Com p letem itochondrialgenome of the ThaiRed Junglefow l(Gallus gallus)and phylogenetic analysis
2018-05-07ChatmongkonSuwannapoom,Ya-JiangWu,XingChen等
DEAR EDITOR,
In this study,we sequenced the complete mitochondrialgenom e(mitogenome)of the Thai Red Jungle fow l(RJF;Gallusgallus)using the next-generation sequencing(NGS)platform of the Ion Torrent PGM.Samples were taken from Mae Wang District,Chiang Mai Province,northern Thailand.Our data showed the comp letemitogenome to be 16 785 bp in length,com posed by 13 protein-coding genes,22 tRNA genes,two rRNA genes,and one control region.The genome nucleotide com position was 30.3%A,23.7%T,32.5%C,and 13.5%G,resulting in a high percentage of A+T(50.4%).Phylogenetic analysis revealed that the mitogenome belonged to haplogroup X,whereas those of all domestic chickens belong to haplogroups A to G.This newly released mitogenome sequence w ill advance further evolutionary and population genetics study of the RJF and domestic chicken.The availability of theG.gallusmitogenome willalso contribute to further conservation genetics research of a unique species,listed as ‘data deficient’in Thailand.
The Red Junglefow l(RJF;Gallusgallus)is a major wild ancestor of the domestic chicken(Darw in,1875;Liu et al.,2006;Miao et al.,2013). Early studies based on mitochondrial DNA(m tDNA)revealed that the Thai RJF has a close relationship with the domestic chicken(Fumihito et al.,1994;Fumihito et al.,1996),imp lying that Thailand is likely a domestication center of the chicken.To the best of our know ledge,no com plete mtDNA genome(i.e.,mitogenom e)sequence of the ThaiRJF has been reported.In this study,we collected a RJF sam ple from the Mae Wang District of Chiang MaiProvince in northern Thailand(permission provided by the Thai Institute of Animals for Scientific Purpose Development(No.U1-01205-2558)).The complete mitochondrial genome was submitted to GenBank(accession No.:MG605671).
Genom ic DNA was extracted from whole blood using the HiPure Tissue DNA Micro Kit(Magen,China). The PCR amp lification,library construction,and next-generation sequencing were in accordance with our earlier study(Chen et al.,2016). We used adenovolong fragment PCR and NGS strategy to obtain high quality m tDNA reads and exclude NTMT pseudogenes.We followed caveats for quality control in m tDNA genom ic studies of domestic animals(Shi et al.,2014).The generated sequence was aligned against a reference sequence AP003321(Nishiboriet al.,2005),with allvariants then output.Using the Integrative Genomics Viewer(Thorvaldsdóttir etal.,2013),we checked the bamfile exported by Torrent Suite 5.0.2 to confirm the scored variants.
Phylogenetic analysis was perform ed using com p lete m tDNA sequences of all major haplogroups and sub-hap logroups,as defined by Miao et al. (2013)and Peng etal.(2015).Allm itogenomes were aligned by ClustalW,then analyzed bymaximum parsimony(MP)in MEGA 7.0 with 1 000 bootstrap rep licates(Tamura etal.,2011).
The com pletemitogenome sequence of the ThaiRJF(16 785 bp;GSA accession No.:PRJCA000287,GenBank accession No.:MG605671)had an overall base composition of 30.3%for A,23.7%for T,32.5%for C,and 13.5%for G,w ith high a A+T content of 54.0%.The m itogenome consisted of 13 protein-coding genes,22 tRNA genes,two rRNA genes,and a displacem ent loop(D-loop).Mostm itogenom e genes were encoded on the heavy strand,except for eight tRNA genes and one protein-coding gene(ND6),which were encoded on the light strand.Some protein-coding genes shared the start and stop codons;for instance,all13 genes began with ATG,except forCOX1,which started with GTG,and of the remaining 12 protein-coding genes,nine(ND1,COX2,ATPase8,ATPase6,ND3,ND4L,ND5,cytb,andND6)shared the stop codon TAA,two(COX3andND4)shard the stop codon “T– –”,andND2used TAG.The lengths of the 12S rRNA and 16S rRNA genes were 976 bp and 1 622 bp,respectively.
As the RJF is a w ild type ofGallusgallus,it differed from allmajor domestic haplogroups(A to G).We calculated the differences between the ThaiRJF and random ly selected individuals with haplogroups A to G(Table 1).Com pared with the common domestic hap logroups,the RJF had 40 different base pairs on average(range:29–53).

Tab le 1 M itochond rialgenom e d ifferences vs.ThaiRed Jung le fow l(PRJA000287)
A phylogenetic tree(Figure 1)of all knownGallusgallusm itogenom es was constructed and tested with 1 000 bootstrap replications using the MEGA 7 software package(Tamura et al.,2013).The tree showed that the Thai RJF mitogenome clustered with GU261692 from Yunnan(Miao et al.,2013)into hap logroup X,as defined by Miao etal.(2013)and Peng etal.(2015).To the bestofourknow ledge,alldomestic chickens are distributed in haplogroups A to G.However,the arrangement of the RJF was identical to hap logroup X,which is only found in wild chickens.In the MP tree,all hap logroups had very high bootstrap values,all larger than 90%.The rootof the MP tree was in haplogroup Y,according to the Chicken Reference Tree in dometree.org(Peng et al.,2015).Thus,our study strongly supported the previously defined reference tree.
Here,we assembled the first complete mitogenome of the Thai RJF.This study will provide useful information for future evolutionary and population genetics analyses of the RJF and domestic chicken.
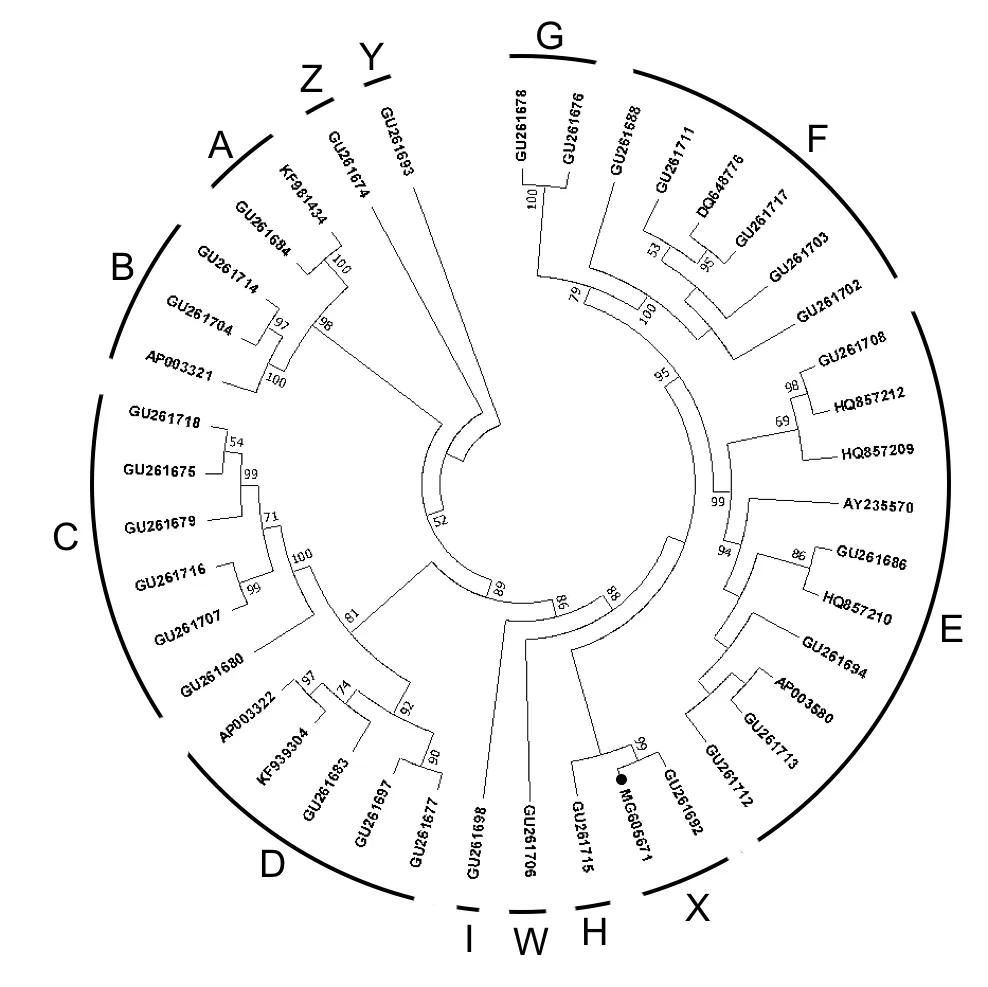
Figu re 1 Phy logenetic tree o f 41 com p lete m itochond rial genom es o f Ga llus gallus cons truc ted w ith m axim um parsim ony
COMPETING INTERESTS
The authors declare that they have no com peting interests.
AUTHORS’CONTRIBUTIONS
The authors a lone are responsible for the contentand w riting of this paper.
Chatmongkon Suwannapoom1,2,#,Ya-Jiang Wu3,#,Xing Chen4,#,AdeniyiC.Adeola4,Jing Chen5,Wen-ZhiWang4,5,6,*
1School of AgricutreandNturalResources,UniversityofPhayao,Phayao56000,Thailand
2SoutheastAsiaBiodiversityResearchInstitute(CAS-SEABRI),ChineseAcademyofSciences,YezinNayPyiTaw05282,Myanmar
3StateKeyLaboratoryforConservationandUtilizationofBio-resourceinYunnan,YunnanUniversity,KunmingYunnan650091,China
4StateKeyLaboratoryofGeneticResourcesandEvolution,KunmingInstituteofZoology,ChineseAcademyofSciences,KunmingYunnan650223,China
5WildlifeForensicsScienceServiceCentre,KunmingYunnan650203,China
6GuizhouAcademyofTestingandAnalysis,GuizhouAcademyof Sciences,GuiyangGuizhou550002,China
#Authors contributed equa lly to this work
Chen X,Ni G,He K,Ding ZL,Li GM,Adeo la AC,Murphy RW,Wang WZ,Zhang YP.2016.An im proved de novo pipeline for enrichment of high diversity mitochondrial genomes from Amphibia to high-throughput sequencing.bioRxiv.080689.
Darw in C.1875.The variation of animals and p lants under domestication.Barrett PH,Freeman RB,editors.New York:New York University Press.
Fumihito A,Miyake T,Sum i SI,Takada M,Ohno S,Kondo N.1994.One subspecies of the red junglefow l(Gallusgallusgallus)suffices as the matriarchic ancestor of all domestic breeds.ProceedingsoftheNational Academyof Sciencesof theUnitedStatesofAmerica,91:12505–12509.
Fumihito A,Miyake T,Takada M,Shingu R,Endo T,Go jobor T,Kondo N,Ohno S.1996.Monophyletic origin and unique dispersal patterns of domestic fowls.Proceedingsof theNational Academyof SciencesoftheUnitedStatesofAmerica,93:6792–6795.
Liu YP,Wu GS,Yao YG,Miao YW,LuikartG,Baig M,Be ja-Pereira A,Ding ZL,Palanicham y MG,Zhang YP.2006.Multip lematernal origins of chickens:out of the Asian jung les.Molecular PhylogeneticsandEvolution,38(1):12–19.Miao YW,Peng MS,Wu GS,Ouyang YN,Yang ZY,Yu N,Liang JP,Pianchou G,Beja-Pereira A,Mitra B,Palanicham y MG,Baig M,Chaudhuri TK,Shen YY,Kong QP,Murphy RW,Yao YG,Zhang YP.2013.Chicken domestication:an updated perspective based on mitochondri algenomes.Heredity,110(3):277–282.
NishiboriM,Shimogiri T,HayashiT,Yasue H.2005.Molecular evidence for hybridization of species in the genusGallusexcept forGallusvarius.Animal Genetics,36:367–375.
Peng MS,Fan L,ShiNN,Ning T,Yao YG,Murphy RW,Wang WZ,Zhang YP.2015.DomeTree:a canonical too lkit form itochondrial DNA analyses in domesticated animals.MolecularEcologyResouces,15:1238–1242.
ShiNN,Fan L,Yao YG,Peng MS,Zhang YP.2014.Mitochondrialgenomes of domestic animals need scrutiny.MolecularEcology,23:5393–5397.
Thorva ldsdóttir H,Robinson JT,Mesirov JP.2013.Integrative Genom ics Viewer(IGV):high-perform ance genom ics data visualization and exp loration.BriefingsinBioinformatics,14:178–192.
Tamura K,Peterson D,Peterson N,Stecher G,Nei M,Kumar S.2011.MEGA5:molecularevolutionary genetics ana lysis usingmaximum likelihood,evolutionary distance,andm aximum parsimonymethods.MolecularBiology andEvolution,28:2731–2739.
Wang YQ,Song FH,Zhu JW,Zhang SS,Yang YD,Chen TT,Tang BX,Dong LL,Ding N,Zhang Q,Bai ZX,Dong XN,Chen HX,Sun MY,Zhai S,Sun YB,Yu L,Lan L,Xiao JF,Fang XD,Lei HX,Zhang Z,Zhao WM.2017.GSA:Genom e Sequence Archive.GenomicsProteomicsBioinformatics,15:14–18.
杂志排行
Zoological Research的其它文章
- In vivo genome editing thrives with diversified CRISPR technologies
- Kidney disease models: tools to identify mechanism s and potential therapeutic targets
- King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates
- Biogeography of the Shimba Hills ecosystem herpetofauna in Kenya
- Discovery of Japalura chapaensis Bourret, 1937(Reptilia: Squamata: Agam idae) from Southeast Yunnan Province, China
- Stereotypy and variability of socialcalls among clustering female big-footed myotis(Myotismacrodactylus)
