Identification of commonly regulated protein targets and molecular pathways in PC-3 and DU145 androgen-independent human prostate cancer cells treated with the curcumin analogue 1,5-bis(2-hydroxyphenyl)-1,4-pentadiene-3-one
2018-10-12KaminiCitalingamFaridahAbasNordinLajisIekhsanOthmanRakeshNaidu
Kamini Citalingam, Faridah Abas, Nordin H. Lajis, Iekhsan Othman*, Rakesh Naidu*✉
1Jeffery Cheah School of Medicine and Health Sciences, Monash University Malaysia, Jalan Lagoon Selatan, 47500 Bandar Sunway, Selangor,Malaysia
2Laboratory of Natural Products, Faculty of Science, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia
3Department of Food Science, Faculty of Food Science and Technology, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia
Keywords:Androgen-independent prostate cancer Diarylpentanoid Proteomics profiling Mass spectrometry Pathway analysis
ABSTRACT Objective: To identify mutually regulated proteins in PC-3 and DU145 androgen-independent prostate cancer cell lines treated with 1,5-bis(2-hydroxyphenyl)-1,4-pentadiene-3-one(MS17), and to study the molecular pathways that contributed to the anticancer activity of MS17. Methods: PC-3 and DU145 cells were treated with 3 × EC50 (15 μM) concentration of MS17 for 24 h and were subjected to protein expression profiling using two-dimensional gel electrophoresis and protein identification by mass spectrometry. Selected differentially expressed proteins with significant P-value of P<0.05 and fold change over 1.5-folds were filtered through and ontologically classified. Mutually regulated proteins were ranked by fold change and identified as common protein targets of MS17. Results: Profiling data revealed that, the mutually down-regulated proteins included ACTB and ACTG associated with structural molecule activity, ACTN1 with cell cycle, ACTN4 with cell migration, HNRPK with apoptosis, PLST with morphogenesis and TERA with proteolysis. However, the expressions of CH60 and HS71A respectively associated with response to unfolded protein demonstrated opposing regulation in PC-3 and DU145 cells. Pathway analysis of the differentially expressed proteins in PC-3 cells demonstrated the modulation of top pathways associated with cell-cell adhesion and cytoskeletal organization while in DU145 cells the pathways were associated with proteosomal degradation, regulation of electrolytes and water, regulation control of germ cells and organization of filament assembly/disassembly. Conclusions: The findings of the present study provide an understanding on the anti-tumorigenic activity of MS17 at the proteome level and warrant further research for its potential application for the management and treatment of androgen-independent prostate cancer.
1. Introduction
The study of proteomics using 2-dimensional gel electrophoresis(2-DE) and mass spectrometry for protein identification has become a powerful tool for the comprehensive analysis of specific protein target along with their interacting partners and networks in order to understand cellular processes. As proteins are the key players that have regulatory roles to influence cell growth in tumorigenesis,proteins may regulate cellular response towards drug treatment.Proteomics study is therefore required to understand the potential regulatory mechanism of a candidate anticancer agent in elucidating its involvement in cancer progression and in the hope of identifying target proteins in response to treatment for therapeutic management of prostate cancer.
Curcumin is the most studied phytochemical extracted from the plant Curcuma longa. Curcumin significantly inhibits prostate cancer cell growth and prevents the progression of the tumour to its hormone-refractory stage by interfering with the signalling pathways in androgen-dependent and -independent prostate cancer cells[1,2]. While phase Ⅰ clinical trials have already proven the safety profile of curcumin even at high doses, the clinical application of this compound remains limited due to its poor pharmacokinetic properties. Analogues of curcumin such as diarylpentanoids, a group of curcumin-like analogues with 5-carbon chain between its aryl rings that demonstrated to be effective at lower concentration and improved growth suppressive activity have been developed in recent years[3]. Diarylpentanoid EF24 [3,5-bis(2-fluorobenzylidene)-4-piperidone] was reported to demonstrate potent anti-tumorigenic effects on cancer cells compared to curcumin[4]. Similarly, other studies on diarylpentanoids ca27 and Ca 37 which are structurally identical to MS17 [1, 5-bis(2-hydroxyphenyl)-1,4-pentadiene-3-one] were reported to exhibit anticancer activity on prostate cancer cells[5,6]. However, the molecular mechanisms that demonstrate its anticancer activity were not determined in prostate cancer cells. In our previous findings, MS17 showed growth inhibition, cytotoxicity and cell death inducing activities on androgen-nonresponsive prostate cancer (PC-3 and DU145) and cervical cancer (HeLa and CaSki) cells[7,8]. MS17 demonstrated significant apoptotic activity at 24 h following treatment with 3 × EC50(15 μM) concentration in PC-3 and DU145 cells[7]. MS17 demonstrated potent cytotoxicity effect on cancer cells with lower toxicity against normal cells compared to curcumin. Therefore, the focus of the current study is to perform protein expression profiling on treated PC-3 and DU145 cells to identify mutually regulated proteins as common targets in both of these cell lines as well as molecular pathways that contribute to the anticancer activity of MS17 using 2-DE and nanoflow liquid choromatography electrospray-ionisation coupled with mass spectrometry/mass spectrometry (LC-MS/MS).
2. Materials and methods
2.1. Cell culture and MS17 diarylpentanoid
The human androgen-independent metastatic prostate cancer cell lines, PC-3 and DU145 were purchased from American Type Culture Collection (ATCC, Rockville, USA) and maintained using appropriate media under standard conditions as previously described[7]. The chemically purified diarylpentanoid, MS17 was synthesized and prepared based on the method described previously[9].
2.2. Treatment and total protein extraction
Cells were plated in a 75 cm2tissue culture flasks, treated with 15 μM dose of MS17 and incubated in a humidified incubator for 24 h. All treatments were performed in triplicates and fresh medium containing 0.5% DMSO was added to the untreated control cells.Following incubation, cells were harvested and washed with ice cold PBS solution twice before protein lysate was extracted from the treated and untreated cells. Cell pellets were resuspended in 1 mL lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 65 mM dithiothreitol (DTT), 2% immobilized pH gradient (IPG) buffer and protease inhibitor mix. Suspensions were vortexed on ice at 2 500 rpm for 2 min and frozen on dry ice for 10 min. The second cycle of freeze-thaw was performed for 5 min. Then, the samples were vortexed on ice at 2 500 rpm for another 15 min. DNase Ⅰ (Qiagen,Valencia, CA., USA) and RNase A (Thermo Scientific, USA) at final concentrations of 20 units/mL and 0.25 mg/mL respectively were added into each sample to eliminate any DNA contaminants and subsequently incubated on ice for another 45 min. Following incubation, the protein lysate was centrifuged at 13 000 × g at 4 ℃for 1 h and the supernatants of the treated and untreated samples were collected in sterile microcentrifuge tubes and placed on ice before protein concentration was quantified with the 2D Quant kit (GE Healthcare, UK) based on the protocol provided by the manufacturer.
2.3. 2-DE
Appropriate amount of individual protein samples was adjusted with the addition of rehydration buffer [7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 65mM DTT, 0.2% Triton X-100, 1% bromophenol blue,0.5% (v/v) IPG buffer] to a final concentration of 1 mg and incubated for 30 min at room temperature. The protein samples were then applied to IPG strips (13 cm, pH 3-10 linear) on Immobiline DryStrip reswelling tray (GE Healthcare, Buckinghamshire, UK) and allowed to rehydrate for 18 h at room temperature. Following rehydration,isoelectric focusing was performed at 20 ℃ in a stepwise voltage ramp using Ettan IPGphorⅢ system (GE Healthcare, UK): 500 V for 2 h 20 min, hold; 1 000 V for 1 h, gradient; 8 000 V for 2 h 30 min,gradient, and 8 000 V for 30 min, hold. Once isoelectric focusing was completed, the IPG strips were equilibrated in equilibration buffer[375 mM Tris-HCL (pH 8.8), 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 0.002% bromophenol blue and 1% (w/v) DTT] for 30 min,followed by the same buffer containing 2.5% (w/v) iodoacetamide instead of DTT for another 30 min. The second dimensional SDSPAGE was carried out on 12% SDS-polyacrylamide gels at 18 ℃ in Laemmli running buffer using a Hoefer SE600 system (GE Healthcare,UK). Separation was performed at 90 V for 30 min and 200 V constant power till the bromophenol dye front reached the bottom of the gels.Upon completion, the gels were fixed in 10% methanol, stained in Coomassie brilliant blue G-250, destained in a mixture of 50% ethanol and 12% acetic acid until the background was clear. The experiment was performed in three biological replicates.
2.4. Protein visualization and image analysis
Gel images were scanned using a ChemiDocTMXRS Imaging System (Bio-Rad, USA) and captured using Quantity One®Software(Bio-Rad,USA). Spot detection and matching between gels were automatically performed using the PDQuest software version 8.0.1(Bio-Rad, USA). The PDQuest software was also used to assign identification number and to calculate intensity of each protein spot. Selected spots were filtered based on sensitivity value of 25.0, minimal peak value of 1 000 with Gaussian modelling. Each spot volume from replicate gel was normalized relative to the total spot volume of the control gel. Relative comparison of the spot intensity abundance between treated and untreated control groups was analyzed with Student’s t-test. Statistical significance was set as P<0.05 and significant protein spots with at least 1.5-fold differences were selected for protein identification.
2.5. In-gel tryptic digestion
Protein spots of interest from the treated and control gels were excised manually and transferred to 0.5 mL microfuge tubes for tryptic digestion. The pooled gel pieces were incubated several times with 200 μL of 200 mM ammonium bicarbonate in 40% acetonitrile(ACN) for 30 min at 37 ℃ to remove excess dye. The solvent was discarded and 20 μL of trypsin solution was then added to the gel plugs and subsequently incubated for 5 min at room temperature.Next, 50 μL of digestion buffer (40 mM ammonium bicarbonate in 9% ACN) were added to the gel plugs and left incubated overnight at 37 ℃. After overnight incubation, the supernatants were collected and saved into separate collection tubes. The gel pieces were then extracted with 50 μL of 5% formic acid followed by a mixture of 5%formic acid / 50% ACN and lastly with 50 μL HPLC grade pure ACN with 15 minutes incubation in between solutions at 37 ℃. Supernatant was collected and the gel pieces were discarded. The pooled extracts were then dried overnight at 60 ℃ using centrifugal evaporator CVE-3100 (Eyela, Japan).
2.6. LC-MS/MS analysis
The dried peptides were reconstituted with 8 μL of 0.1% formic acid in water and loaded into Agilent C18 300A Large Capacity Chip (Agilent Technologies, USA) and equilibrated with 0.1% formic acid in water.The following gradient: 5%-75% formic acid in water from 0-30 min followed by 75%-75% formic acid in water from 30-39 min was used to elute the peptides from the column. Q-TOF polarity was set at positive with capillary and fragmentor voltage being set at 1 790 V and 175 V respectively and 5 L/min of gas flow at 325 ℃. Protein spectra were analyzed in auto MS mode ranging from 110-3 000 m/z for MS scan and 50-3 000 m/z for LC-MS/MS scan. Data obtained from LC-MS/MS was processed with PEAKS Studio 7.0 software (Bioinformatics Solution,Waterloo, Canada).
2.7. Protein identification and data analysis
Protein identification by automated de nova sequencing was determined using PEAKS studio 7.0 software. Protein homology of each sample was searched against Homo sapiens protein database in the UniProtKB/Swiss-Prot database (December 2015) by comparing the de nova sequence tag. Parent mass and fragment mass error tolerance were set at 0.1Da with monoisotopic as precursor mass search type. Carbamidomethylation was set as fixed modification and maximum mixed cleavages at 3. Trypsin was used for protein digestion. The search results were filtered based on the following criteria to minimize false positive results: i) False Discovery rate(FDR) < 1.0% and ii) PEAK Score (-logP) > 20. Peptides that did not satisfy the set parameters were excluded from analysis. LC-MS/MS spectra were also searched against cRAP protein database in The Global Proteome Machine (Version 2012.01.01) to eliminate possible common contaminants. The list of proteins that satisfy the above criteria was considered as differentially expressed proteins(DEPs). In addition, DEPs with a protein score (-10logP) cut-off> 50 and identified with at least two peptides, which imposed a percentage coverage of > 15% and fold change values > 1.0-fold were considered for further analysis.
2.8. Bioinformatics analysis
The lists of DEPs were analysed based on gene ontology classification using PANTHERTMclassification system (http://pantherdb.org/) to classify them according to their biological processes/molecular function. Accession numbers of the DEPs were imported into Ingenuity®Pathway Analysis (IPA) software(IPA®; QIAGEN Redwood City) for identification of top significant canonical pathways. Using right-tailed Fisher Exact test P-value set at P<0.05, canonical pathways were considered as statistically significant when it passed through a threshold of 1.3 [calculated by –log (P-value)]. In addition, the DEPs within the top five identified pathways were extracted and overlaid to view their protein interaction network using the MyPathway design tool.
3. Results
3.1. Protein expression profiling and gene ontology classification analysis
PC-3 and DU145 cells were treated with 3 × EC50(15 μM) dose of MS17 and DMSO (control) respectively for 24 h. Using the PDQuest software, protein spots with significant P-value of P<0.05 and a fold change over 1.5-fold in MS17 treated cells compared to control cells in replicate gels were averaged and quantified. Overall, a total of 20 significant protein spots were detected in PC-3 cells while 24 spots were detected in DU145 cells, as indicated by the specific spot number (SSP number). Representative 2-DE gel images of control and MS17-treated cells in PC-3 and DU145 cells are respectively shown in Figures 1 and 2. Protein identification by PEAKS software and UniProt/SwissProt database identified a total of 46 DEPs in PC-3 cells and 47 DEPs in DU145 cells. Amongst these DEPs, a total of 10 up- and 36 down-regulated (Table 1) were obtained in PC-3 cells while 24 up- and 23 down-regulated (Table 2) DEPs were selected in DU145 cells. The significant regulated proteins were then ontologically classified based on their biological processes or molecular functions.

Figure 1. Representative 2-DE gel images of protein expression in (A)control and (B) 15 μM-treated PC-3 cells.
Based on the PANTHER classification analysis, a total of 12 functional categories were identified in PC-3 and 11 in DU145 cells.Among these categories in PC-3 cells, metabolic process constitutes for the major proportion (23%) followed by apoptosis and cell cycle each comprised of 13%, protein folding 9% while proteolysis,regulation of transcription, structural molecule activity and transporter activity account for 7% respectively. However, the other functional categories including cell migration, morphogenesis and response to unfolded protein constitute 12%, each with 4% and cell adhesion accounts for 2% of the DEPs. Similarly, among the 11 functional categories associated with DU145 cells, the largest category was metabolic process (20%) followed by response to unfolded protein(17%). Apoptosis, cytoskeletal organization and proteolysis account for approximately 11% respectively while regulation of transcription,structural molecule activity and transporter activity account for 6%respectively. However, the other functional categories including cell migration, morphogenesis and signal transduction activity represent 12% of the DEPs, each with 4%.
Amongst the total DEPs, 9 regulated proteins in PC-3 (Table 1)and DU145 (Table 2) cells were identified as mutually expressed proteins and common protein targets of MS17 in both the cell lines.The mutually down-regulated proteins included, ACTB and ACTG which were associated with structural molecule activity, ACTN1 with cell cycle, ACTN4 with cell migration, HNRPK with apoptosis,PLST with morphogenesis and TERA with proteolysis. Interestingly,the expressions of CH60 and HS71A respectively associated with response to unfolded protein demonstrated opposing regulation in both the cell lines. The expression of CH60 was up-regulated in PC-3 cells but down-regulated in DU145 cells while HS71A was downregulated in PC-3 but up-regulated in DU145 cells. The selected proteins along with their corresponding functional categories were illustrated in Tables 1 and 2.
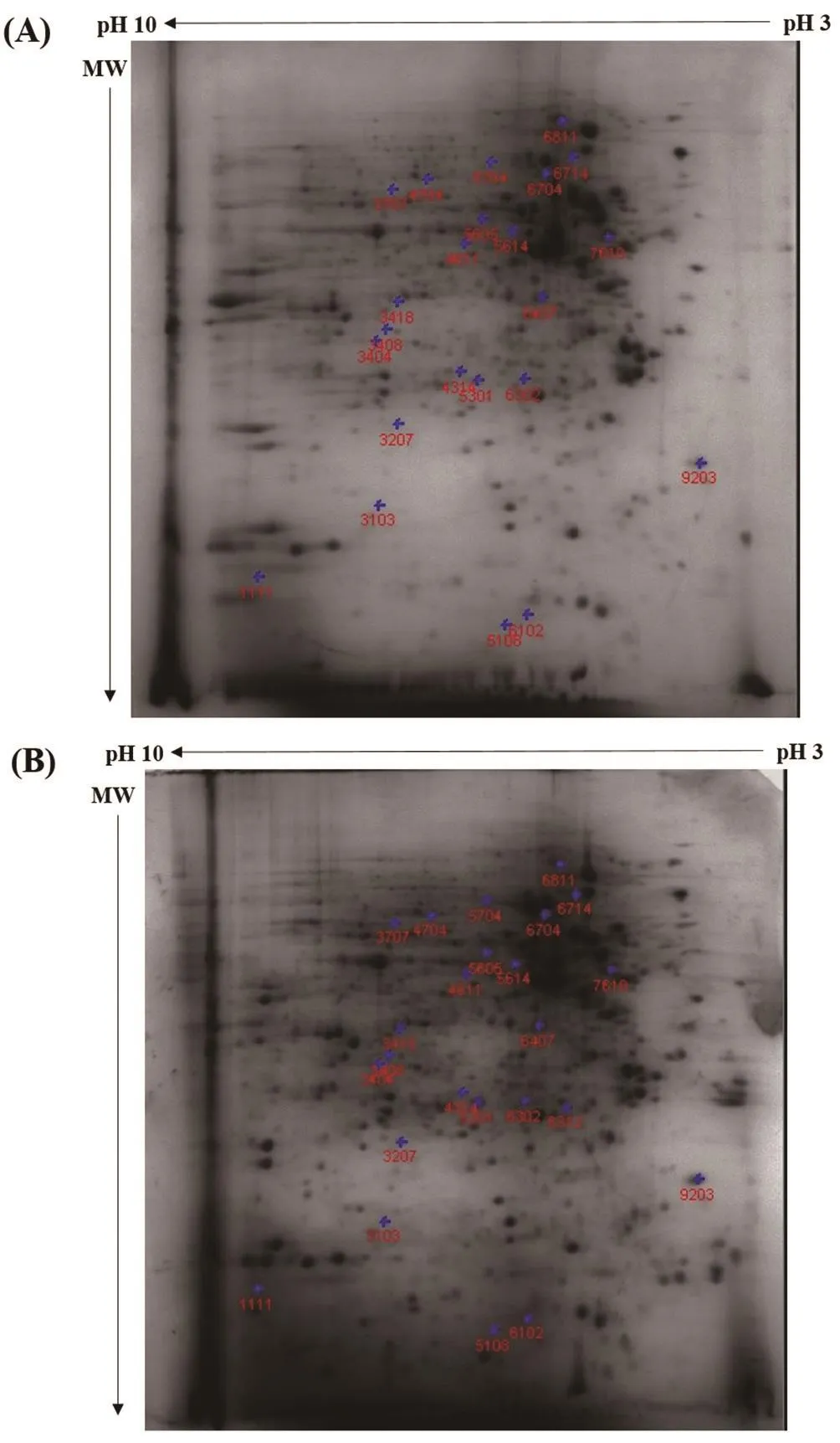
Figure 2. Representative 2-DE gel images of protein expression in (A) control and (B) 15 μM-treated DU145 cells.

Table 2 Selected differentially expressed proteins (DEPs) regulated following 15 μM treatment of MS17 in DU145 cells at 24 h.

Table 3 Top 5 canonical pathways significantly modulated by the DEPs in 15 μM-treated PC-3 and DU145 cells.
3.2. Pathway analysis
The deregulated DEPs were mapped to biological canonical pathways to identify and understand the cellular pathways that were significantly modulated by MS17 treated prostate cancer cells. The top 5 canonical pathways of PC-3 and DU145 cells that were significantly activated by treatment were selected for further analysis. For each pathway, the nominal P-value along with all the input proteins that are encoded by its respective genes was presented in Table 3.
In PC-3 cells, the pathways, “Remodeling of Epithelial Adherens Junctions”, “Epithelial Adherens Junction Signaling”, “Sertoli Cell-Sertoli Cell Junction Signaling” and “Germ Cell-Sertoli Cell Junction Signaling” were associated with the down-regulation of TUBB4B, ACTB, VCL, ACTN4, ACTG1 and ACTN1 while “Paxillin Signaling” pathway was modulated by the down-regulation of ACTB, VCL, ACTN4, ACTG1 and ACTN1 (Table 3). On the other hand, the two most significantly activated pathways in DU145 cells;“Protein Ubiquitination Pathway” and “Aldosterone Signaling in Epithelial Cells” were associated with the up-regulation of HSPA1A/HSPA1B, HSP90AA1, HSP90AB1 and HSPB1 and the repressed expression of HSPA8, HSPA9 and HSPD1. In addition, “Protein Ubiquitination Pathway” was also modulated by the up-regulation of UCHL1 and PSMA6 and down-regulation of USP14 and PSMB1.The pathways, “Germ Cell-Sertoli Cell Junction Signaling”, “Sertoli Cell-Sertoli Cell Junction Signaling” and “Actin Cytoskeleton Signaling” were modulated by the down-regulation of ACTB,ACTN4, ACTG1 and ACTN1. In addition, “Germ Cell-Sertoli Cell Junction Signaling” and “Sertoli Cell-Sertoli Cell Junction Signaling” pathways were also associated with the down-regulation of CLINT1 and PLS1. Conversely, “Actin Cytoskeleton Signaling”was modulated by the induced expression of PFN1 and CFL1 while“Germ Cell-Sertoli Cell Junction Signaling” was modulated by the up-regulation of CFL1 expression (Table 3).
4. Discussion
In this study, PC-3 and DU145 cells were used as a model of androgen-independent human prostate cancer cells. PC-3 and DU145 cells were metastatic that harbour wild-type and mutant p53, respectively. Despite the biological differences, we identified mutually regulated DEPs as common targets in both cell lines and DEPs mapped to relevant cellular pathways, and their role in anticancer activity was discussed.
ACTB and ACTG associated with structural molecule activity are both actin cytoplasmic proteins implicated in the actin cytoskeletal network that are associated with cell motility and maintenance of the cytoskeleton[10]. Over-expression of ACTG in non-small cell lung cancer cells significantly enhanced cancer cell migration[11] while its down-regulation in neuroblastoma cells resulted in decreased cancer cell migration and motility[12]. Increased expression of ACTB was detected in numerous cancers and was correlated with tumour growth and metastasis[13]. The repressed expression of the two alphaactinin proteins (ACTN1 and ACTN4) involved in cell migration was also observed in PC-3 and DU145 cells. Alpha-actinins are ubiquitously expressed cytoskeletal proteins that are associated with cytoskeletal integrity and regulation of cell movement. Overexpression of ACTN4 in colorectal cancer cells induced lymph node metastasis in immune-deficient mice while its down-regulation suppressed migration and invasion of cancer cells[14]. Increased ACTN1 expression detected in prostate cancer cells was associated with resistance to zoledronic acid treatment and cancer cells invasion while its decreased expression abolished their resistance to zoledronic acid and inhibited epithelial to mesenchymal transition,reduced focal adhesion kinase and metalloproteases expressions in the cancer cells[15]. The data suggests that the down-regulation of these proteins may have functional roles that impair the tumorigenic responses in the treated prostate cancer cells, thus supporting the anti-cancer role of MS17.
CH60 encoded by HSPD1 gene, is a highly conserved molecular chaperone found mainly in the mitochondria and some in the cytosol. Interestingly, the expression of CH60 was up-regulated in PC-3 cells while repressed in DU145 cells. It has a dual behaviour,acting either as a tumour suppressor or oncogene depending on the tumour microenvironment. Low level of CH60 expression detected in bladder cancer cells correlated with high tumour stage and was associated with the risk of developing an infiltrating recurrence[16]. Elevated expression of CH60 in gastric cancer was associated with lymph node metastasis, increased cell invasion and poor patient prognosis[17]. In addition, HS71A associated with the similar category also demonstrated opposing regulation in the prostate cancer cells. The expression of HS71A was observed to be repressed in PC-3 cells while up-regulated in DU145 cells.HS71A belongs to the family of heat shock proteins that functions as ATP-dependent molecular chaperone which supports the folding of newly synthesized polypeptides, transport of protein across cellular membrane and targeting of proteins for lysosomal degradation[18]. Inhibition of this protein stimulates a profound apoptotic response through caspase-3 activation in cancer cells[19].In contrast, over-expression of HS71A protein expression was also reported to act as cellular chaperone that modulates apoptotic cell death pathways[20,21]. It may be inferred that the induction of the abovementioned protein expressions reflects a similar mechanism by which the exposure of MS17 in the cancer cells induces stress response that may trigger apoptotic activation and impair tumour metastasis in the treated cells.
The mutual down-regulation of HNRPK, PLST and TERA respectively associated with apoptosis, morphogenesis and proteolysis was also noted in both the prostate cancer cells. HNRPK is a RNA binding protein that binds to DNA and RNA in a sequence specific manner, regulates transcription and mRNA metabolism.Up-regulation of HNRPK was associated with cellular migration of cancer cells leading to metastasis while down-regulation of this protein caused decreased migratory activity of the cancer cells[22]. PLST from the family of Plastins is a conserved, versatile modulator of the actin cytoskeleton, which is an important protein in cell migration, adhesion and endocytosis. Increased expression of PLST in colorectal cancer correlated with cancer cell metastasis[23].Conversely, up-regulation of TERA in colorectal carcinoma repressed apoptosis and sensitivity to chemotherapeutic treatment as well as increased cancer metastasis while its down-regulation inhibited proliferation and invasion, reduced chemoresistance,induced apoptosis and suppressed carcinogenesis in vivo[24]. The inhibition of the abovementioned protein expressions by MS17 may similarly modulate its anti-tumorigenic activity in the treated prostate cancer cells.
In the present study, several DEPs modulated by MS17 treated PC-3 and DU145 cells were identified and mapped to relevant biological canonical pathways. In addition to the common proteins,DEPs associated with pathways that were exclusively noted in either PC-3 or DU145 cells were discussed. The top 5 canonical pathways that were significantly activated in PC-3 were associated with cellcell adhesion and cytoskeletal organization. Notably, the pathways“Remodeling of Epithelial Adherens Junctions”, “Epithelial Adherens Junction Signaling”, “Sertoli Cell-Sertoli Cell Junction Signaling”and “Germ Cell-Sertoli Cell Junction Signaling” were modulated by the down-regulation of TUBB4B, VCL, ACTB, ACTG1, ACTN4 and ACTN1 while “Paxillin Signaling” was modulated by the downregulation of VCL, ACTB, ACTG1, ACTN4 and ACTN1. TUBB4B and VCL down-regulation in cancer cells coincides with enhanced drug sensitivity to Eribulin[25] and suppressed cancer cell invasion[26-28] respectively. Similarly, the down-regulation of ACTN1 and ACTN4 expressions has demonstrated involvement in the disruption of cancer progression via induction of anti-metastatic activity and reduced chemotherapy drug resistance[14,15]. Meanwhile, ACTB and ACTG1 are both implicated in the cytoskeletal network. Overexpression of ACTB and ACTG1 in cancer cells was correlated with tumour growth, increased cancer cell migration and metastasis[11,13]while its down-regulation impaired cell migration and motility[12].The modulation of the abovementioned pathways suggests that MS17 regulates the expression of the cytoskeletal proteins to mediate anti-metastatic, anti-proliferative and anti-invasive activities as well as increase sensitivity to drug treatment in treated PC-3 cells.
Likewise, the modulation of the top five canonical pathways in DU145 cells demonstrated the activation of pathways associated with proteasomal degradation, regulation of electrolytes and water,regulation control of germ cells and organization of filament assembly/disassembly. The pathways, “Protein Ubiquitination Pathway” and “Aldosterone Signaling in Epithelial Cells” were modulated by the induced expressions of HSPA1A/1B, HSP90AA1,HSP90AB1 and HSPB1 and repressed expression of HSPA8,HSPA9 and HSPD1. HSPA1A/1B up-regulation in osteosarcoma cells enhanced drug sensitivity to mitomycin C treatment[29]. While heat shock proteins act as protein chaperones in cellular processes,HSP90AB1 and HSP90AA1 which encode for Hsp90B and Hsp90A respectively are commonly induced during hypoxia which promotes tissue repair mechanism. Hsp90B functions to stabilise the low density lipoprotein receptor-related protein-1 receptor at the cell surface while Hsp90A protein is released into the extracellular space and signals to induce cell migration leading to wound closure[30]. As impaired responses to hypoxia is correlated with cancer progression,this may therefore reflect a similar mechanism by which MS17 upregulates HSP90AB1 and HSP90AA1 expressions to modulate tissue repair mechanism in treated cells thus contributing to antitumorigenesis. Conversely, down-regulation of HSPB1 in vivo model of lung carcinoma induced endothelial-to-mesenchymal transition,promoting cancer while[31] its up-regulation inhibited endothelial-tomesenchymal transition leading to suppressed lung tumorigenesis[32].Furthermore, these pathways were also modulated by the downregulation of HSPA8, HSPA9 and HSPD1 protein expressions.Increased HSPA8 expression is commonly associated with tumorigenesis while its inhibition confers anti-proliferative, antimetastatic and pro-apoptotic properties in endometrial carcinoma cells[33]. Meanwhile, HSPA9 contributes to cell proliferation upon up-regulation but promoted apoptosis and growth arrest following its down-regulation in medullary thyroid carcinoma cells[34]. HSPD1 encodes for CH60 is essential for cell physiology and survival. Upregulation of CH60 protein in gastric cancer cells was correlated with tumour aggressiveness and poor prognosis[17]. In addition to these proteins, “Protein Ubiquitination Pathway” was also modulated by the up-regulation of UCHL1 and PSMA6 and downregulation of USP14 and PSMB1 expressions. UCHL1 expression mediates tumour suppressive function and its up-regulation inhibited cell proliferation and demonstrated apoptotic activity in several cancer cells including prostate cancer[35-37]. USP14,a proteasomal deubiquitinating enzyme, was reported to exhibit oncogenic functions in tumour cells upon its induction while its down-regulation suppressed tumour cell proliferation, invasion and promoted apoptosis[38-40]. Induced PSMA6 expression in prostate cancer cells correlated with chemotherapy sensitivity[41] while PSMB1 down-regulation in colorectal cancer cells upon treatment with anticancer agent was associated with apoptotic induction[42].Hence, the modulation of UCHL1, USP14, PSMB1 and PSMA6 in association with “Protein Ubiquitination Pathway” and “Aldosterone Signaling in Epithelial Cells” pathways may similarly be regulated by MS17 to exhibit apoptotic and anti-proliferative effects observed in the treated cells.
Meanwhile, the pathways “Germ Cell-Sertoli Cell Junction Signaling” and “Sertoli Cell-Sertoli Cell Junction Signaling” were modulated by the repressed expressions of ACTB, ACTG1, ACTN4 and ACTN1 which were also noted in PC-3 cells. In addition,“Germ Cell-Sertoli Cell Junction Signaling” and “Sertoli Cell-Sertoli Cell Junction Signaling” pathways were also modulated by repressed expressions of CLINT1 and PLS1, which were not identified in PC-3 cells. While literature regarding the anti-cancer role of CLINT1 is limited, the expression of CLINT1 was identified in human osteosarcoma cells and was postulated to be related to cancer cell proliferation[43]. Plastin-1 encoded by PLS1 is an actinbundling protein which is important to determine architecture of the actin cytoskeleton[44]. Inhibition of PLS1 expression in prostate cancer cells led to reduction of tumour growth and metastasis[45].Meanwhile, induced expression of CFL1 which encodes for Cofilin-1 was also mapped to the “Germ Cell-Sertoli Cell Junction Signaling” pathway. CFL1 was shown to suppress non-small cell lung cancer cell motility and invasion while selective inhibition of CFL1 conferred cancer cell growth and invasion[46]. Interestingly,activation of “Actin Cytoskeleton Signaling” was also modulated by induced expression of CFL1 and PFN1. PFN1 is a tumour suppressor protein that binds to proline-rich ligands to perform cellular processes such as actin assembly, endocytosis and gene transcription[47]. Over-expression of PFN1 in cancer cells suppressed cell proliferation, motility and invasion resulting in a nontumorigenic phenotype[48,49]. Taken together, the evidence suggests that the interaction of these proteins in association with the pathways inhibits cell growth via deactivation of proliferative responses and potentially underlies the anti-proliferative and apoptotic activity in MS17 treated DU145 cells.
PC-3 and DU145 are androgen-independent prostate cancer cells with different biological properties, and we identified several mutually regulated proteins by MS17 which could be potential therapeutic targets for these cells. These commonly regulated proteins may inhibit cell proliferation, migration, metastases and induce apoptosis in prostate cancer cells. The pathway analysis revealed genes mapped to the relevant biological pathways were mainly associated with anti-proliferative, anti-metastatic activity and/or apoptosis when treated with MS17. Thus the findings of the current study provide an insight into the antitumor activity of MS17 as a potential chemotherapeutic agent for androgen-independent prostate cancer.
Conflict of interest statement
All the authors declare no conflict of interest.
Acknowledgments
The first author would like to acknowledge the support from the Ministry of Higher Education Malaysia for providing scholarship under the MyBrain (MyPhD) scheme.
Funding
This study was financially supported by the Fundamental Research Grant Scheme, (FRGS/1/2016/SKK08/MUSM/02/1) under the Ministry of Higher Education (MOHE), Malaysia.
杂志排行
Asian Pacific Journal of Tropical Biomedicine的其它文章
- Spatial distribution of sand flies (Diptera: Psychodidae; Larroussius group), the vectors of visceral leishmaniasis in Northwest of Iran
- Effects of physicochemical factors on development and survival of Opisthorchis viverrini uterine eggs
- Add-on therapy of herbal formulation rich in standardized fenugreek seed extract in type 2 diabetes mellitus patients with insulin therapy: An efficacy and safety study
- Anticancer activity of crude acetone and water extracts of Tulbaghia violacea on human oral cancer cells
- Conocarpus erectus L., a plant with a high content of structural sugars, ions and phenolic compounds, shows antioxidant and antimicrobial properties promoted by different organic fractions
