In vivo genome editing thrives with diversified CRISPR technologies
2018-05-07XunMaAverySumYuWongHeiYinTamSamuelYungKinTsuiDittmanLaiShunChungBoFeng
Xun Ma, Avery Sum-Yu Wong, Hei-Yin Tam, Samuel Yung-Kin Tsui, Dittman Lai-Shun Chung, Bo Feng,2,3,*
1 Key Laboratory for Regenerative M edicine in M inistry of E ducation, School of B iomedical Sciences, Faculty of M edicine, The Chinese University of Hong Kong, Hong Kong SAR, China
2 Guangzhou Institute of Biomedicine and Health, Chinese Academ y of Sciences, Guangzhou Guangdong 510530, China
3 SBS Core Laboratory, CUHK Shenzhen Research Institute, Shenzhen Guangdong 518057, China
INTRODUCTION
Genome editing by manipulating functional DNA sequences in the host genome is a fundamental strategy for biomedical research. Starting from the discovery of the basic principles of DNA structure and genome organization, scientists have investigated various strategies for many decades to improve genome editing technology for different research and application purposes.
In the 1980s, gene targeting methods emerged together with a deepening understanding of DNA repair mechanisms. Back then, DNA conversion was found to occur between homology sequences, often termed homologous recombination (HR) (Zinn& Butow, 1985). Early studies took advantage of this finding to rep lace a selected endogenous genome DNA segment with a foreign DNA donor carrying homology sequences in living cells(Vasquez et al., 2001). Subsequently, by combining this with mouse em bryonic stem cell (ESC) technology established at the same time, traditional gene targeting technology was developed to generate genetically modified mice (Koller et al.,1989). Since 1989, genetic modification by HR-based gene targeting in living mammals has become a fundamental approach to analyze gene functions and has revolutionized our understanding of mammalian development, metabolism, and genetic diseases (Capecchi, 2005;Koller et al., 1989).
Traditional HR-based gene targeting is associated with low efficiency and requires laborious clonal expansions and sophisticated selections to identify target cells carrying the desired modifications (Koller et al., 1989). With pioneering studies finding that the introduction of double-strand breaks (DSBs) in target DNA by rare-cutting endonuclease I-Sce-1 could increase HR efficiency by several orders of magnitude in the subsequent DNA repair process (Rouet et al., 1994), extensive effort has been made to develop programmable endonucleases.1
Zinc finger nuclease (ZFN), which was first reported in 1986 as an artificial nuclease to carry a zinc finger domain and a catalytic domain from restriction enzyme FokI, was suitable for introducing DNA cleavage and enhancing HR-dependent gene targeting(Bibikova et al., 2002;Kim et al., 1996). However, the laborious work involved in the design and identification of an efficient ZFN to a new ly selected target sequence significantly limited its utility.Transcription activator-like effector protein (TALE), which originated in plant pathogenXanthomonassp.,was found to recognize target DNA with highly conserved yet variable repetitive elements, each showing a preference to bind to specific nucleotides (Boch et al., 2009;Moscou & Bogdanove, 2009).Fusion of the programmable TALE domains and FokI catalytic domain thus yielded TALE-nuclease (TALEN), which is easier to construct and can introduce DNA cleavage and targeted genome modification equally efficiently as ZFN (Christian et al., 2010).
More recently, an RNA-guided DNA-targeting approach was developed from the type II prokaryotic clustered regularly interspaced short palindrom ic repeats (CRISPR) adaptive immune system (Bhaya et al., 2011;Wiedenheft et al., 2012). In this system, a programmable small guide RNA (sgRNA)com plexes with Cas9 nuclease and anneals with a 20-nt target DNA sequence, at the presence of the adjacent NGG PAM(proto-spacer adjacent motif) sequence in a base-pairing manner. This process allows Cas9 to introduce DSB at the target region and enables genome modification in a site-specific manner (Jinek et al., 2012). The ease of constructing a sequence-specific sgRNA and the highly specific RNA-DNA recognition has made the CRISPR/Cas9 system superior to ZFN and TALEN, becom ing the most popular tool for introducing programmed DNA cleavage as well as site-specific genome modifications in cells and animals (Barrangou &Doudna, 2016;Mali et al., 2013;Ran et al., 2013).
These recent advances in engineered nucleases, especially the CRISPR/Cas system, have opened new prospects for accomplishing robust gene targeting in previously non-perm issive cell contexts. More importantly, it has widely revolutionized biomedical research by promoting quick generation of various animal models, which either carry complex genome modifications or are derived from species that could not be genetically modified previously (Dow et al., 2015;Swiech et al., 2015;Yin et al., 2016).Such progress has provided a wide range of methods as well as advanced animal models to study gene function and biological processes, significantly promoting research underinvivoconditions. Hence, in this review, we focus on summarizing the recent developments and applications of CRISPR-based technology in generating various animal models.
OVERVIEW OF RECENT DEVELOPMENTS IN CRISPRBASED ANIMAL MODELS
Since early 2013, when the first successful CRISPR-based genome editing was demonstrated in mammalian cells (Mali et al., 2013), the number of studies using the CRISPR system has grown dramatically. Among the CRISPR-basedinvivostudies,the majority (61.2%) have been conducted using mouse models(Figure 1, left panel). With the comprehensive know ledge and technologies established so far, research investigations using CRISPR technology in mouse models have covered various areas of biomedical research, including inherited metabolic disorders (Xue et al., 2014;Yang et al., 2016), cancer (Maddalo et al., 2014;Platt et al., 2014), neurology and neuroscience (Li et al., 2015c;Sw iech et al., 2015), and virus-related studies(Jiang et al., 2017;Zhu et al., 2016).
In addition to mouse models, CRISPR-based genome editing has been demonstrated in large mammals such as pigs and monkeys to establish disease or genetic models for organ transplantation (Niu et al., 2014;Yu et al., 2016). At the same time,CRISPR/Cas9 technology has also been applied in various lower vertebrate and invertebrate models (Irion et al., 2014;Shi et al.,2015;Wen et al., 2016). The success of CRISPR technology is particularly valuable in lower vertebrate models, such asXenopusand zebrafish (Irion et al., 2014;Shi et al., 2015), in which targeted genome editing could not be achieved previously.
Molecular mechanism s for various genome editing strategies
Sequence-specific DNA cleavage induced by any of the above engineered nucleases will elicit endogenous cellular responses to repair the damaged DNA in target cells. Utilizing various DNA repair mechanisms to induce mutations/deletions or to incorporate insertions of foreign DNA lays the foundation for genome editing. Cellular repair of DNA damage is mediated by two main pathways, namely, homology-directed repair (HDR) and nonhomologous end joining (NHEJ). Despite their varied activities in different cell types and species, both pathways are highly conserved, from yeasts to mammals (Taylor & Lehmann, 1998).
The HDR pathway mediates a strand-exchange process to repair DNA damage based on existing hom ologous DNA sequences (Heyer et al., 2010), allowing precise insertion of foreign DNA at target regions by replacing endogenous genom ic segments with donor DNA. CRISPR/Cas9-introduced site-specific DNA cleavage triggers DNA repair and greatly promotes HR at nearby regions, thus enhancing the efficiency of HDR-based genome editing (Yang et al., 2013). In contrast,the conventional NHEJ pathway initiates DNA repair with quick occupation by the Ku70/Ku80 complex at DNA broken ends,followed by recruitment of other components for end processing and subsequently DNA ligase IV for ligation. NHEJ-based DNA repair is a homology-independent and mechanistically flexible process, which often results in random insertions or deletions(indels) of a small number of nucleotides (Lieber, 2010). Hence,CRISPR/Cas9-induced NHEJ repair has been em ployed to generate loss-of-function alleles in protein-coding genes (Wang et al., 2013). In general, the NHEJ pathway mediates rapid DNA repair and plays an important role in various cellular contexts. Therefore, CRISPR/Cas9-induced NHEJ repair offers high efficiency and has been exploited to develop a variety of targeting strategies.
More recently, in addition to the conventional HDR and NHEJ pathways, studies have discovered the microhomology-mediated end joining (MMEJ) pathway, which is also termed as alternative NHEJ (Alt-NHEJ) pathway (Lieber, 2010;McVey & Lee, 2008).This MMEJ pathway repairs DNA damage by initiating singlestrand resection similar to the HDR process, followed by microhomology-based alignment and ligation by DNA ligase III. In general, the MMEJ pathway mediates an error-prone repair process and plays a minor role to complement DNA repair by HDR and NHEJ (McVey & Lee, 2008). Collectively, the coupling of different DNA repair mechanisms with various strategies to design donor templates or select target sites in genomes, has resulted in a variety of targeting approaches, each having distinct advantages in different species (Table 1).
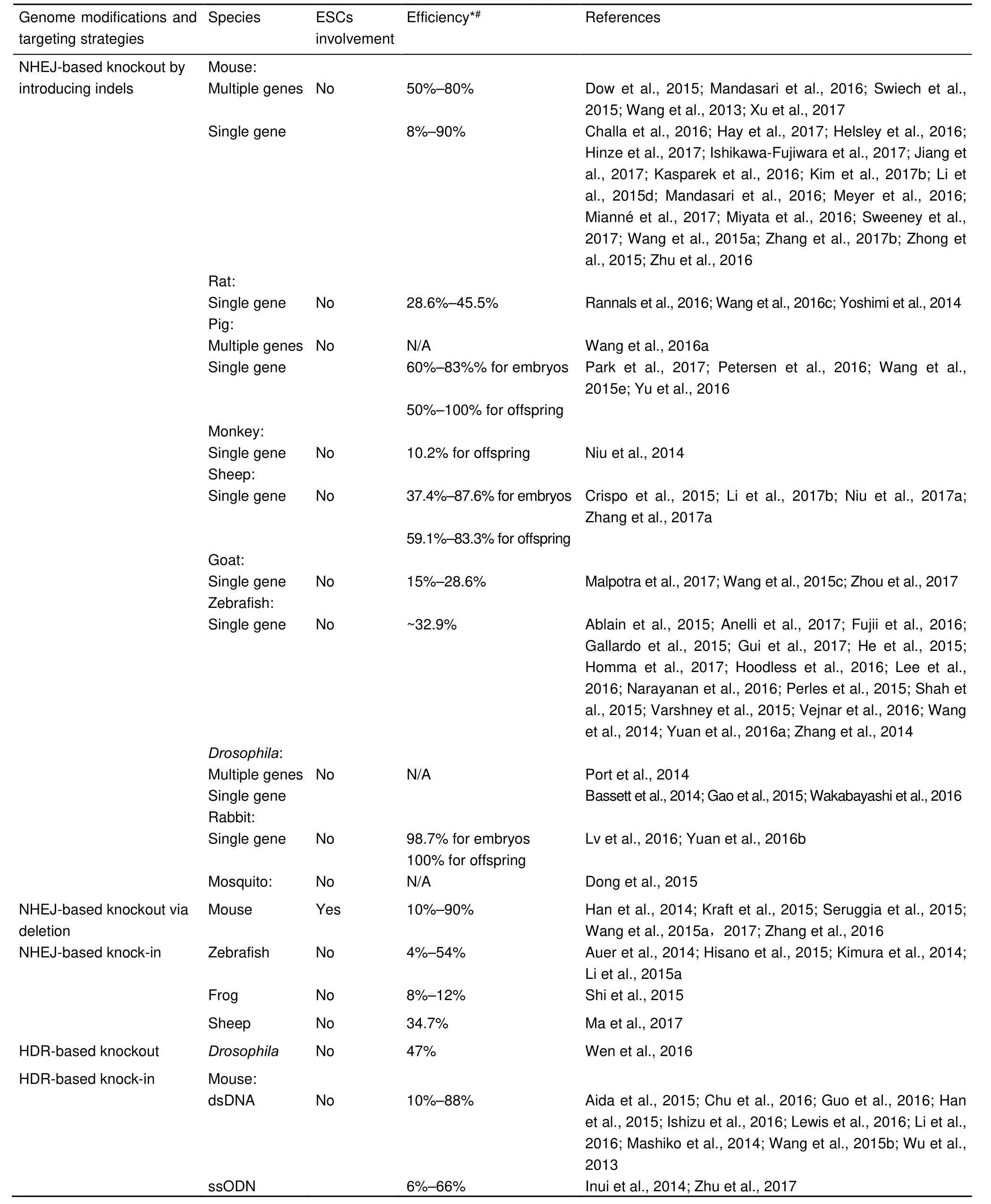
Tab le 1 Summ ary o f CRISPR-based in vivo genom e ed iting in zygo tes via d ifferen t DNA repair m echanism sind ifferen t species
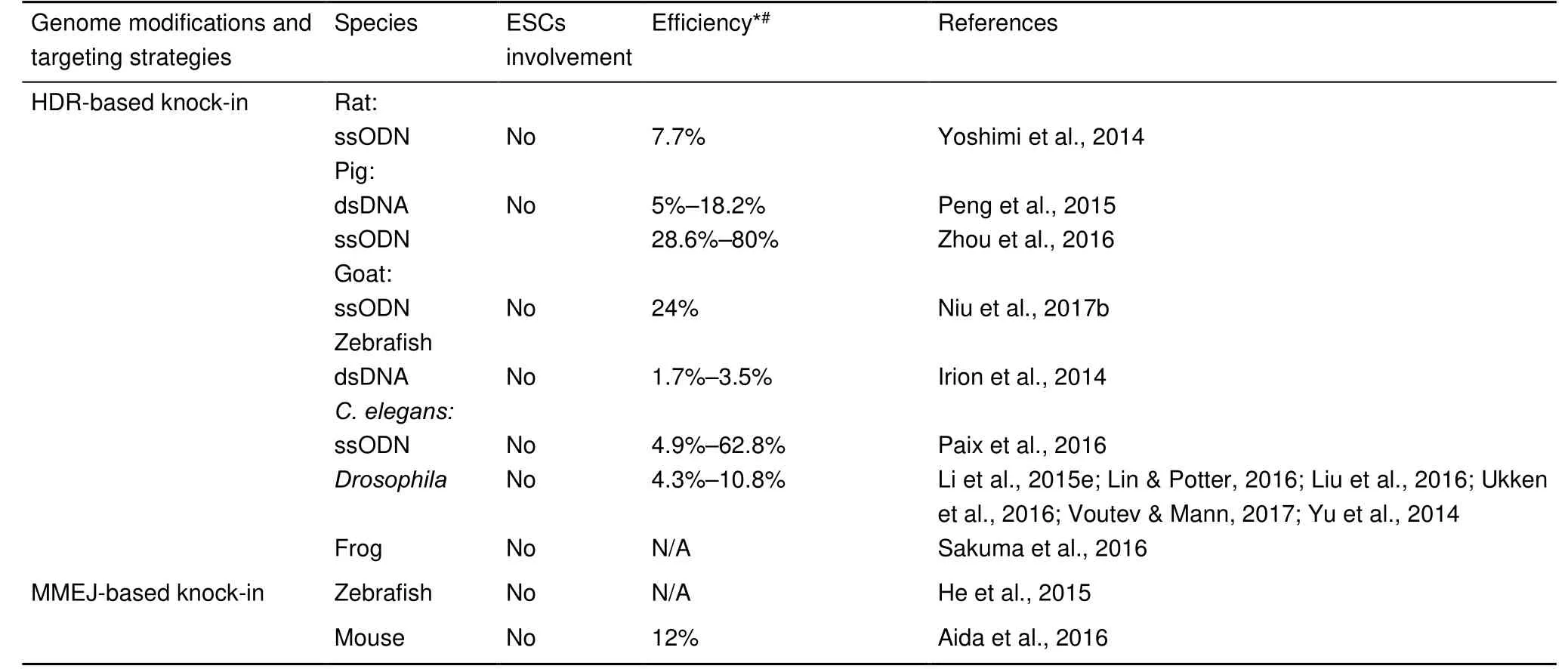
Continued
Enhanced genome editing via CRISPR-induced HDR
HDR is a major DNA repair mechanism broadly employed in CRISPR-based genome editing (Heyer et al., 2010). In the presence of Cas9 nuclease and specific sgRNA targeting a selected sequence in the genome, site-specific DNA cleavage is introduced at the target genomic locus, which then will trigger DNA repair. When the target cells are given a large quantity of donor templates carrying homology sequences, HDR-based repair will utilize the donors as templates to repair the damaged genome, thus introducing foreign DNA included in the donor construct into the recipient genome (Heyer et al., 2010).
The traditional gene targeting approach succeeded before the establishment of engineered nucleases. To accom plish sequence replacement in the genome, this approach relies on the HDR repair process triggered by spontaneous DNA damage that random ly occurs near target regions, (Koller et al., 1989).The desired targeting events occur at low frequency. Hence,successful genome targeting requires long homology arm s in donor constructs, and needs sophisticated selection and clonal expansion in m ouse ESCs before generating chimeric animals and genetically modified offspring (Koller et al., 1989;Thomas &Capecchi, 1987). It often takes more than one year to establish a knock-in or knockout strain of mouse.
Site-specific DNA breaks trigger DNA repair around a target region. Hence, coupling this to the CRISPR system can greatly enhance the efficiency of HDR-based genome targeting and result in a high success rate of desired targeting. This improvement has bypassed the usage of ESC cells, allowing direct genome targeting in mouse zygotes (Yang et al., 2013).The direct genome targeting in zygotes via CRISPR-coup led HDR can produce a high percentage of chimeric animals and genetically modified mouse strains within 3–6 months, a much shortened period of time (Yang et al., 2013). Moreover, direct genome targeting in zygotes has also overcome the lim itations of ESC unavailability, and made genome editing possible in many previously inaccessible organisms, such as pigs and monkeys (Peng et al., 2015). Furthermore, the introduction of site-specific DNA breaks allows the use of much shorter homology arm s to achieve successful genetic modifications.Around 1 000 bp homology fragments are usually sufficient, and around 100 bp single-stranded oligodeoxynucleotides (ssODN)carrying a 50–60 nt hom ology sequence at each side are effective in introducing small mutations/insertions to produce genetically modified animals (Inui et al., 2014;Zhou et al.,2016).
CRISPR-coupled HDR-mediatedinvivogenome editing has been broadly used to introduce knock-in or knockout in the genome of various animal models for studying gene functions,modeling diseases, or developing novel treatment by correcting disease-associated mutations. Direct injection of Cas9 m RNA,sgRNA targeting only the mutant allele, and donor ssODN carrying a w ild-type allele sequence into mouse zygotes carrying a heterozygous dom inant-negative cataract-causing mutation in theCrygcgene resulted in cataract-free progeny(Wu et al., 2013). Besides rodents, large animals like pigs have also been used for disease modeling (Peng et al., 2015;Wang et al., 2015d;Zhou et al., 2016). In these studies, together with the use of the single blastocyst genotyping system and/or ssODN donors, researchers can assess sgRNA efficiency at the embryonic stage and achieve up to 80% targeting efficiency in producing animals carrying the desired genetic modification.Furthermore, successful targeting has also been reported in lower vertebrates and invertebrates (Irion et al., 2014;Li et al.,2015e;Lin & Potter, 2016;Liu et al., 2016;Paix et al., 2016;Sakuma et al., 2016;Ukken et al., 2016;Voutev & Mann, 2017;Yu et al., 2014). Targeted gene modification and tagging has been achieved inDrosophilabased on the CRISPR/Cas9-coupled HDR approach (Li et al., 2015e;Lin & Potter, 2016;Liu et al., 2016;Ukken et al., 2016;Voutev & Mann, 2017;Yu et al.,2014), with a similar method also app lied in zebrafish,producing up to 50% targeted mutations in larvae (Irion et al.,2014). With modified ssODN temp lates and CRISPR com ponents, gene editing efficiency has reached 85% inC.elegans(Paix et al., 2016). Targeted genes or long noncoding RNA (lncRNA) can be precisely replaced with fluorescence reporters to deplete target genes by inserting visible markers(Platt et al., 2014;Wen et al., 2016).
Gene correction in somatic tissues has also been performed using the CRISPR system and donor DNA (Table 2). Targeting of deficient ornithine transcarbamylase in the mouse model showed more than 10% correction of the deficient gene in liver cells and significantly improved the survival rate in target groups(Yang et al., 2016). Sim ilarly, somatic correction of Duchenne muscular dystrophy (DMD) caused by a mutation in the gene encoding dystrophin has been reported, showing a 70%increase in functional dystrophin and apparent im provement in the mouse model (Bengtsson et al., 2017).

Tab le 2 Summ ary o f CRISPR-based in vivo genome editing in somatic tissues
Diverse targeting strategies th rough CRISPR-induced NHEJ-m ed iated DNA repair
Double-strand DNA breaks due to the disruption of phosphodiester bonds between adjacent nucleotides in doublehelix DNA. While HDR repairs a broad range of DNA damage,NHEJ is the primary mechanism for repairing DSBs in mammalian cells. W ith site-specific DSBs able to be introduced at almost any target site in the genome with high efficiency and accuracy using the CRISPR system, the NHEJ repair mechanism has been broadly employed to introduce random mutations at selected target sites. This CRISPR-coupled NHEJ-based mutagenesis approach can disrupt protein coding potential of a target gene by causing frame shift or premature term ination, and therefore dep lete functional proteins and introduce loss-of-function effects (Figure 1). To date, most animal models established using CRISPR technology have em ployed this strategy to knockout a specific gene, especially model organism s that are incom patible with the traditional HDR-based strategy, such as zebrafish orXenopus(broadly noticed via personal communications) (Table 1 and 2) (Auer & Del Bene, 2014;Irion et al., 2014;Won & Daw id, 2017).Furthermore, due to its simp le principles and procedures,CRISPR-NHEJ-based mutagenesis has been applied in highthroughput studies. Xuetal.reported successful loss-offunction screening to identify genes essential to tumorigenesis in mice using pre-constructed sgRNA libraries (Xu et al., 2017).Interestingly,invivoapp lication of a sgRNA library has also been reported in zebrafish (Shah et al., 2015). Combining CRISPR-based high-throughput screening with excellent accessibility to embryonic development, straight-forward phenotyping has allowed large scale analysis of gene function.Shawn M. Burgess and colleagues have verified more than 50 genes by this method (Varshney et al., 2015), and Stefania Nicolia’s team has succeeded in a similar screening using the sgRNA pool-targeting miRNA family (Narayanan et al., 2016).
In addition, NHEJ repair has been found to be highly efficient in re-ligating DNA ends from DSBs concurrently produced by the CRISPR system at two different genome loci, despite the long distance in genome. In support of these observations, the CRISPR-coupled NHEJ repair mechanism has also been emp loyed to delete selected large DNA fragments by targeting two regions in the same chromosome (Dow et al., 2015;Han et al., 2014;Wang et al., 2015b) or catalyzing the desired genom ic rearrangements by targeting two selected regions from different chromosomes (Blasco et al., 2014). These strategies have succeeded in generating mouse models carrying a 353-kb intragenic deletion ofLaf4,which recapitulates a human malformation syndrome (Kraft et al., 2015), and engineering mouse models that harbor chromosomal rearrangements recurrently found in lung cancer to model carcinogenesis(Blasco et al., 2014;Maddalo et al., 2014). The functional study of lncRNA genes is another im portant application of NHEJ-mediated large fragment deletion. Knockout of the lncRNA geneRianthrough a large deletion of up to 23 kb demonstrated efficiency as high as 33% (Han et al., 2014) can be achieved,w ith similar results reported for the tyrosinase (Tyr) associated lncRNA gene (Seruggia et al., 2015).
Rather strikingly, CRISPR-coupled NHEJ repair has also enabled high-efficiency knock-in of exogenous DNA at preselected locations. This is consistent with common observations that NHEJ is the predom inant repair mechanism in mammalian cells. Since the early 1980s, transgenic technology has been established and applied broadly to render stable ectopic expression by introducing foreign DNA fragments carrying complete gene cassettes into host genomes (Palm iter et al.,1982). Later studies have found that the NHEJ repair mechanism is responsible for capturing foreign DNA fragments at spontaneously occurring DSBs in the genome, resulting in random integrations (Lin & Waldman, 2001). Consistently,traditional gene targeting studies have also shown that the frequency of random DNA integration via the NHEJ repair mechanism is significantly higher (over 1 000-fold) than targeted insertion mediated by the HDR pathway (Vasquez et al., 2001).Due to the unavailability of programmable site-specific nucleases and their erroneous nature, the potential of the NHEJ mechanism in targeted DNA knock-in was largely neglected for a long time.
Until recently, after ZFN was successfully established, short oligonucleotides (<100 bp) were able to be inserted efficiently at ZFN-induced DSBs via NHEJ repair (Orlando et al., 2010).Subsequently, inclusion of a ZFN or TALEN target sequence in donor vectors showed that simultaneous cleavage of donor and genome DNA could enable targeted integration via NHEJ repair(Cristea et al., 2013;Maresca et al., 2013). Using promoterless fluorescence reporters followed by direct quantification using fluorescence-activated cell sorting (FACS), we com pared the frequencies of NHEJ- and HDR-mediated knock-in after coupling with the CRISPR system (He et al., 2016). We found that knock-in via CRISPR/Cas9-induced NHEJ is superior to the commonly used HDR-based m ethod in all human cell lines exam ined (He et al., 2016). This NHEJ-based knock-in approach has been applied in precise reporter knock-in in zebrafish (Auer et al., 2014;Hisano et al., 2015;Irion et al.,2014;Kimura et al., 2014;Li et al., 2015a) andXenopus(Shi et al., 2015), with such gene targeting previously impeded by the deficiency of the HDR pathway. More recently, CRISPR/Cas9-induced NHEJ has been shown to mediate high efficiency knock-in in mouse somatic tissues (Suzuki et al., 2016), but success in targeting zygotes or blastocysts to generate genetically modified mice has not yet been reported.
Through CRISPR-coupled NHEJ repair, various genome targeting strategies have been established and utilized in generating genetically modified animal models. From studies published since early 2013, 75.9% (110/145) ofinvivogenome editing studies have employed NHEJ-based targeting strategies.Extensive evidence has shown that NHEJ-based genome targeting is simpler, more flexible, and more efficient compared with HDR-based approaches. W ithout homology sequences involved, the design and system construction for NHEJ-based strategies are less laborious. On the other hand, however, the random nature of NHEJ repair incurs disadvantages including the unpredictability of indel-based mutagenesis as well as offtarget cleavage and insertion.
Genom e ed iting by CRISPR-induced MMEJ repair
Distinct from NHEJ and HDR, the two common form s of DNA repair, MMEJ requires microhomologous sequences of only 5–25 bp for the repair of DSBs in DNA. Sakuma et al. devised a detailed protocol for CRISPR-based gene knock-in using MMEJ, termed Precise Integration into Target Chromosomes(PITCh) (Sakuma et al., 2016).
In this system, DSBs are needed in both the genom ic DNA and donor vector to insert a DNA fragment from the donor into the genome. As MMEJ repair requires the presence of m icrohomology both upstream and downstream of the DSB site, two microhomologous sequences need to be added to the donor vector at both sides of the purpose sequence (Sakuma et al., 2016). For the CRISPR system, two sgRNAs are required to generate DNA cleavages near the m icrohomology sequences on both sides, while one sgRNA is used to induce DSBs on the genome DNA (Figure 2). Longer m icrohomologies of around 20 bp are currently used to improve accuracy. After alignment between microhomologous sequences, the unmatched nonhomologous sequences at the 3'-parts on both sides of the donor appear as single-strand tails and are removed. This results in the loss of a small part of the genome sequence at the target sites. Therefore, MMEJ-based genome editing is associated with deletion/insertions that are often larger than NHEJ-introduced in dels (Villarreal et al., 2012).
Targeted integration mediated by CRISPR-coupled MMEJ has been demonstrated in cultured cells and the generation of genetically modified zebrafish (He et al., 2015;Hisano et al.,2015;Nakade et al., 2014). Moreover, one-step knock-in of gene cassettes and floxed alleles has also been achieved in human cells and mouse zygotes through MMEJ (Aida et al.,2016). Recently, precisely targeted gene integration in somatic tissues to correct mutation of theFahgene and rescue liver failure inFah-/-mice has also been demonstrated (Yao et al.,2017).
Comparison between different targeting strategies
Conventional NHEJ repair does not require the presence of homology sequences and involves minimal processing of DNA broken ends. The activity of the NHEJ pathway is high and stable throughout the cell cycle. Distinctly, the HDR repair mechanism relies on long homology sequences (> 500 bp in general) to repair DNA lesions, and is only active from the late S phase to G2 phase during the cell cycle. The MMEJ pathway depends on microhomology sequences (5–25 bps) for DSB repair and is active during the M to early S phase (Taleei &Nikjoo, 2013). These differences explain why the activities of the different DNA repair pathways vary in different cell contexts.
The intrinsic activities of the two major pathways, HDR and NHEJ, also vary in different species, despite high conservation of these pathways across a broad range of organism s. Lower vertebrates, such as zebrafish andXenopus,are deficient in HDR-based DNA repair. Hence, modification of genome sequences in these models has mainly succeeded with NHEJ-based strategies, such as transgenesis, indel-based targeted mutagenesis/deletion, or the recent knock-in approach based on coup ling TALEN- or CRISPR-induced DNA cleavage to the NHEJ repair mechanism (Auer et al., 2014;Hisano et al., 2015;Irion et al., 2014;Kimura et al., 2014;Li et al., 2015a;Shi et al.,2015) (Table 1). In mammalian systems, although HDR was first em ployed to produce genetically modified mice, evidence shows that the NHEJ repair mechanism is predominant(Vasquez et al., 2001). Thus, the efficiency of NHEJ-based genome editing is generally superior to HDR-based approaches(He et al., 2016).
Scientists have attempted to manipulate the balance between the HDR and NHEJ pathways. Through inhibiting DNA ligase IV,a key component of the NHEJ pathway, studies have shown that the efficiency of HDR-based gene targeting can be increased substantially (Chu et al., 2015). Similarly, silencing KU70, KU80, or DNA ligase IV largely suppressed NHEJ-mediated introduction of indels at the junction and enhanced HDR-mediated genome editing (Pierce et al., 2001). To date,this type of approach has not been app lied forinvivogene targeting.
Besides efficiency, accuracy is another major concern. The HDR-based targeting strategy requires homology sequences as a template for DNA replication to repair induced DNA cleavage.It involves the cloning of homologous DNA and multi-step construction of donor plasm ids. In return, the designed modifications can be introduced into the genome with high accuracy and off-target integrations can be largely reduced compared to other knock-in strategies. MMEJ-based targeting requires microhomologous sequences, which can be easily introduced into donor vectors through synthesized oligos, or during PCR amplification of the desired DNA for insertion.Although the intrinsic MMEJ pathway often plays a m inor role in overall DNA repair, the MMEJ-based targeting strategy has shown efficiency up to 10-fold higher than that of the HDR-based approach (Yao et al., 2017). Lastly, the NHEJ repair mechanism, which is com pletely independent of any homology sequences, offers the easiest path to modify an existing design for a new target site in the genome. In our recent study, a universal donor was established with the use of artificial sgRNA,which did not target any sequence in mice and humans (He et al., 2016). With the m inimum work involved in constructing the new sgRNA to the genome, the whole system was easily orientated for targeting a new locus (He et al., 2016). However,the random errors potentially present at the integration/ repair junctions with NHEJ-based targeting approaches should be considered during the design.
CONCLUSIONS
The recent advent of CRISPR technology has offered the simplest and possibly ultimate solution for introducing sitespecific DSBs in genome DNA, which was once an insurmountable challenge in genome editing. Through coupling with different DNA repair mechanisms present in the endogenous cellular system, various targeting strategies have been developed to introduce a wide range of modifications in the genome through sequence-based editing. While further research is needed to evaluate the off-target issues and overcome the risks by developing improved CRISPR system s,the above technological advances have undoubtedly revolutionized biomedical research. The CRISPR-based genome editing approaches have significantly promoted studies on gene function via the rapid generation of animal models that carry genetic deficiencies of single or multiple genes. In addition, they have also enabled modeling of genetic diseases caused by chromosomal rearrangement or large deletions. Therefore,rapid progress could be foreseen in establishing various animal models for disease modeling or therapeutic intervention, which w ill significantly im prove our understanding of human diseases and promote the development of new therapeutic strategies.
COMPETING INTERESTS
The authors declare that they have no com peting interests.
AUTHORS’ CONTRIBUTIONS
X.M. A.S.W., H.Y.T., S.Y.T, and D.L.C. w rote different parts of the manuscript;B.F. com piled and revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank Xiang-Jun He and Chen-Zi Zhang for critical comments on the manuscript.
Ab lain J, Durand EM, Yang S, Zhou Y, Zon LI. 2015. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish.Developmental Cell,32(6): 756–764.
Aida T, Chiyo K, Usam i T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF,Sakuma T, Yam amoto T, Tanaka K. 2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice.Genome Biology,16:87.
Aida T, Nakade S, Sakuma T, Izu Y, Oishi A, Mochida K, Ishikubo H, Usam i T, Aizawa H, Yamam oto T, Tanaka K.2016. Gene cassette knock-in in mammalian cells and zygotes by enhanced MMEJ.BMC Genomics,17:979.
Anelli V, Villefranc JA, Chhangawala S, Martinez-McFaline R, Riva E,Nguyen A, Verma A, Bare ja R, Chen Z, Scognam iglio T, Elemento O,Houvras Y.2017. Oncogenic BRAF disrupts thyroid morphogenesis and function via tw ist expression.ELife, doi: 10.7554/eLife.20728.
Auer TO, Del Bene F. 2014. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish.Methods,69(2): 142–150.
Barrangou R, Doudna JA. 2016. App lications of CRISPR technologies in research and beyond.Nature Biotechnology,34(9): 933–941.
Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, McGowan S,Stanger N, Ewe ls PA, Taylor S, Ponting CP, Liu JL, Sauka-Spengler T,Fulga TA. 2014. Understanding functional m iRNA-target interactionsin vivoby site-specific genom e engineering.Nature Communications,5: 4640.
Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD,Hauschka SD, Chamberlain JR, Chamberlain JS. 2017. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysio logy in a mouse model for Duchenne muscular dystrophy.Nature Communications,8: 14454.
Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas system s in bacteria and archaea: versatile small RNAs for adaptive defense and regulation.Annual Review of Genetics,45(1): 273–297.
Bibikova M, Golic M, Go lic KG, Carro ll D. 2002. Targeted chromosoma l cleavage and mutagenesis in Drosophila using zinc-finger nucleases.Genetics,161(3): 1169–1175.
Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG,Voena C, Chiarle R. 2014. Sim p le and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology.Cell Reports,9(4): 1219–1227.
Boch J, Scho lze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T,Nickstadt A, Bonas U. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors.Science,326(5959): 1509–1512.
Capecchi MR. 2005. Gene targeting in mice: functional analysis of the m ammalian genome for the twenty-first century.Nature Reviews,6(6):507–512.
Challa AK, Boitet ER, Turner AN, Johnson LW, Kennedy D, Downs ER,Hymel KM, Gross AK, Kesterson RA. 2016. Novel hypom orphic alleles of the mouse tyrosinase gene induced by CRISPR-Cas9 nucleases cause non-albino pigmentation phenotypes.PLoSOne,11(5): e0155812.
Chen S, Lee B, Lee AYF, Modzelewski AJ, He L. 2016. Highly e fficient m ouse genome editing by CRISPR ribonucleoprotein e lectroporation of zygotes.Journal of Biological Chemistry,291(28): 14457–14467.
Cheng RR, Peng J, Yan YH, Cao PL, Wang JW, Qiu C, Tang LC, Liu D,Tang L, Jin JP, Huang XX, He FC, Zhang PM. 2014. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9.FEBS Letters,588(21): 3954–3958.
Chiou SH, W inters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, Brady JJ, Yang D, Grüner BM, Chuang CH, Caswell DR, Zeng H, Chu P, Kim GE,Carpizo DR, Kim SK, W inslow MM. 2015. Pancreatic cancer mode ling using retrograde viral vector de livery and in vivo CRISPR/Cas9-mediated somatic genome editing.Genes & Development,29(14): 1576–1585.
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Humm el A,Bogdanove AJ, Voytas DF. 2010. Targeting DNA double-strand breaks with TAL effector nucleases.Genetics,186(2): 757–761.
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R.2015. Increasing the efficiency of homology-directed repair for CRISPRCas9-induced precise gene editing in mamm alian cells.Nature Biotechnology,33(5): 543–548.
Chu VT, Weber T, Gra f R, Sommermann T, Petsch K, Sack U, Volchkov P,Ra jewsky K, Kühn R. 2016. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes.BMC Biotechnology,16: 4.
Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EHA, Pedrio li DML,McLean WHI, Nesbit MA, Moore CBT. 2016. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables bothin vitroandin vivoKRT12mutationspecific targeting.Gene Therapy,23(1): 108–112.
Crispo M, Mu let AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC,Nguyen TH, Crénéguy A, Brusselle L, Anegón I, Menchaca A. 2015.Efficient generation of m yostatin knockout sheep using CRISPR/Cas9 technology and m icroinjection into zygotes.PLoSOne,10(8): e0136690.
Cristea S, Freyvert Y, Santiago Y, Ho lmes MC, Urnov FD, Gregory PD,Cost GJ. 2013. In vivo cleavage of transgene donors promotes nucleasem ediated targeted integration.Biotechnology and Bioengineering,110(3):871–880.
de Solis CA, Ho A, Holehonnur R, Ploski JE. 2016. The deve lopment of a viral m ediated CRISPR/Cas9 system with doxycycline dependent gRNA expression for induciblein vitroandin vivogenome editing.Frontiers in Molecular Neuroscience,9: 70.
Ding QR, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA,Rader DJ, Musunuru K. 2014. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing.Circulation Research,115(5): 488–492.
Dong SZ, Lin JY, Held NL, Clem RJ, Passare lli AL, Franz AWE. 2015.Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito,Aedes aegypti.PLoS One,10(3): e0122353.
Dow LE, Fisher J, O'Rourke KP, Muley A, Kastenhuber ER, Livshits G,Tschaharganeh DF, Socci ND, Lowe SW. 2015. Induciblein vivogenom e editing with CRISPR-Cas9.Nature Biotechnology,33(4): 390–394.
El Fatim y R, Subramanian S, Uhlmann EJ, Krichevsky AM. 2017. Genome editing revea ls glioblastoma addiction to MicroRNA-10b.Molecular Therapy,25(2): 368–378.
Fujii T, Tsunesum i S, Sagara H, Munakata M, Hisaki Y, Sekiya T, Furukawa Y, Sakamoto K, Watanabe S. 2016. Smyd5 plays pivotal roles in both prim itive and de finitive hematopoiesis during zebrafish em bryogenesis.Scientific Reports,6: 29157.
Gallardo VE, Varshney GK, Lee M, Bupp S, Xu L, Shinn P, Craw ford NP,Inglese J, Burgess SM. 2015. Phenotype-driven chem ical screening in zebrafish for com pounds that inhibit collective cell m igration identifies mu ltip le pathways potentially involved in metastatic invasion.Disease Models & Mechanisms,8(6): 565–576.
Gao JL, Fan YJ, Wang XY, Zhang Y, Pu J, Li L, Shao W, Zhan S, Hao J, Xu YZ. 2015. A conserved intronic U1 snRNP-binding sequence prom otestrans-sp licing inDrosophila.Genes & Develpoment,29(7): 760–771.
Gui HS, Schriemer D, Cheng WW, Chauhan RK, Antiňo lo G, Berrios C,Bleda M, Brooks AS, Brouwer RWW, Burns AJ, Cherny SS, Dopazo J,Eggen BJL, Griseri P, Ja lloh B, Le TL, Lui VCH, Luzón -Toro B, Matera I,Ngan ESW, Pelet A, Ruiz-Ferrer M, Sham PC, Shepherd IT, So MT,Sribudiani Y, Tang CSM, van den Hout MCGN, van der Linde HC, van Ham TJ, van IJcken WFJ, Verheij JBGM, Am iel J, Borrego S, Ceccherini I,Chakravarti A, Lyonnet S, Tam PKH, Garcia-Barce ló MM, Hofstra RMW.2017. Whole exome sequencing coup led with unbiased functiona l analysis reveals new Hirschsprung disease genes.Genome Biology,18(1): 48.
Guo H, Cooper S, Friedm an AD. 2016.In vivode letion of thecebpa+37 kb enhancer m arked ly reducescebpam RNA in m yeloid progenitors but not in non-hematopoietic tissues to im pair granu lopoiesis.PLoS One,11(3):e0150809.
Guo YX, VanDusen NJ, Zhang LN, Gu WL, Sethi I, Guatimosim S, Ma Q,Jardin BD, Ai YL, Zhang DH, Chen BY, Guo A, Yuan GC, Song LS, Pu WT.2017. Analysis of cardiac m yocyte maturation using CASAAV, a p latform for rapid dissection of cardiac myocyte gene function in vivo.Circulation Research,120(12): 1874–1888.
Han JX, Zhang J, Chen L, Shen B, Zhou JK, Hu B, Du YN, Tate PH, Huang XG, Zhang WS. 2014. Efficient in vivo deletion of a large im p rinted lncRNA by CRISPR/Cas9.RNA Biology,11(7): 829–835.
Han Y, Slivano OJ, Christie CK, Cheng AW, Miano JM. 2015. CRISPRCas9 genome editing of a single regulatory e lem ent nearly abolishes target gene expression in mice--brie f report.Arteriosclerosis, Thrombosis, and Vascular Biology,35(2): 312–315.
Hay EA, Khalaf AR, Marini P, Brown A, Heath K, Sheppard D, MacKenzie A.2017. An ana lysis of possible off target effects following CAS9/CRISPR targeted deletions of neuropep tide gene enhancers from the mouse genome.Neuropeptides,64: 101–107.
He MD, Zhang FH, Wang HL, Wang HP, Zhu ZY, Sun YH. 2015. Efficient ligase 3-dependent m icrohomo logy-mediated end joining repair of DNA double-strand breaks in zebrafish embryos.Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis,780:86–96.
He XJ, Tan CL, Wang F, Wang YF, Zhou R, Cui DX, You WX, Zhao H, Ren JW, Feng B. 2016. Knock-in of large reporter genes in human ce lls via CRISPR/Cas9-induced homology-dependent and independent DNA repair.Nucleic Acids Research,44(9): e85.
Heckl D, Kowalczyk MS, Yudovich D, Be lizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. 2014. Generation of mouse models of m ye loid m alignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing.Nature Biotechnology,32(9): 941–946.
Helsley RN, Sui YP, Park SH, Liu Z, Lee RG, Zhu BB, Kern PA, Zhou CC.2016. Targeting IκB kinase β in adipocyte lineage cells for treatment of obesity and m etabolic dysfunctions.StemCells,34(7): 1883–1895.
Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes.Annual Review of Genetics,44(1): 113–139.
Hinze SJ, Jackson MR, Lie S, Jolly L, Fie ld M, Barry SC, Harvey RJ,Shoubridge C. 2017. Incorrect dosage of IQSEC2, a known inte llectual disability and epilepsy gene, disrupts dendritic spine morphogenesis.Translational Psychiatry,7: e1110.
Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S, Okamoto H, Yamamoto T,Kawahara A. 2015. Precise in-fram e integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish.Scientific Reports,5: 8841.
Homma N, Harada Y, Uchikawa T, Kamei Y, Fukamachi S. 2017.Protanopia (red color-blindness) in medaka: a sim p le system for producing color-blind fish and testing their spectral sensitivity.BMC Genetics,18: 10.Hood less LJ, Lucas CD, Duffin R, Denvir MA, Haslett C, Tucker CS, Rossi AG. 2016. Genetic and pharm acological inhibition of CDK9 drives neutrophil apoptosis to resolve in flammation in zebrafishin vivo.Scientific Reports,5: 36980.
Hung SSC, Chrysostomou V, Li F, Lim JK, Wang JH, Powell JE, Tu L,Daniszewski M, Lo C, Wong RC, Crowston JG, Pébay A, King AE, Bui BV,Liu GS, Hewitt AW. 2016. AV-mediated CRISPR/Cas gene editing of retinal cellsin vivo.Investigative Ophthalmology & Visual Science,57(7): 3470–3476.
Inui M, Miyado M, Igarashi M, Tamano M, Kubo A, Yamashita S, Asahara H,Fukam i M, Takada S. 2014. Rapid generation of mouse m odels with defined point mutations by the CRISPR/Cas9 system.Scientific Reports,4:5396.
Irion U, Krauss J, Nusslein-Volhard C. 2014. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system.Development,141(24):4827–4830.
Ishikawa-Fujiwara T, Shiraishi E, Fujikawa Y, Mori T, Tsu jimura T, Todo T.2017. Targeted inactivation of DNA photo lyase genes in m edaka fish(Oryzias latipes).Photochemistry and Photobiology,93(1): 315–322.
Ishizu N, Yui DS, Hebisawa A, Aizawa H, Cui WP, Fu jita Y, Hashim oto K,Ajioka I, Mizusawa H, Yokota T, Watase K. 2016. Im paired striatal dopam ine re lease in hom ozygous Vps35 D620N knock-in mice.Human Molecular Genetics,25(20): 4507–4517.
Jiang C, Mei M, Li B, Zhu XR, Zu WH, Tian YJ, Wang QN, Guo Y, Dong YZ,Tan X. 2017. A non-vira l CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA andpcsk9in vivo.Cell Research,27(3): 440–443.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012.A programmable dual-RNA-guided DNA endonuclease in adap tive bacterial immunity.Science,337(6096): 816–821.
Kalebic N, Taverna E, Tavano S, Wong FK, Suchold D, W inkler S, Huttner WB, Sarov M. 2016. CRISPR/Cas9-induced disruption of gene expression in m ouse embryonic brain and single neural stem cellsin vivo.EMBO Reports,17(3): 338–348.
Kam inski R, Bella R, Yin C, Otte J, Ferrante P, Gendelman HE, Li H, Booze R, Gordon J, Hu W, Khalili K. 2016. Excision of HIV-1 DNA by gene editing:a proof-of-conceptin vivostudy.Gene Therapy,23(8–9): 690–695.
Kasparek P, Ileninova Z, Haneckova R, Kanchev I, Jenickova I, Sed lacek R.2016. A viab le mouse mode l for Netherton synd rome based on m osaic inactivation of theSpink5gene.Biological Chemistry,397(12): 1287–1292.Katigbak A, Cencic R, Robert F, Senecha P, Scuoppo C, Pelletier J. 2016. A CRISPR/Cas9 functional screen identifies rare tumor suppressors.Scientific Reports,6: 38968.
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S,Kim JH, Kim JH, Kim JS. 2017a.In vivogenome editing with a small Cas9 orthologue derived fromCampylobacterjejuni.Nature Communications,8: 14500.
Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, Kim KE, Kim JH, Kim JS.2017b. Genom e surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration.Genome Research,27(3): 419–426.
Kim YG, Cha J, Chandrasegaran S. 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain.Proceedings of the National
Academy of Sciences of the United States of America,93(3): 1156–1160.
Kimura Y, Hisano Y, Kawahara A, Higashijim a S. 2014. Efficient generation of knock-in transgenic zebrafish carrying reporter/d river genes by CRISPR/Cas9-m ediated genom e engineering.Scientific Reports,4: 6545.Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, W illiam s PJ, First NL, Maeda N, Sm ithies O. 1989. Germ-line transm ission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homo logous recom bination in embryonic stem ce lls.Proceedings of the National Academy of Sciences of the United States of America,86(22):8927–8931.
Kraft K, Geuer S, W ill AJ, Lee Chan W, Paliou C, Borschiwer M, Harabula I,W ittler L, Franke M, Ibrahim DM, Kragesteen BK, Spielmann M, Mund los S,Lupiáñez DG, Andrey G. 2015. Deletions, inversions, duplications:engineering of structural variants using CRISPR/Cas in mice.Cell Reports,10(5): 833–839.
Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Com itato A,Marigo V, Recchia A. 2016.In vivoediting of the human mutant rhodopsin gene by electroporation of p lasm id-based CRISPR/Cas9 in the Mouse Retina.Molecular Therapy Nucleic Acids,5: e389.
Lee RT, Ng AS, Ingham PW. 2016. Ribozyme mediated gRNA generation forin vitroandin vivoCRISPR/Cas9 mutagenesis.PLoS One,11(11):e0166020.
Lewis WR, Malarkey EB, Tritschler D, Bower R, Pasek RC, Porath JD,Birket SE, Saunier S, Antignac C, Know les MR, Leigh MW, Zariwa la MA,Challa AK, Kesterson RA, Rowe SM, Drummond IA, Parant JM,Hildebrandt F, Porter ME, Yoder BK, Berbari NF, Dutcher SK. 2016.Mutation of grow th arrest specific 8 reveals a role in motile cilia function and human disease.PLoS Genetics,12(7): e1006220.
Li J, Zhang BB, Ren YG, Gu SY, Xiang YH, Huang C, Du JL. 2015a. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system.Cell Research,25(5): 634–637.Li L, Song LJ, Liu XW, Yang X, Li X, He T, Wang N, Yang S, Yu C, Yin T,Wen YZ, He ZY, Wei XW, Su WJ, Wu QJ, Yao SH, Gong CY, Wei YQ.2017a. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice.ACSNano,11(1): 95–111.
Li MC, Feng B, Wang L, Guo S, Zhang P, Gong J, Zhang Y, Zheng AK, Li HL. 2015c. Tollip is a critical mediator of cerebra l ischaem ia-reperfusion injury.The Journal of Pathology,237(2): 249–262.
Li MY, Huang R, Jiang X, Chen YX, Zhang Z, Zhang XY, Liang PP, Zhan SQ, Cao SB, Zhou SY, Huang JJ. 2015d. CRISPR/Cas9 promotes functional study of testis specific X-linked genein vivo.PLoS One,10(11):e0143148.
Li QY, Barish S, Okuwa S, Volkan PC. 2015e. Exam ination of endogenous rotund exp ression and function in developingDrosophilao lfactory system using CRISPR-Cas9-m ediated protein tagging.G3(Bethesda),5(12):2809–2816.
Li WR, Liu CX, Zhang XM, Chen L, Peng XR, He SG, Lin JP, Han B, Wang LQ, Huang JC, Liu MJ. 2017b. CRISPR/Cas9-mediated loss of FGF5 function increases wool staple length in sheep.FEBS Journal,284(17):2764–2773.
Li XL, Li YQ, Han GY, Li XR, Ji YS, Fan ZR, Zhong YL, Cao J, Zhao J,Mariusz G, Zhang MZ, Wen JG, Nesland JM, Suo ZH. 2016. Establishment of mitochondrial pyruvate carrier 1 (MPC1) gene knockout mice with preliminary gene function analyses.Oncotarget,7(48): 79981–79994.
Liang WC, Liang PP, Wong CW, Ng TB, Huang JJ, Zhang JF, Waye MMY,Fu WM. 2017. CRISPR/Cas9 technology targeting fas gene protects mice from concanavalin-a induced fulminant hepatic failure.Journal of Cellular Biochemis try,118(3): 530–536.
Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway.Annual Review of Biochemistry,79: 181–211.
Lin CC, Potter CJ. 2016. Non-mendelian dominant maternal effects caused by CRISPR/Cas9 transgenic com ponents inDrosophilamelanogaster.G3(Bethesda),6(11): 3586–3691.
Lin Y, Waldman AS. 2001. Capture of DNA sequences at double-strand breaks in mammalian chromosomes.Genetics,158(4): 1665–1674.
Liu Y, Lin JJ, Zhang MJ, Chen K, Yang SX, Wang Q, Yang HQ, Xie SS,Zhou YJ, Zhang X, Chen F, Yang YF. 2016. PINK1 is required for timely cell-type specific mitochondrial clearance duringDrosophilamidgut metamorphosis.Developmental Biology,419(2): 357–372.
Long CZ, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E,Bhattacharyya S, Shelton JM, Basse l-Duby R, Olson EN. 2016. Postnata l genom e editing partially restores dystrophin expression in a m ouse mode l of muscular dystrophy.Science,351(6271): 400–403.
Lv QY, Yuan L, Deng JC, Chen M, Wang Y, Zeng J, Li ZJ, Lai LX. 2016.Efficient generation ofMyostatingene mutated rabbit by CRISPR/Cas9.Scientific Reports,6: 25029.
Ma T, Tao JL, Yang MH, He CJ, Tian XZ, Zhang XS, Zhang JL, Deng SL,Feng JZ, Zhang ZZ, Wang J, Ji PY, Song YK, He PL, Han HB, Fu JC, Lian ZX, Liu GS. 2017. AnAANAT/ASMTtransgenic anim al model constructed with CRISPR/Cas9 system serving as the mamm ary gland bioreactor to produce melatonin-enriched m ilk in sheep.Journal of Pineal Research,63(1): e12406.
Madda lo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC,Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. 2014.In vivoengineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system.Nature,516(7531): 423–427.
Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE,Church GM. 2013. RNA-guided human genome engineering via Cas9.Science,339(6121): 823–826.
Malpotra S, Vats A, Kumar S, Gautam D, De S. 2017. Generation of genomic deletions (ofRig-IGENE) in goat primary cell culture using CRISPR/CAS9method.Animal Biotechnology,doi: 10.1080/10495398.2017.1331915.
Mandasari M, Sawangarun W, Katsube K, Kayamori K, Yamaguchi A,Sakamo to K. 2016. A facile one-step strategy for the generation of conditional knockout mice to exp lore the role of Notch1 in oroesophageal tumorigenesis.Biochemical and Biophysical Research Communications,469(3): 761–767.
Maresca M, Lin VG, Guo N, Yang Y. 2013. Ob ligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining.Genome Research,23(3):539–546.
Maresch R, Mueller S, Veltkam p C, Öllinger R, Friedrich M, Heid I, Steiger K, Weber J, Engleitner T, Barenboim M, Klein S, Louzada S, Banerjee R,Strong A, Stauber T, Gross N, Geumann U, Lange S, Ringelhan M, Varela I,Unger K, Yang FT, Schm id RM, Vassiliou GS, Braren R, Schneider G,Heikenwalder M, Brad ley A, Saur D, Rad R. 2016. Mu ltip lexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice.Nature Communications,7: 10770.
Mashiko D, Young SAM, Muto M, Kato H, Nozawa K, Ogawa M, Noda T,Kim YJ, Satouh Y, Fu jihara Y, Ikawa M. 2014. Feasibility for a large scale mouse m utagenesis by injecting CRISPR/Cas p lasm id into zygotes.Development, Growth & Differentiation,56(1): 122–129.
McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director's cut): de leted sequences and a lternative endings.Trends in Genetics,24(11): 529–538.
Meyer MB, Benkusky NA, Onal M, Pike JW. 2016. Selective regulation ofMmp13by 1,25(OH)2D3, PTH, and Osterix through distal enhancers.The Journal of Steroid Biochemistry and Molecular Biology,164: 258–264.
Mianné J, Codner GF, Caulder A, Fe ll R, Hutchison M, King R, Stewart ME,Wells S, Tebou l L. 2017. Ana lysing the outcome of CRISPR-aided genome editing in embryos: Screening, genotyping and quality control.Methods,121–122: 68–76.
Miyata H, Castaneda JM, Fujihara Y, Yu ZF, Archambeault DR, Isotani A,Kiyozum i D, Kriseman ML, Mashiko D, Matsumura T, Matzuk RM, Mori M,Noda T, O ji A, Okabe M, Prunskaite-Hyyrylainen R, Ram irez-Solis R,Satouh Y, Zhang Q, Ikawa M, Matzuk MM. 2016. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice.Proceedings of the National Academy of Sciences of the United States of America,113(28): 7704–7710.
Monteys AM, Ebanks SA, Keiser MS, Davidson BL. 2017. CRISPR/Cas9 editing of the mutant huntingtin alle le in vitro and in vivo.Molecular Therapy,25(1): 12–23.
Moscou MJ, Bogdanove AJ. 2009. A sim p le cipher governs DNA recognition by TAL effectors.Science,326(5959): 1501.
Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T,Sezutsu H, Yamamoto T, Sakuma T, Suzuki KIT. 2014. Microhomologymediated end-joining-dependent integration of donor DNA in ce lls and animals using TALENs and CRISPR/Cas9.NatureCommunications,5:5560.
Narayanan A, Hill-Teran G, Moro A, Ristori E, Kasper DM, Roden CA, Lu J,Nicoli S. 2016. In vivo mutagenesis of m iRNA gene fam ilies using a scalable multiplexed CRISPR/Cas9 nuclease system.Scientific Reports,6:32386.
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan XF, Ran FA, Yan WX, Asokan A, Zhang F,Duan DS, Gersbach CA. 2016. In vivo genome editing improves muscle function in a m ouse m odel of Duchenne m uscular dystrophy.Science,351(6271): 403–407.
Niu Y, Jin M, Li Y, Li P, Zhou J, Wang X, Petersen B, Huang X, Kou Q,Chen Y. 2017a. Biallelicβ-caroteneoxygenase2 knockout results in yellow fat in sheep via CRISPR/Cas9.AnimalGenetics,48(2): 242–244.
Niu YY, Shen B, Cui YQ, Chen YC, Wang JY, Wang L, Kang Y, Zhao XY, Si W, Li W, Xiang AP, Zhou JK, Guo XJ, Bi Y, Si CY, Hu B, Dong GY, Wang H,Zhou ZM, Li TQ, Tan T, Pu XQ, Wang F, Ji SH, Zhou Q, Huang XX, Ji WZ,Sha JH. 2014. Generation of gene-modified cynom olgus m onkey via Cas9/RNA-mediated gene targeting in one-cell embryos.Cell,156(4): 836–843.
Niu YY, Zhao XE, Zhou JK, Li Y, Huang Y, Cai B, Liu YT, Ding Q, Zhou SW,Zhao J, Zhou GX, Ma BH, Huang XX, Wang XL, Chen YL. 2017b. Efficient generation of goats with defined point mutation (I397V) inGDF9through CRISPR/Cas9.Reproduction, Fertilityand Development,doi:10.1071/RD17068.
Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, Miller JC, Holmes MC, Gregory PD, Urnov FD, Cost GJ. 2010. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with lim ited chromosoma l homology.Nucleic Acids Research,38(15): e152.
Ortinski PI, O'Donovan B, Dong XY, Kantor B. 2017. Integrase-deficient lentiviral vector as an all-in-one p latform for highly efficient CRISPR/Cas9-mediated gene editing.Molecular Therapy. Methods & Clinical Development,5: 153–164.
Ou ZH, Niu XH, He WY, Chen YC, Song B, Xian YX, Fan D, Tang DL, Sun XF. 2016. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassem ia in mice.Scientific Reports,6: 32463.Paix A, Schm idt H, Seydoux G. 2016. Cas9-assisted recombineering inC.elegans: genome editing usingin vivoassemb ly of linear DNAs.Nucleic Acids Research,44(15): e128.
Palm iter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG,Birnberg NC, Evans RM. 1982. Dramatic grow th of mice that develop from eggs m icroin jected with m etallothionein-grow th hormone fusion genes.Nature,300(5893): 611–615.
Pankow icz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA,Ravishankar M, Sun Q, Yang D, Borow iak M, Sumazin P, Elsea SH, Bissig-Choisat B, Bissig KD. 2016. Rep rogramm ing metabolic pathwaysin vivowith CRISPR/Cas9 genome editing to treat hereditary tyrosinaem ia.Nature Communications,7: 12642.
Park KE, Kaucher AV, Powe ll A, Waqas MS, Sandmaier SES, Oatley MJ,Park CH, Tibary A, Donovan DM, Blomberg LA, Lillico SG, White law CB,Mileham A, Telugu BP, Oatley JM. 2017. Generation of germ line ablated male pigs by CRISPR/Cas9 editing of theNANOS2gene.Scientific Reports,7: 40176.
Peng J, Wang Y, Jiang JY, Zhou XY, Song L, Wang LL, Ding C, Qin J, Liu LP, Wang WH, Liu JQ, Huang XX, Wei H, Zhang PM. 2015. Production of hum an a lbum in in pigs through CRISPR/Cas9-mediated knockin of human cDNA into sw ine album in locus in the zygotes.Scientific Reports,5: 16705.Perles Z, Moon S, Ta-Shma A, Yaacov B, Francescatto L, Edvardson S,Rein AJ, Elpeleg O, Katsanis N. 2015. A human laterality disorder caused by a homozygous deleterious mutation inMMP21.Journal of Medical Genetics,52(12): 840–847.
Petersen B, Frenze l A, Lucas-Hahn A, Herrmann D, Hassel P, Klein S,Zieg ler M, Hade ler KG, Niemann H. 2016. Efficient production of biallelicGGTA1knockout pigs by cytop lasm ic m icroinjection of CRISPR/Cas9 into zygotes.Xenotransplantation,23(5): 338–346.
Pierce AJ, Hu P, Han MG, Ellis N, Jasin M. 2001. Ku DNA end-binding protein modu lates homo logous repair of double-strand breaks in mammalian cells.Genes & Development,15(24): 3237–3242.
Platt RJ, Chen SD, Zhou Y, Yim MJ, Swiech L, Kem pton HR, Dahlman JE,Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S,Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A,Feng GP, Sharp PA, Zhang F. 2014. CRISPR-Cas9 knockin mice for genome editing and cancer modeling.Cell,159(2): 440–455.
Port F, Chen HM, Lee T, Bullock SL. 2014. Optim ized CRISPR/Cas tools for efficient germ line and som atic genom e engineering inDrosophila.Proceedings of the National Academy of Sciences of the United States of America,111(29): E2967–E2976.
Ran FA, Hsu PD, W right J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system.Nature Protocols,8(11):2281–2308.
Rannals MD, Page SC, Campbell MN, Gallo RA, Mayfield B, Maher BJ.2016. Neurodevelopm ental m odels of transcription factor 4 deficiency converge on a comm on ion channel as a potential therapeutic target for Pitt Hopkins syndrome.Rare Diseases,4(1): e1220468.
Rouet P, Sm ih F, Jasin M. 1994. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells.Proceedings of the National Academy of Sciences of the United States of America,91(13):6064–6068.
Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. 2016. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh system s.Nature Protocols,11(1): 118–133.
Seruggia D, Fernández A, Cantero M, Pelczar P, Montoliu L. 2015.Functional validation of mousetyrosinasenon-coding regulatory DNA elements by CRISPR-Cas9-mediated mutagenesis.Nucleic Acids Research,43(10): 4855–4867.
Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. 2015. Rapid reverse genetic screening using CRISPR in zebrafish.Nature Methods,12(6): 535–540.
Shi ZY, Wang FQ, Cui Y, Liu ZZ, Guo XG, Zhang YQ, Deng Y, Zhao H,Chen YL. 2015. Heritable CRISPR/Cas9-mediated targeted integration inXenopustropicalis.FASEB Journal,29(12): 4914–4923.
Shinm yo Y, Tanaka S, Tsunoda S, Hosom ichi K, Tajima A, Kawasaki H.2016. CRISPR/Cas9-mediated gene knockout in the mouse brain usingin uteroelectroporation.Scientific Reports,6: 20611.
Straub C, Granger AJ, Saulnier JL, Sabatini BL. 2014. CRISPR/Cas9-mediated gene knock-down in post-m itotic neurons.PLoS One,9: e105584.
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ,Hatanaka F, Yam amoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo SC, Chen S, Goebl A, Soligalla RD, Qu J, Jiang TS, Fu X, Jafari M,Esteban CR, Berggren WT, Lajara J, Nuñez -Delicado E, Guillen P,Cam pisto l JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM,Zhang K, Belmonte JCI. 2016.In vivogenome editing via CRISPR/Cas9 mediated homo logy-independent targeted integration.Nature,540(7631):144–149.
Sweeney CL, Choi U, Liu C, Koontz S, Ha SK, Malech HL. 2017. CRISPR-mediated knockout of cybb in NSG mice establishes a model of chronic granulomatous disease for human stem-cell gene therapy transp lants.human gene therapy,28(7): 565–575.
Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M,Zhang F. 2015.In vivointerrogation of gene function in the mamm alian brain using CRISPR-Cas9.Nature Biotechnology,33(1): 102–106.
Tabebordbar M, Zhu KX, Cheng JK, Chew WL, W id rick JJ, Yan WX,Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH,Church GM, Wagers AJ. 2016. In vivo gene editing in dystrophic m ouse muscle and muscle stem cells.Science,351(6271): 407–411.
Taleei R, Nikjoo H. 2013. Biochem ical DSB-repair mode l for m amma lian cells in G1 and early S phases of the cell cycle.Mutation Research/Genetic Toxicology and Environmental Mutagenesis,756(1–2): 206–212.
Taylor EM, Lehmann AR. 1998. Conservation of eukaryotic DNA repair m echanism s.International Journal of Radiation Biology,74(3): 277–286.
Thomas KR, Capecchi MR. 1987. Site-directed m utagenesis by gene targeting in mouse em bryo-derived stem-cells.Cell,51(3): 503–512.
Ukken FP, Bruckner JJ, Weir KL, Hope SJ, Sison SL, Birschbach RM,Hicks L, Taylor KL, Dent EW, Gonsalvez GB, O'Connor-Giles K. 2016.
BAR-SH3 sorting nexins are conserved interacting proteins of Nervous w reck that organize synapses and promote neurotransm ission.Journal of Cell Science,129(1): 166–177.
Varshney GK, Pei WH, LaFave MC, Idol J, Xu LS, Ga llardo V, Carrington B,Bishop K, Jones M, Li MY, Harper U, Huang SC, Prakash A, Chen WB,Sood R, Ledin J, Burgess SM. 2015. High-throughput gene targeting and phenotyping in zebra fish using CRISPR/Cas9.Genome Research,25(7):1030–1042.
Vasquez KM, Marburger K, Intody Z, W ilson JH. 2001. Manipulating the mammalian genome by homologous recombination.Proceedings of the National Academy of Sciences of the United States of America,98(15):8403–8410.
Véron N, Qu ZD, Kipen PAS, Hirst CE, Marcelle C. 2015. CRISPR m ediated somatic cell genome engineering in the chicken.Developmental Biology,407(1): 68–74.
Ve jnar CE, Moreno-Mateos MA, Cifuentes D, Bazzini AA, Giraldez AJ. 2016.Optim ized CRISPR-Cas9 system for genome editing in zebrafish.Cold Spring Harbor Protocols,2016(10): pdb.prot086850.
Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, Lee SE. 2012.Microhomo logy directs diverse DNA break repair pathways and chromosom al translocations.PLoS Genetics,8(11): e1003026.
Voutev R, Mann RS. 2017. Bxb1 phage recombinase assists genome engineering in Drosophila melanogaster.Biotechniques,62(1): 37–38.
Wakabayashi S, Sawamura N, Voelzmann A, Broemer M, Asahi T, Hoch M.2016. Ohgata the singleDrosophilaortholog of human cereblon regu lates insulin signaling-dependent organism ic grow th.Journal of Biological Chemistry,291(48): 25120–25132.
Wang BL, Li MY, Qu C, M iao WY, Yin Q, Liao JY, Cao HT, Huang M, Wang K, Zuo EW, Peng GD, Zhang SX, Chen GD, Li Q, Tang K, Yu Q, Li ZJ,Wong CCL, Xu GL, Jing NH, Yu X, Li JS. 2017. CRISPR-Cas9-mediated genome editing in one blastomere of two-cell embryos reveals a novel Tet3 function in regulating neocortical deve lopment.Cell Research,27(6): 815–829.
Wang D, Mou HW, Li SY, Li YX, Hough S, Tran K, Li J, Yin H, Anderson DG,Sontheimer EJ, Weng ZP, Gao GP, Xue W. 2015a. Adenovirus-mediated somatic genome editing ofPtenby CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses.Human Gene Therapy,26(7): 432–442.
Wang HY, Yang H, Shiva lila CS, Daw laty MM, Cheng AW, Zhang F,Jaenisch R. 2013. One-step generation of mice carrying m utations in multiple genes by CRISPR/Cas-mediated genome engineering.Cell,153(4): 910–918.
Wang LR, Shao YJ, Guan YT, Li L, Wu LJ, Chen FR, Liu MZ, Chen HQ, Ma YL, Ma XY, Liu MY, Li DL. 2015b. Large genom ic fragm ent de letion and functional gene cassette knock-in via Cas9 protein mediated genome editing in one-cell rodent em bryos.Nautre Scientific Reports,5: 17517.
Wang S, Sengel C, Emerson MM, Cepko CL. 2014. A gene regulatory network controls the binary fate decision of rod and bipolar cells in the vertebrate retina.Developmental Cell,30(5): 513–527.
Wang X, Raghavan A, Chen T, Qiao L, Zhang YX, Ding QR, Musunuru K.2016b. CRISPR-Cas9 targeting ofPCSK9in human hepatocytes in vivobrief report.Arteriosclerosis, Thrombosis, and Vascular Biology,36(5):783–786.
Wang X, Tang Y, Lu J, Shao YJ, Qin X, Li YM, Wang LR, Li DL, Liu MY.2016c. Characterization of novel cytochrome P450 2E1 knockout rat m odel generated by CRISPR/Cas9.Biochemical Pharmacology,105: 80–90.
Wang XL, Zhou JW, Cao CW, Huang JJ, Hai T, Wang YF, Zheng QT, Zhang HY, Qin GS, Miao XN, Wang HM, Cao SZ, Zhou Q, Zhao JG. 2015d.Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin a fter rapid selection of highly active sgRNAs in pigs.Scientific Reportsi,5: 13348.
Wang XL, Yu HH, Lei AM, Zhou JK, Zeng WX, Zhu HJ, Dong ZM, Niu YY,Shi BB, Cai B, Liu JW, Huang S, Yan HL, Zhao XE, Zhou GX, He XL, Chen XX, Yang YX, Jiang Y, Shi L, Tian XE, Wang YJ, Ma BH, Huang XX, Qu L,Chen YL. 2015c. Generation of gene-modified goats targetingMSTNandFGF5via zygote in jection of CRISPR/Cas9 system.Scientific Reports,5:13878.
Wang XL, Cao CW, Huang JJ, Yao J, Hai T, Zheng QT, Wang X, Zhang HY,Qin GS, Cheng JB, Wang YF, Yuan ZQ, Zhou Q, Wang HM, Zhao JG.2016a. One-step generation of trip le gene-targeted pigs using CRISPR/Cas9 system.Scientific Reports,6: 20620.
Wang Y, Du YA, Shen B, Zhou XY, Li J, Liu Y, Wang JY, Zhou JK, Hu B,Kang NN, Gao JM, Y LQ, Huang XX, Wei H. 2015e. Efficient generation of gene-modified pigs via in jection of zygote with Cas9/sgRNA.Scientific Reports,5: 8256.
Weber J, Öllinger R, Friedrich M, Ehmer U, Barenboim M, Steiger K, Heid I,Mue ller S, Maresch R, Eng leitner T, Gross N, Geum ann U, Fu BY, Segler A,Yuan DT, Lange S, Strong A, de la Rosa J, Esposito I, Liu PT, Cadiñanos Jan, Vassiliou GS, Schm id RM, Schneider G, Unger K, Yang FT, Braren R,Heikenwälder M, Varela I, Saur D, Bradley A, Rad R. 2015. CRISPR/Cas9 somatic multip lex-mutagenesis for high-throughput functional cancer genom ics in mice.Proceedings of the National Academy of Sciences of the United States of America,112(45): 13982–13987.
Wen KJ, Yang LJ, Xiong TL, Di C, Ma DH, Wu MH, Xue ZY, Zhang XD,Long L, Zhang WM, Zhang JY, Bi XL, Dai JB, Zhang QF, Lu ZJ, Gao GJ.2016. Critical roles of long noncoding RNAs inDrosophilasperm atogenesis.Genome Research,26(9): 1233–1244.
W iedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing system s in bacteria and archaea.Nature,482(7385): 331–338.
Won M, Daw id IB. 2017. PCR artifact in testing for homo logous recombination in genom ic editing in zebrafish.PLoS One,12(3): e0172802.Wu WB, Lu ZW, Li F, Wang WJ, Qian NN, Duan JZ, Zhang Y, Wang FC,Chen T. 2017. Efficient in vivo gene editing using ribonucleoproteins in skin stem cells of recessive dystrophic epidermolysis bu llosa m ouse model.Proceedings of the National Academy of Sciences of the United States of America,114(7): 1660–1665.
Wu YX, Liang D, Wang YH, Bai MZ, Tang W, Bao SM, Yan ZQ, Li DS, Li JS.2013. Correction of a genetic disease in m ouse via use of CRISPR-Cas9.Cell Stem Cell,13(6): 659–662.
Xie C, Zhang YP, Song L, Luo J, Qi W, Hu JL, Lu DB, Yang Z, Zhang J,Xiao J, Zhou B, Du JL, Jing NH, Liu Y, Wang Y, Li BL, Song BL. 2016.Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac synd rome.Cell Research,26(10): 1099–1111.
Xu CL, Qi XL, Du XG, Zou HY, Gao F, Feng T, Lu HX, Li SL, An XM, Zhang LJ, Wu YY, Liu Y, Li N, Capecchi MR, Wu S. 2017.piggyBacmediates efficient in vivo CRISPR library screening for tumorigenesis in mice.Proceedings of the National Academy of Sciences of the United States of America,114(4): 722–727.
Xue W, Chen SD, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai WX, Yang G, Bronson R, Crow ley DG, Zhang F, Anderson DG, Sharp PA,Jacks T. 2014. CRISPR-mediated direct mutation of cancer genes in the mouse liver.Nature,514(7522): 380–384.
Yang H, Wang HY, Shivalila CS, Cheng AW, Shi LY, Jaenisch R. 2013.One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering.Cell,154(6): 1370–1379.
Yang Y, Wang LL, Bell P, McMenam in D, He ZN, White J, Yu HW, Xu CY,Morizono H, Musunuru K, Batshaw ML, W ilson JM. 2016. A dual AAV system enables the Cas9-mediated correction of a metabo lic liver disease in newborn mice.Nature Biotechnology,34(3): 334–338.
Yao X, Wang X, Liu JL, Hu XD, Shi LY, Shen XW, Ying WQ, Sun XY, Wang X, Huang PY, Yang H. 2017. CRISPR/Cas9 - mediated precise targeted integration in vivo using a double cut donor with short homo logy arm s.EBioMedicine,20: 19–26.
Yin CR, Zhang T, Qu XY, Zhang YG, Putatunda R, Xiao X, Li F, Xiao WD,Zhao HQ, Dai S, Qin XB, Mo XM, Young WB, Khalili K, Hu WH. 2017. In vivo excision of HIV-1 provirus by saCas9 and m ultip lex single-guide RNAs in anima l models.Molecular Therapy,25(5): 1168–1186.
Yin H, Song CQ, Dorkin JR, Zhu LJ, Li YX, Wu QQ, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao GP,Weng ZP, Dong YZ, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG. 2016. Therapeutic genome editing by com bined viral and non-viral de livery of CRISPR system com ponentsin vivo.Nature Biotechnology,34(3): 328–333.
Yoshim i K, Kaneko T, Voigt B, Mashimo T. 2014. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas p latform.Nature Communications,5: 4240.
Yu HH, Zhao H, Qing YB, Pan WR, Jia BY, Zhao HY, Huang XX, Wei HJ.2016. Porcine zygote injection with Cas9/sgRNA results inDMD-modified pig with muscle dystrophy.International Journal of Molecular Sciences,17(10): 1668.
Yu ZS, Chen HQ, Liu JY, Zhang HT, Yan Y, Zhu NN, Guo YW, Yang B,Chang Y, Dai F, Liang XH, Chen YX, Shen Y, Deng WM, Chen JM, Zhang B,Li CQ, Jiao RJ. 2014. Various app lications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify theDrosophilagenom e.Biology Open,3(4): 271–280.
Yuan BL, Wang XS, Fan CY, You J, Liu YC, Weber JD, Zhong HB, Zhang YD. 2016a. DHX33 transcriptionally controls genes involved in the cell cycle.Molecular and Cellular Biology,36(23): 2903–2917.
Yuan L, Sui TT, Chen M, Deng JC, Huang YY, Zeng J, Lv QY, Song YN, Li ZJ, Lai LX. 2016b. CRISPR/Cas9-mediatedGJA8knockout in rabbits recapitulates hum an congenital cataracts.Scientific Reports,6: 22024.
Zhang DH, Golubkov VS, Han WL, Correa RG, Zhou Y, Lee S, Strongin AY,Dong PD. 2014. Identification of Annexin A4 as a hepatopancreas factor involved in liver ce ll surviva l.Developmental Biology,395(1): 96–110.
Zhang T, Yin YJ, Liu H, Du WL, Ren CH, Wang L, Lu HZ, Zhang ZY. 2016.Generation of VDR knockout mice via zygote injection of CRISPR/Cas9 system.PLoS One,11(9): e0163551.
Zhang XM, Li WR, Liu CX, Peng XR, Lin JP, He SG, Li XJ, Han B, Zhang N,Wu YS, Chen L, Wang LQ, MaYila, Huang JC, Liu MJ. 2017a. Alteration of sheep coat color pattern by disruption ofASIPgene via CRISPR Cas9.Scientific Reports,7: 8149.
Zhang YH, Wu LZ, Liang HL, Yang Y, Qiu J, Kan Q, Zhu W, Ma CL, Zhou XY. 2017b. Pu lmonary surfactant synthesis in m iRNA-26a-1/m iRNA-26a-2 double knockout mice generated using the CRISPR/Cas9 system.American Journal of Translational Research,9(2): 355–365.
Zhong H, Chen YY, Li YM, Chen R, Mardon G. 2015. CRISPR-engineered mosaicism rapid ly reveals that loss ofKcnj13function in mice m im ics human disease phenotypes.Scientific Reports,5: 8366.
Zhou WJ, Wan YJ, Guo RH, Deng MT, Deng KP, Wang Z, Zhang YL, Wang F. 2017. Generation of beta-lactoglobulin knockout goats using CRISPR/Cas9.PLoS One,12(10): e0186056.
Zhou XY, Wang LL, Du YN, Xie F, Li L, Liu Y, Liu CH, Wang SQ, Zhang SB,Huang XX, Wang Y, Wei H. 2016. Efficient generation of gene-modified pigs harboring precise orthologous hum an mutation via CRISPR/Cas9-induced hom ology-directed repair in zygotes.Human Mutation,37(1): 110–118.
Zhu QM, Ko KA, Ture S, Mastrangelo MA, Chen MH, Johnson AD,O'Donnell CJ, Morrell CN, Miano JM, Lowenstein CJ. 2017. Novel thrombotic function of a human SNP inSTXBP5revea led by CRISPR/Cas9 gene editing in mice.Arteriosclerosis, Thrombosis, and Vascular Biology,37(2): 264–270.
Zhu W, Xie K, Xu YJ, Wang L, Chen KM, Zhang LZ, Fang JM. 2016.CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse.Virus Research,217: 125–132.
Zinn AR, Butow RA. 1985. Nonreciprocal exchange between alleles of the yeast m itochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break.Cell,40(4): 887–895.
杂志排行
Zoological Research的其它文章
- Kidney disease models: tools to identify mechanism s and potential therapeutic targets
- King cobra peptide OH-CATH30 as a potential candidate drug through clinic drug-resistant isolates
- Biogeography of the Shimba Hills ecosystem herpetofauna in Kenya
- Discovery of Japalura chapaensis Bourret, 1937(Reptilia: Squamata: Agam idae) from Southeast Yunnan Province, China
- Stereotypy and variability of socialcalls among clustering female big-footed myotis(Myotismacrodactylus)
- AutoSeqMan:batch assembly of contigs for Sanger sequences
