Detection and characterization of an isolate of Tomato mottle mosaic virus infecting tomato in China
2018-05-08ZHANBinhuiCAONingWANGKainaZHOUXueping
ZHAN Bin-hui, CAO Ning, WANG Kai-na, ZHOU Xue-ping,
1 State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, China Academy of Agricultural Sciences, Beijing 100193, P.R.China
2 State Key Laboratory of Rice Biology, Institute of Biotechnology, Zhejiang University, Hangzhou 310058, P.R.China
1. Introduction
Tomato (Solanum lycopersicumL.) is one of the most economically important vegetable crops throughout the world. The annual tomato production showed an increasing tendency year by year, while the incidence and severity of diseases have limited the yield and quality of tomato causing serious losses. Viral diseases are the major constraints in tomato production among the known pathogen-induced diseases (Hanssenet al. 2010).
There are at least 136 characterized viruses that have been reported infecting tomato in the world which are much greater than the other vegetables (Bruntet al. 1996; Xuet al. 2017). Tobamoviruses are a kind of economically important virusesin the familyVirgaviridae, which contains 37 members according to the taxonomy by the International Committee on Taxonomy of Viruses (https://talk.ictvonline.org/taxonomy/). The tobamoviruses were divided into different subgroups according to their genomic structure, host range, the amino acid composition of the coat proteins (CPs), etc. The phylogenetic relationship of the tobamoviruses conducted by Liet al. (2017) revealed that in the Solanaceae-infecting group,Tobacco mosaicvirus(TMV),Tomato mosaic virus(ToMV),Tomato mottle mosaic virus(ToMMV) andTomato brown rugose fruit virus(ToBRFV) are the four reported tomato-infecting tobamoviruses and are classi fied into one clade based on the complete genome and amino acid sequences. TMV and ToMV are the most epidemic viruses among the world,ToBRFV is a new species that has been characterized in Jordan (Salemet al. 2016) and Israel (Luriaet al.2017), while ToMMV was also a recently characterized species and showed fast spread in the world. ToMMV was first described in 2013 infecting tomato in Mexico (Liet al. 2013) and subsequently was detected on tomato in different parts of the world including the USA (Websteret al. 2014; Fillmeret al. 2015; Padmanabhanet al.2015; Suiet al. 2017), Israel (Turinaet al. 2016), Brazil(KT222999) and Spain (Ambróset al. 2017). Additionally,Pirovanoet al.(2014) showed its presence inCicer arietinumL. in Italy and Liet al. (2014, 2017) established two isolates infecting pepper from Yunnan Province and Tibet Autonomous Region in China.
The genome of ToMMV contains four open reading frames expressing four proteins like the other tobamoviruses in the familyVirgaviridae, ~126 kDa protein and ~180 kDa read-through protein involved in virus replication, ~30 kDa movement protein and ~18 kDa coat protein. The sequence characteristic of ToMMV genome like the other tobamoviruses represents that the genome contains a Ω fragment in the 5´ untranslated region (UTR) which had no G residues except the most-proximal m7G cap(Richardset al. 1977; Zhanget al. 2008), a conserved region TCCCTCCACTTAAATCGAAGGGTT located in the 3´ UTR with the CCCA ending sequence (Chnget al. 1996)and the ‘4404-50 motif’ reported as the tobamovirusesspecific nucleotide motif which indicated that the 29 sites of the 47 nucleotides (nt) are invariant in all tobamoviruses sequences (Gibbset al. 2004). Liet al. (2017) showed that the remaining 18 variable sites are also conserved among the previously reported ToMMV isolates and 9 of the 18 variable sites can distinguish ToMMV from ToMV. The ToMMV-specific sequence is as follows:GGTGATGTTACAACTTTCATAGGAAATACTGTTATTATA GCCGCGTG.
In a survey of viral diseases in 2016, a devastating disease infecting tomato crops was observed in the greenhouses and natural fields in Hainan Province in China. The tomato plants showing virus-like symptoms were collected for virus detection. The pathogen has been identified as ToMMV and characterized the full sequence of ToMMV Hainan isolate-infecting tomato.
2. Materials and methods
2.1. Plant growth conditions and inoculations
Tobacco, pepper and tomato plants were grown in a growth room (28°C day and 24°C night, 16 h/8 h photoperiod).Virus-infected leaf tissues were homogenized in 0.01 mol L–1phosphate buffer (PBS, 0.01 mol L–1KH2PO4:0.01 mol L–1Na2HPO4=49:51 (v/v), pH=7.0) at 1:10 ratio (w/v). The crude extracts were rub-inoculated to the newly grown leaves of 4-week-old tobacco, pepper and tomato plants,respectively. The virus-inoculated plants were grown in the same conditions.
2.2. Dot enzyme-linked immunosorbent assay(dot-ELISA)
The dot-ELISA procedures were conducted as described previously with some modification (Liet al. 2015). Briefly, the 3 μL plant sample supernatants in 0.01 mol L–1PBS (pH 7.4)were spotted onto the nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ, USA). The TMV monoclonal antibody solution was diluted by 1:5 000-fold with blocking buffer (5% skimmed milk in PBS with 0.5% Tween-20). The membranes were developed with the substrate nitro-blue tetrazolium (NBT, 0.083 mg mL–1) and 5-bromo-4-chloro-3-indolyl phosphate salt (BCIP, 0.05 mg mL–1).
2.3. RNA extraction, RT-PCR, full-length genome amplification and sequencing
The total RNA was extracted from tomato leaves with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and treated with DNaseI. A total of 1 μg of total RNA, random primer, and M-MLV reverse transcriptase were used for synthesis of first-strand cDNA using Prime-ScriptTMRT Reagent Kit(TaKaRa Bio. Inc., Dalian, China). RT-PCR was performed according to the manufacturer’s protocols. The primer pairs used in the RT-PCR experiments were listed in Appendix A. The RT-PCR products were puri fied with E.Z.N.A. Gel Extraction Kit (Omega, Norcross, GA, USA) and cloned into pGEM-T vector (Promega, Madison, WI, USA), then transfected into DH5α competent cells. The recombinant clones were sequenced with universal primer pairs M13F/M13RviaSanger sequencing using ABI 3730XL DNA Analyzers (Applied Biosystems, Foster City, CA, USA). At least three independent clones were sequenced.
The 5´- and 3´-terminal sequence of the ToMMV were obtained through 5´- and 3´-rapid amplication of cDNA ends (RACE) using SMARTer RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. The 5´ and 3´ gene-specific primer (GSP) (both were designed according to the known ToMMV fragment and listed in Appendix A) paired with the Universal Primer Mix (UPM) (the long primer provided by kits) were used in 5´- and 3´-RACE, respectively.
2.4. Sequence assembly and analysis
The complete sequence of ToMMV was assembled and analyzed with the software DNAMAN version 5.0 (Lynnon Biosoft, Quebec, QC, Canada). The sequence fragment and the whole genome were compared against the database from the National Center for Biotechnology Information(NCBI) using a BLAST search.
The phylogenetic trees derived from the alignment of the multiple sequences were constructed using the neighborjoining method with 1 000 bootstrap replications by MEGA 6.0. The sequences used for comparison were obtained from the GenBank database.
3. Results
3.1. Virus detection with dot-ELISA and RT-PCR
In December 2016, a severe tomato disease broke out in the greenhouse and field condition in Hainan Province. The diseased tomato plants showed severe virus-like symptoms with leaves distortion, mosaic and mottle of light and dark green, systemic crinkling which seriously reduced the production (Fig. 1-A). The infected tomato samples were collected and Identification of the pathogens was conducted by serological analysis and biological research.
As the disease symptoms developed very quickly in the greenhouse and under field condition, the pathogen was speculated as a tobamovirus other than a tospovirus or a geminivirus. The collected samples were subjected to dot-ELISA using TMV monoclonal antibody (provided by Professor Zhou Xueping’s lab in Zhejiang University in China). The sap from leaves of the diseased tomato plants reacted strongly to TMV antibody showing purple colors as the positive control, which suggested that the tobamovirus is likely associated with the disease (Fig. 1-B). Both TMV and ToMV are widely distributed tobamoviruses in China and the two viruses show positive reaction with TMV antibody,to further investigate which virus caused the disease, the total RNA was extracted and RT-PCR was conducted using the TMV- or ToMV-specific primer pairs (Appendix A). The primer pairs TA/TB and TMf1/TMr1 were used as TMV-specific primers, while the primer pairs ToA/ToB and ToMf1/ToMr1 were designed as ToMV-specific primers. RT-PCR results indicated that no bands were obtained with primer pairs TA/TB, TMf1/TMr1 or ToA/ToB, while a specific band of ~500 bp was generated with ToMf1/ToMr1. Sequencing and BLAST analysis revealed that the sequence was 99%identical to ToMMV coat protein gene. For the confirmation of the result, the tobamovirus degenerate primers TobamoF/TobamoR (Liet al. 2014) and the designed ToMMV-specific primers ToMMVF/ToMMVR were used in RT-PCR. The approximately ~1 000 bp products were obtained with the two primer pairs (Fig. 1-C). Sequence analysis showed that the acquired PCR products shared the highest identity with ToMMV isolate YYMLJ (GenBank no. KR824950.1),which indicated that the symptomatic tomatos from Hainan Province were infected by the ToMMV.
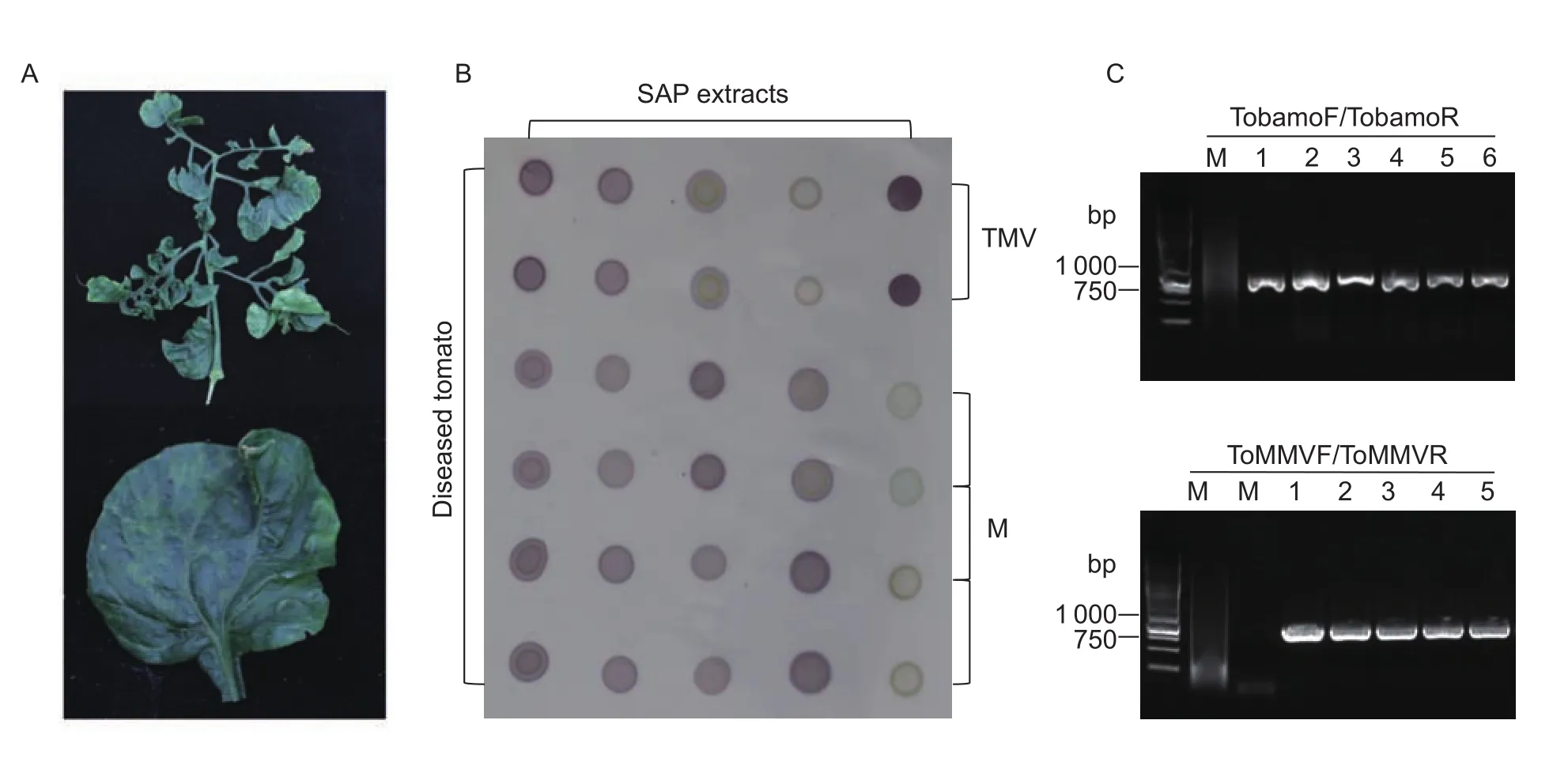
Fig. 1 Dot enzyme-linked immunosorbent assay (dot-ELISA) and RT-PCR detection of the diseased tomato plants. A, symptoms on tomato plants infected with Tomato mottle mosaic virus (ToMMV) in Hainan in China. B, detection of ToMMV by dot-ELISA with Tobacco mosaic virus (TMV) monoclonal antibody. C, detection of ToMMV by RT-PCR using tobamovirus degenerate primers and ToMMV-specific primers. M, mock-inoculated plant; 1–6, tomato samples in Hainan.
3.2. Complete sequence amplification and analysis
The complete genome sequence of ToMMV was divided into six fragments and amplified by RT-PCR using designed primers. Every fragment overlapped with the adjacent one or two fragments. The six pairs of primers listed in Appendix A were designed based on the conserved sequences of all ToMMV sequence from NCBI database. To obtain whole sequence of the genome, we amplified the ~770 bp of the most proximal 5´ regions and the ~550 bp of the 3´ regions(primers listed in Appendix A) of the genome. The whole genome sequence of ToMMV Hainan was assembled and determined to be 6 397 nt (GenBank no. MG171192), the full-length nucleotide sequence of ToMMV Hainan isolate was compared with the other isolates and tobamoviruses.The results showed that ToMMV Hainan shared the highest identity with ToMMV TiLhaLJ (GenBank no. KR824951) of 99.7% and subsequently with ToMMV YYMLJ (GenBank no. KR824950) of 99.55%, ToMMV MX-5 (GenBank no.KF477193) of 99.44%, ToMMV NY-13 (GenBank no.KT810183) of 99.30%, ToMMV 10-100 (GenBank no.KT810183) of 99.28% and ToMMV VLC-1 (GenBank no.KU594507) of 98.81%, respectively. ToMMV Hainan shared the similarity of 85% with ToMV, 81% with TBRFV and 80% with TMV. To investigate genetic similarity between the ToMMV isolates and the other tobamoviruses, the phylogenetic tree was constructed using the neighbor-joining method analysis based on the complete genome, which showed that seven ToMMV isolates differentiated from the other three tobamoviruses. Within the phylogenetic ToMMV cluster, it is observed that ToMMV Hainan isolate is more closely related with ToMMV TiLhaLJ and ToMMV YYMLJ which are isolated from peppers in China, the ToMMV MX-5 from Mexico and ToMMV NY-13 from New York are in one subgroup and are close to ToMMV 10-100 from the USA,while the isolate ToMMV LVC-1 from Spain is in a different clade with the other six ToMMV isolates (Fig. 2-A).
ToMMV Hainan isolate contains the specific genomic structure like the other tobamoviruses in the familyVirgaviridae. The whole genome included four open reading frames expressing four proteins. The 5´-most open reading frame (ORF) started at 74 nt encoding a 126-kDa protein and its read-through resulted a 180-kDa protein which are involved in virus replication. The ~30-kDa movement protein and ~18-kDa CP were 4 908–5 714 and 5 717–6 196 nt,respectively. Sequence analyses revealed that the 5´ UTR was 73 nt possessing a Ω fragment, a conserved region6270TCCCTCCACTTAAATCGAAGGGTT6293located in the 3´ UTR and the ToMMV specific ‘4404-50 motif’ located in 4 409–4 455 nt.
3.3. Biological assay and fulfillment of Koch’s postulates
To investigate the biological characterization of the new isolate ToMMV Hainan, we used the crude sap extracts of tomato sample infected by ToMMV Hainan to inoculate tomato plants, pepper andN.benthamianaplants. The virus caused mosaic and mottle symptoms in the infected tomato plants at 7 days post inoculation (dpi) and later the leaves began narrowing and crinkling, the whole plants showed stunting compared with the uninfected control (Fig. 2-B).On inoculated seedlings of pepper, there were crinkling and mosaic at 7 dpi in the upper systematic leaves, later the necrotic lesions appeared in the growth point (Fig. 2-B). TheN.benthamianawas systematically infected and showed crinkling and yellow on upper leaves at 4 dpi. At 6 dpi,the apical shoot of theN.benthamianaseedlings started showing serious necrosis symptoms (Fig. 2-B).
4. Discussion
These results presented here show that the distortion and crinkling symptoms on tomato plants in Hainan Province in China are associated with the infection of ToMMV. In this study, we characterized a new isolate ToMMV Hainan and described the phylogenetic relationship with the other ToMMV isolates and tobamoviruses.
In our dot-ELISA analysis, the ToMMV-infected tomato samples without TMV infection reacted positively with the TMV monoclonal antibody, which was coincided with the previous reports (Websteret al. 2014; Turinaet al. 2016).The dot-ELISA results suggested that ToMMV had the serological relation with TMV. Given the high amino acids sequence similarity between TMV, ToMV and ToMMV,the infections being diagnosed as TMV or ToMV using serological methods solely might actually be ToMMV.Before ToMMV was characterized as a new species, many isolates were deposited in GenBank as ToMV, for example,the sequence from Iran (JX112024, JX112025, JX121570,JX121574, JX121575, JX121576, HQ593616), from Brazil (AF411922, AM411425, AM411430) and from China(JX025564). The sequence from Brazil (AF411922) may be the first sequence recorded corresponding to ToMMV(Moreiraet al. 2003).
ToMMV Hainan isolate has the highest identity and the closest evolutionary relationship to ToMMV TiLhaLJ and ToMMV YYMLJ which were isolated from peppers in China,while are in a different subgroup with the isolates from Mexico and USA or the isolate from Spain.

Fig. 2 Phylogenetic tree and infectivity of the Tomato mottle mosaic virus (ToMMV) Hainan isolate. A, neighbor-joining phylogenetic tree showing the relationship between ToMMV isolates and other tobamoviruses. B, infectivity of ToMMV Hainan isolate in tomato,pepper and tobacco plants. Left, non-infected plants; middle, ToMMV-infected plants; right, details of the leaf or apical shoot from the infected plants.
ToMMV was firstly identified as a novel tobamovirus species infecting tomatoes in 2013 in Mexico (Liet al. 2013),and now it has been identified in several countries and areas in the world. In China, ToMMV was firstly reported infecting peppers in Yunnan Province and Tibet Autonomous Region in 2014 (Liet al. 2014). Here, we are reporting the first case of ToMMV infection on tomato in Hainan Province of China and characterized the whole sequence. The ToMMV need more attention because of its rapid spread and devastating damage to tomato industry. Therefore, further studies need to be performed to identify the real occurrence and epidemiology of the ToMMV.
5. Conclusion
A severe disease on tomatoes broke out in Hainan Province in China. We identified thatTomato mottle mosaic viruswas the pathogen causing the mottle symptoms by dot-ELISA and RT-PCR. The full-length sequence of the new ToMMV Hainan isolate was determined and the biological characterization has been investigated. This was the first Identification and characterization of ToMMV-infecting tomato in Hainan in China.
Acknowledgements
This research was supported by the Agricultural Science and Technology Innovation Program, China (ASTIP). We thank technicians and students from Hainan Runda Modern Agriculture Corporation, China for helping in field surveys and samples collections.
Appendixassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Ambrós S, Martínez F, Ivars P, Hernández C, Iglesia F D L,Elena S F. 2017. Molecular and biological characterization of an isolate ofTomato mottle mosaic virus(ToMMV)infecting tomato and other experimental hosts in eastern Spain.European Journal of Plant Pathology, 149, 261–268.
Brunt A A, Crabtree K, Dallwitz M J, Gibbs A J, Watson L.1996.Plant Viruses Online:Descriptions and Lists from the Vide Database. 16th ed. Australian National University,Canberra, Australia.
Chng C G, Wong S M, Mahtani P H, Loh C S, Goh C J, Kao M C, Chung M C, Watanabe Y. 1996. The complete sequence of a Singapore isolate of odontoglossum ringspot virus and comparison with other tobamoviruses.Gene, 171, 155–161.
Fillmer K, Adkins S, Pongam P, D’Elia T. 2015. Complete genome sequence of a tomato mottle mosaic virus isolate from the United States.Genome Announcements, 3,e00167–e00182.
Gibbs A J, Armstrong J S, Gibbs M J. 2004. A type of nucleotide motif that distinguishes tobamovirus species more efficiently than nucleotide signatures.Archives of Virology, 149,1941–1954.
Hanssen I M, Lapidot M, Thomma B P. 2010. Emerging viral diseases of tomato crops.Molecular Plant-Microbe Interactions, 23, 539–548.
Li N, Chen Z, Liu Y, Liu Y, Zhou X, Wu J. 2015. Development of monoclonal antibodies and serological assays specific forbarley yellow dwarf virus GAV strain.Virology Journal,12, 136.
Li R, Gao S, Fei Z, Ling K S. 2013. Complete genome sequence of a new tobamovirus naturally infecting tomatoes in Mexico.Genome Announcements, 1, e00794–e00807.
Li Y, Wang Y, Hu J, Xiao L, Tan G, Lan P, Liu Y, Li F. 2017. The complete genome sequence, occurrence and host range ofTomato mottle mosaic virusChinese isolate.Virology Journal, 14, 15.
Li Y Y, Wang C L, Xiang D, Li R H, Liu Y, Li F. 2014. First report of tomato mottle mosaic virus infection of pepper in China.Plant Disease, 98, 1447.
Luria N, Smith E, Reingold V, Bekelman I, Lapidot M, Levin I,Elad N, Tam Y, Sela N, Abu-Ras A. 2017. A new Israelitobamovirusisolate infects tomato plants harboringTm-22resistance genes.PLoS ONE, 12, e0170429.
Moreira S R, Eiras M, Chaves A L R, Galleti S R, Colariccio A. 2003. Characterization of a newTomato mosaic virusstrain isolated from tomato in the State of São Paulo, Brazil.Fitopatologia Brasileira, 28, 602–607. (in Portuguese)
Padmanabhan C, Yi Z, Li R, Martin G B, Fei Z, Ling K S. 2015.Complete genome sequence of a tomato-infecting tomato mottle mosaic virusin New York.Genome Announcements,3, e01523–e01538.
Pirovano W, Miozzi L, Boetzer M, Pantaleo V. 2014.Bioinformatics approaches for viral metagenomics in plants using short RNAs: model case of study and application to aCicer arietinumpopulation.Frontiers in Microbiology, 5, 790.
Richards K, Jonard G, Guilley H, Keith G. 1977. Leader sequence of 71 nucleotides devoid of G in tobacco mosaic virus RNA.Nature, 267, 548–550.
Salem N, Mansour A, Ciuffo M, Falk B W, Turina M. 2016. A new tobamovirus infecting tomato crops in Jordan.Archives of Virology, 161, 503–506.
Sui X, Zheng Y, Li R, Padmanabhan C, Tian T, Groth-Helms D, Keinath A P, Fei Z, Wu Z, Ling K S. 2017. Molecular and biological characterization ofTomato mottle mosaic virusand development of RT-PCR detection.Plant Disease,101, 704–711.
Turina M, Geraats B P J, Ciuffo M. 2016. First report ofTomato mottle mosaic virusin tomato crops in Israel.New Disease Reports, 33, 1.
Webster C G, Rosskopf E N, Lucas L, Mellinger H C, Adkins S. 2014. First report of tomato mottle mosaic virus infecting tomato in the United States.Plant Health Progress, 15,151–152.
Xu C, Sun X, Taylor A, Jiao C, Xu Y, Cai X, Wang X, Ge C, Pan G, Wang Q, Fei Z, Wang Q. 2017. Diversity, distribution,and evolution of tomato viruses in China uncovered by small RNA sequencing.Journal of Virology, 91, e00173–e00190.
Zhang Z C, Lei C Y, Zhang L F, Yang X X, Chen R, Zhang D S. 2008. The complete nucleotide sequence of a novelTobamovirus, Rehmannia mosaic virus.Archives of virology, 153, 595–599.
杂志排行
Journal of Integrative Agriculture的其它文章
- Yield and water use responses of winter wheat to irrigation and nitrogen application in the North China Plain
- A simulation of winter wheat crop responses to irrigation management using CERES-Wheat model in the North China Plain
- ldentification of the strain-specifically truncated nonstructural protein 10 of porcine reproductive and respiratory syndrome virus in infected cells
- Effect of dietary supplementation with flavonoid from Scutellaria baicalensis Georgi on growth performance, meat quality and antioxidative ability of broilers
- ldenti fication of miRNAs and target genes regulating catechin biosynthesis in tea (Camellia sinensis)
- Variability in total antioxidant capacity, antioxidant leaf pigments and foliage yield of vegetable amaranth
