCRV GD108 and about which differences are most closely related to the observed variation in virulence of GCRVs.Determination of such differences and investigation of the ability of GCRVs to induce cross immunity in grass carp will be of particular importance for the development of GCRV vaccines.
In the current study,replication of GCRVs was detected 4,8,and 13 h post infection in CIK cells(Fig.4).In experimental samples,in which GCRV 096 VP6 protein or antiserum was added to the cell culture media,the replication of GCRV 096 decreased relative to that in untreated controls when CIK cells were challenged with GCRV 096(Fig.4).There are two possible explanations for these results.First,the expressed VP6 protein may bind to a receptor or protein in the CIK cell membrane.When challengedwith GCRV 096,binding of the expressed VP6 protein to a receptor or protein on the CIK cell membrane may prevent entry of GCRV into cells,leading to a decrease in GCRV 096 replication.Second,after addition of the antiserum to the cell medium,the combination of antibody with GCRV 096 may affect its entry into cells and decrease its replication.
In the GCRV GD108 experimental samples(Fig.4),addition of GCRV 096 VP6 recombinant protein or antiserum against GCRV 096 VP6 to the culture medium did not affect its replication after challenge with GCRV GD108.We speculate that there may be two possible reasons for this result.First,it is possible that binding of the GCRV 096 VP6 protein with its putative receptor is unable to prevent cell entry of GCRV GD108;therefore,there was no effect on GCRV GD108.Second,antiserum against the GCRV 096 VP6 protein might not be able to generate resistance to challenge with GCRV GD108.This indicates that the anti-serum was not able to induce cross-protection between GCRV 096 and GCRV GD108.It has previously been reported that there is no serological cross-reaction or cross-protection between GCRV JX01 and JX02(Wang,Li,&Lv,2013).GCRV JX01 and GCRV 096 belong to GCRV genotype І,while GCRV JX02 and GD108 are genotypeШ.We deduce based on our results and previous results that it might not be possible to induce cross-protection among GCRVs of different genotypes.

Fig.4.The levels of the vp6 gene(A)and the VP6 protein(B)in CIK cells after challenge with GCRVs.1:Control group(GCRV 096);2:Medium C(GCRV 096);3:Medium A(GCRV 096);4:Control group(GCRV GD108);5:Medium C(GCRV GD108);6:Medium A(GCRV GD108).Notes:h is the time after CIK cells infected GCRV 096 or GCEV GD108.a stands for signi ficant difference.Medium A:mixture of the puri fied protein VP6 100μg with 5 ml of M199;Medium C:mixture of antiserum with M199 in volume ratio 2:1.
To date,evaluation of GCRV vaccines has been limited to few GCRV strains(Tian,Ye,Zhang,Deng,&Bai,2013;Wang et al.,2015;Xue et al.,2013;Zeng et al.,2016;Zhu et al.,2014).Studies of the differences between GCRVs genotypes are infrequent(Huang et al.,2016;Wang et al.,2013),and even fewer studies of cross-protection of GCRV vaccines among GCRVs of different genotypes have been reported.Determining whether cross-protection occurs among GCRV strains of different genotypes will be very important for the development of effective GCRV vaccines.A multiple shRNA expression system could be used to generate a cross-reactive antiviral agent for the treatment of hemorrhagic disease of grass carp caused by multiple GCRV genotypes(Ma et al.,2014).Effective therapies against multiple GCRV genotypes should be developed.In the future,recombinant vaccines will have an important role in the prevention of grass carp hemorrhagic disease.
The proteins predicted to interact with VP6 by bioinformatics analysis have roles in cell migration and regulation of Rho-like GTPases in hematopoietic cells.We speculate that the VP6 protein may participate in cell entry,replication,or pathogenic mechanisms of GCRV.What's more,previous studies have shown that some immune-related genes in grass carp and CIK cells are involved in signaling pathways with GCRV challenge(Chen et al.,2013;Heng,Su,Huang,Dong,&Chen,2011;Huang et al.,2012;Huang et al.,2016;Jiang et al.,2015;Lv et al.,2012;Pei et al.,2015;Su,Yuan,&Su,2016;Su,Yang,Xiong,Wang,&Zhu,2009;Su et al.,2012).Functional exploration of GCRV proteins and the interactions of GCRV proteins with host proteins will make an important contribution to understanding the pathogenic mechanism of GCRV.
5.Conclusions
We found that the vp6 gene was conserved among GCRV strains of the same genotype,while homology levels among vp6 genes of GCRV strains of different genotypes were very low.No crossprotection was observed among GCRVs of different genotypes.Multiple GCRV genotypes should be fully considered in the development of GCRV vaccines and effective therapies for grass carp hemorrhagic disease.Bioinformatics analysis results suggested that the protein VP6 probably plays a role during GCRV infection.This study lays the foundation for the prevention of grass carp hemorrhagic disease and for in-depth studies of the pathogenic mechanisms of GCRV.
Conflicts of interest
The author declare that they have no conflict of interest.
Ethical approval
All the applicable international and national guidelines for the care and use of animals were followed.
Acknowledgements
This study was supported by National Natural Science Foundation of China(No.31602199),the Natural Science Foundation of Guangdong Province in China (No.2015A030313622),the Foundation of Guangdong Ocean University(K15246),the Foundation of Zhanjiang(No.2015A03027),the National 973 Plan Project in China(No.2009CB118704),and the project of DepartmentofEducation ofGuangdongprovincein China(No.2012LYM_0075).
杂志排行
Aquaculture and Fisheries的其它文章
- Polymorphisms in the FOXO gene are associated with growth traits in the Sanmen breeding population of the razor clam Sinonovacula constricta
- Hemocytes transcriptome pro file of the Chinese mitten crab(Eriocheir sinensis)
- Cloning and expression analysis of the nuclear factor erythroid 2-related factor 2(Nrf2)gene of grass carp(Ctenopharyngodon idellus)and the dietary effect of Eucommia ulmoides on gene expression
- Positive impacts of Nile tilapia and predatory sahar on carp polyculture production and pro fits
- Assessment of post-harvest fish losses Croaker Pseudotolithus elongatus,(Bowdich,1825),Cat fish Arius heudeloti,(Valenciennes,1840)and Shrimp Nematopalaemon hastatus(Aurivillius,1898)in Ondo State,Nigeria
