Genetic effects of historical anthropogenic disturbance on a longlived endangered tropical tree Vatica mangachapoi
2018-03-19ZhicongDaiChuncanSiDeliZhaiPingHuangShanshanQiYingLinRuipingWangQiongxinZhongDaolinDu
Zhicong Dai•Chuncan Si•Deli Zhai•Ping Huang•Shanshan Qi•Ying Lin•Ruiping Wang•Qiongxin Zhong•Daolin Du,3
Introduction
Forests protect land and water resources,support biological diversity,and mitigate climate change(Mayer et al.2005).However,tropical rainforests are shrinking throughout the world(Gaino et al.2010)because of extensive deforestation,agricultural cultivation,and tourism development;for example,tropical forests on Hainan Island were found to be continuously shrinking(Zhai et al.2014).Hainan Island possesses high plant diversity with about 4200 species,630 of which are endemic to the island,including 50 endangered species(Lopez-Pujol et al.2006).However,the forest cover in Hainan Island has undergone three major change stages since 1950(see Supplementary Appendix A:Fig.S1)because of deforestation and degradation caused by excessive logging,human habitation,and large-scale agricultural activities(Zhang et al.2000;Lopez-Pujol et al.2006).The loss of tropical rainforests,especially Dipterocarp forests,the typical tropical forests in Asia(Bunyavejchewin et al.2003),has led to a decrease in the size and number of plant populations over the past fi ve decades on Hainan.

Table 1 Detailed information and genetic diversity of eleven investigated Vatica mangachapoi populations on Hainan Island
Genetic diversity is on the core form of biodiversity.High genetic diversity increases the adaptive potential of species to the ongoing environmental changes(Jump et al.2009).Loss of genetic diversity could make those small populations more vulnerable to extinction in fragmented habitats(Aguilar et al.2008).Moreover,current remnant populations might re fl ect historical forest composition.Suf fi cient information,such as the level of genetic diversity among populations,is helpful for identifying priority regions,reinforcement of remnant populations,re-introduction,ex situ culture and seed collection(Luo et al.2011).
Over-exploitation of forests have accelerated biodiversity losses and genetic erosion(Lopez-Pujol et al.2006;Gaino et al.2010;Manohara et al.2010).Based on previous research on the effects of forest fragmentation derived from excessive deforestation on plant genetic diversity(Aguilar et al.2008;Dai et al.2013)and the fact of long-term natural forest conversion on Hainan Island,we hypothesize that excessive deforestation over the past decades on Hainan Island could result in negative effects on the genetic diversity of long-lived trees.
To test the assumption,Vatica mangachapoiBlanco(Dipterocarpaceae)was used in the present study.V.mangachapoiis a typical slow-growing and long-lived perennial tree(Chen 1964),and the mixedV.mangachapoiforest is the dominant type of tropical evergreen monsoon rainforest on Hainan Island(Ding et al.2006;Dai et al.2008).As one of dominant family of tropical rainforest and the foremost coastal protective forest in Hainan Island,V.mangachapoiis notable for its economic and ecological signi fi cance.
The tree is widespread in various habitat types on Hainan Island and is common on dry ridges(Ashton 1998).However,the population ofV.mangachapoion Hainan Island has been drastically reduced by overexploitation in the past century.The genetic variability ofV.mangachapoiand the negative impacts of excessive deforestation onV.mangachapoiforests are closely linked to the changes of the total structure of rainforests on Hainan Island.

Fig.1 Sample sites of the 11 populations of V.mangachapoi investigated on Hainan Island.The code and origin of populations refers to Table 1
Inter-simple sequence repeat(ISSR)markers are widely used for population genetic studies because they are rapid,ef fi cient,and hyper variable,and can produce massively polymorphic and repeatable loci(Li and Jin 2008;Matesanz et al.2011;Rucinska and Puchalski 2011).Therefore,in the present study we used ISSR markers to evaluate the status of genetic diversity and the genetic differentiation among populations ofV.mangachapoiin Hainan Island.
Materials and methods
Species studied and sampling
Vatica mangachapoi,endemic to tropical Asia,has been listed as an ‘A1’endangered tree species on the IUCN red list of threatened species(Ashton 1998),and has also been classi fi ed on the Second-class State Key Protected Wild Plants List in China(State Forestry Administration 1999).Long-lived Dipterocarps are mainly outcrossing,self-incompatible,and winged fruits(Sist et al.2003)and they are both anemochory and insect-pollinated, as isV.mangachapoi.
According to its natural distribution and population size,11 distinct wild populations(without being replanted by humans)ofV.mangachapoiwere identi fi ed in nine key forest regions throughout the species’main distribution range on Hainan Island in the present study(Fig.1;Table 1).Except for the populations in Dongfang County 1(DF1),Tongshi County 1(TS1),and Wenchang County 1(WC1),the other eight populations are located in nature reserves.However,all studied populations have suffered some degree of historical disturbances in the past 50 years.
To evaluate the status of genetic diversity under different disturbance pressures,the populations were divided into three disturbance levels(Table 1)according to their protection status(being protected or not)and suffering human activities (tourism/villages/roads,hydrological projects,or logging trace):low disturbance(LD),moderate disturbance(MD),and high disturbance(HD)(Please see Supplementary Appendix B ‘Information of all studied populations’).
Fresh leaves from all 11 populations ofV.mangachapoiwererandomly collected from 320 individualtrees(DBH≥1.5 cm)(Table 1).To avoid the possibility of sampling from the same mother tree or from the same individual twice,the distance between any two sampled individuals was set at>30 m(Sist et al.2003).The fresh leaves were collected from the end of July 2006 to August 2007.In all cases,the tissue samples were enclosed in plastic bags under 4°C and immediately transported to the laboratory stored at-70°C before further molecular analysis.
DNA extraction and ISSR-PCR ampli fi cation
The total DNA of leaves was isolated,using the CTAB method protocol(Stewart and Via 1993;Dai et al.2015),and dissolved in 0.5×TE(pH 8.0).After adjusting the concentration,the total DNA was subjected to PCR ampli fi cation(Dai et al.2013)using UBC ISSR Primer(Set No.9,Biotechnology Laboratory,University of British Columbia,Canada).A 20 ng total DNA template was ampli fi ed in a Rapid-cycler(Mastercyclergradient,Eppendorf China Ltd.),with an initial setting of 5 min at 94°C,followed by 32 cycles of 60 s for denaturing at 94 °C,45 s annealing at 50 °C,90 s extension at 72 °C,and ended with 7 min at 72°C.
Reactions were carried out in 20 μL volume containing 1×Reaction Buffer(including 1.5 mM Mg2+);1 U Taq polymerase(Tiangen Biotech,China);0.2 mmol/LdNTPs(Sangon Biotech,China),0.2 μmol/L ISSR primers(synthesized by Sangon Biotech).Ampli fi cation products were analyzed by electrophoresis(1×TBE buffer)at 100 V for 30 min through a 2%agarose gel and stained with ethidium bromide and then imaged using Gel Imaging System(Alpha Imager 2200,Alpha Innotech Corporation).
Statistical analysis
Since ISSR markers are dominant,we assumed that each band represented the phenotype at a single biallellic locus(Matesanz et al.2011).The ampli fi ed fragments of the homologous bands were scored for each individual as present(1)or absent(0)(Matesanz et al.2011;Rucinska and Puchalski 2011).Only the distinct,reproducible and well-resolved fragments were scored.A matrix of ISSR phenotypes for each sample was constructed and analyzed using POPGENE version 1.32 software(Yeh et al.1999).Genetic parameters were estimated as follows:the mean number of alleles(Na),the percentage of polymorphic bands(PPB);the Nei gene diversity test(h)(Nei 1973);and the Shannon information index(I)(Lewontin 1972).The total gene diversity among populations(HT);the gene diversity within populations(HS);(Nei 1972),and the coef fi cient of genetic differentiation among populations within species(GST)(Nei 1973)were included.The value of gene fl ow among populations was calculated in accordance with the following formula:Nm=0.5(1-GST)/GST(McDermott and McDonald 1993).
Analysis of the molecular variance(AMOVA)of the 999 permutations,with the populations nested within disturbance groups(LD,MD,and HD),was conducted Gen-AlEX software,version 6.41(Peakall and Smouse 2006).Then,we examined the distribution of variation among disturbance groups(ΦRT),populations within disturbance groups(ΦPR),and populations(ΦPT)(an analogue ofFst,e.g.genetic diversity among populations)(Breinholt et al.2009).
To examine population structure,we used the standardized covariance method(Fatemi and Gross 2009)in GenAlEX version 6.4.1,with principal coordinates analysis(PCoA)plot based on the pair-wise genetic difference(Binary distance)of population level and individuals level(Dai et al.2013).
Vatica mangachapoigrows slowly(Chen 1964),and it was once widespread and continuous throughout Hainan Island(Ashton 1998).Combining the natural forest changes history since the early 1950s(Appendix A:Fig.S1),we inferred that geographical pattern among the present populations ofV.mangachapoi(Fig.1)was driven by the longterm historical anthropogenic landscape fragmentation,but not by the natural geographical isolation.Therefore,present geographic distance ofV.mangachapoican re fl ect the human disturbance level of the existing populations.To test thegeneticisolationbydistance(IBD),theMantelstatistical test(Breinholtetal.2009;FatemiandGross2009)ofthe999 permutations by GenAlEX was conducted,using pair-wise population genetic distance(ΦPT)and pair-wise populations geographic distance matrices.
One-way ANOVA was further used to test the in fl uence of disturbance levels on genetic diversity by Tukey’s studentized range(HSD)test,settingp≤0.05 as criterion of statistical signi fi cance.Data analyses were performed using the statistical software SAS version 9.1(SAS Institute Inc.2004).
Results
Genetic diversity within populations
A total of 98 polymorphic loci were ampli fi ed from 320 sampled individuals from the 11 natural populations ofV.mangachapoiacross Hainan Island,using 10 pairs of ISSR primers(Supplementary Appendix C:ISSR data;Supplementary Appendix A:Fig.S3),which were screened from 51 pairs of ISSR primers.The number of bands of each polymorphicprimervariedfrom six(UBC828and UBC836)to 13(UBC818,UBC834 and UBC840)(Supplementary Appendix A:Table S1).Most of the ISSR primers,except UBC836(PPB=83.3%),ampli fi ed 100%polymorphic bands.The genetic diversity(Na,h,I,andPPB)within eleven populations and at species level was showed in Table 1.

Fig.2 Principal coordinates analysis(PCoA)plot(GenAlEx 6.4.1)based on the genetic diversity among the 10 markers used in this study for population identi fi cation
Genetic divergence among populations
The AMOVA of the ISSR data set for the 11 populations revealed most of the variation(76%,p=0.001)resided within the population;2%variation(p=0.001)was among disturbance levels;and 22%variation(p=0.001)was among populations within the disturbance levels.A two-dimensional scatter plot of PCoA analysis revealed three major clusters for population level,which conformed roughly to the geographical distribution of the 11 populations(Fig.2a).Especially,the northernmost population WC1 and the central population TS1 formed a cluster.Axis 1 and axis 2 explained,respectively,35.44 and 25.90% of the variation distribution.
Effects of disturbance on genetic variation
The PCoA analysis for the individual level revealed two clusters:the left one mainly included HD and MD groups,while the right one included LD and MD groups(Fig.2b).The Mantel tests showed that there were strongly positive relations between the pair-wise population geographic isolation and the pair-wise population gene distance(ΦPT)matrix discovered between the 11 populations(p=0.0005;Fig.3).As shown in Table 2,the populations of high disturbance(HD)had greater genetic differentiation(GST),resulting in lower gene fl ow(Nm).Tukey’s multiple comparisontestshowedthatthegeneticvariation(Na,I,andPPB)oflowdisturbance(LD)populationsweregreaterthanthatof the high disturbance(HD)populations(Fig.4;Table 3).
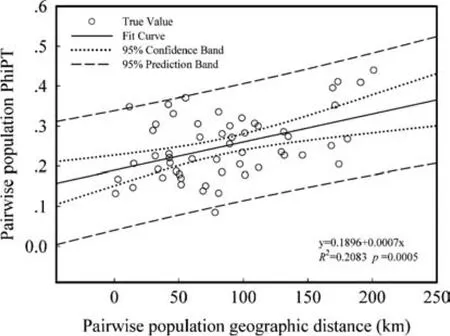
Fig.3 The Mantel test for pair-wise population ΦPTgenetic distance matrix and pair-wise populations distance matrices of the geography for 11 populations
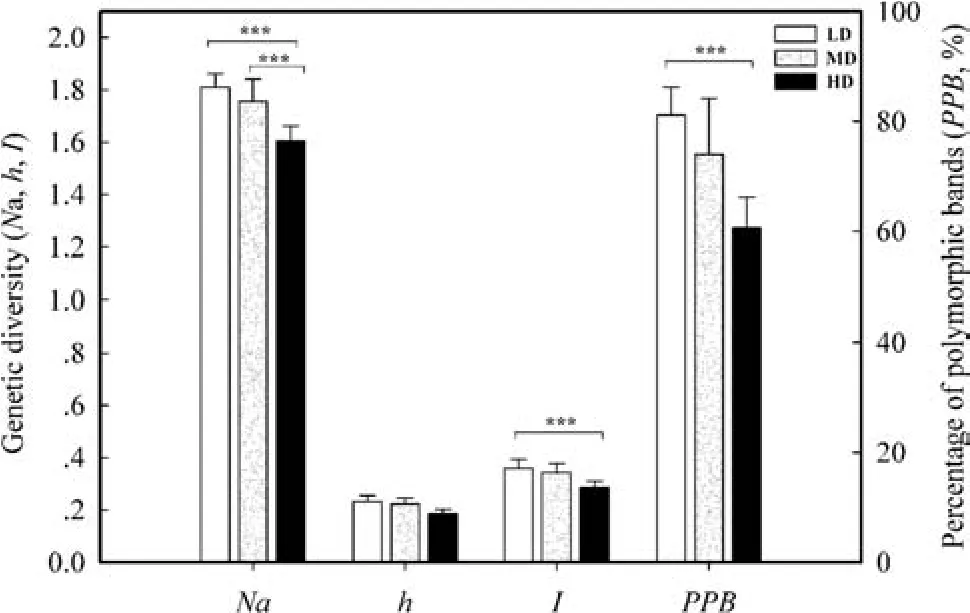
Fig.4 Comparison of population genetic diversity for V.mangachapoi among three disturbance levels
Discussion
Genetic diversity of V.mangachapoipopulations ofV.mangachapoiacross Hainan Island.We compared the results of the mean genetic diversity within populations—summarized by Nybom(2004)(meanhfor ISSR of long-lived perennial plants,wind dispersal plants,and out-crossing plants were 0.25,0.27,and 0.27,respectively)and compared to other tropical endangered trees,e.g.Hagenia abyssinica(long-lived perennial,outcrossing,anemochory;h=0.30)(Feyissa et al.2007)and we found a relatively low variation within populations ofV.mangachapoi(meanh=0.217).However,genetic diversity ofV.mangachapoiat species level(h=0.294,I=0.450)was higher than that at the population level,and was also higher than other endangered trees,e.g.Emmenopterys henryi(long-lived perennial,outcrossing,anemochory;h=0.191,I=0.287)(Li and Jin 2008).These results suggest that the genetic variation at species level ofV.mangachapoiis relatively high,with a considerable proportion of the genetic variation among populations.
This study provided a measurement of the genetic diversity present in 320 individual plants among the 11 natural
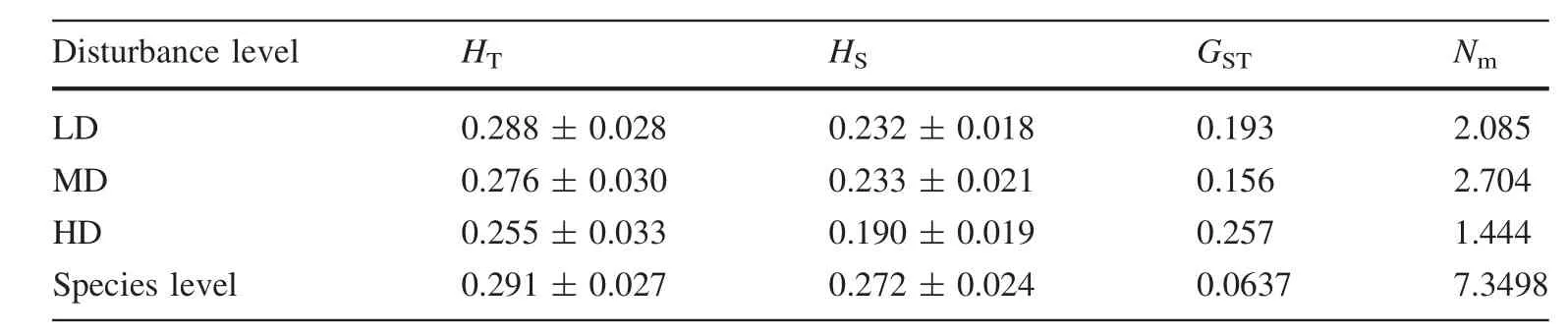
Table 2 Genetic variation of V.mangachapoi within disturbance level

Table 3 Effects of disturbance levels on population genetic diversity of V.mangachapoi
Isolation of population caused genetic reduction and differentiation
Anthropogenic disturbances accelerated and aggravated the fragmentation and shrinkage of remnant populations ofV.mangachapoi.Genetic drift may reduce genetic diversity and increase differentiation in smaller and isolated populations(Li and Jin 2008;Gaino et al.2010).The low disturbance(LD)populations ofV.mangachapoipossessed higher value for all genetic diversity indices(Fig.4).In comparison,the higher disturbance(HD)populations possessed lower genetic diversity.In addition,the results of PCoA analysis,AMOVA analysis,and Mantel tests strongly con fi rm signi fi cant gene differentiation of populations among different disturbance levels,showing a tendency to get larger.
输水管道防腐常用两种方法:一种是隔离法,采用非金属材料(环氧煤沥青、水泥砂浆)涂抹在管道表面;另一种是电化学保护法(牺牲阳极阴极保护)。输水管道工程考虑盐渍土和杂散电流干扰的腐蚀环境,综合采用这两种方法,对抑制管道腐蚀能达到很好的效果。
Therefore,the above results indicate that the anthropogenic disturbances would decrease genetic diversity and cause genetic differentiation among populations ofV.mangachapoi,especially highly disturbed populations.Our previous study also found genetic diversity of endangeredMadhuca hainanensiswas higher in the protected populations with less human disturbance in Hainan(Dai et al.2013).
Due to the long-term isolation derived from anthropogenic forest fragmentation on Hainan(Zhai et al.2014),the outcrossing and self-incompatible mating system ofV.mangachapoimay be attributed to the increase of genetic differentiation among remnant populations.This is because anthropogenic disturbances(especially large-scale land transition)will increase geographical isolation of the populations,which can decrease pollen immigration among populations(Bittencourt and Sebbenn 2007)and increase seed dispersal limitation across the fragmented populations(Herrera and Garcia 2010).All of this results in reducing genetic diversity and effective population size(Gaino et al.2010).The following evidence further supports these fi ndings.
A striking genetic divergence with the increasing geographic isolation driven by historical human disturbances is indeed discovered between our populations(Fig.3).The natural forests in Hainan suffered massive anthropogenic disturbance in history(Appendix A:Fig.S1)and werebroken into fragments.The remaining populations ofV.mangachapoiwere separated from each other spontaneously(Dai et al.2008)and now formed the geographical distribution pattern of isolation(Fig.1).The genetic gap of individuals appeared between low disturbance group and high disturbance group(Fig.2b).
In addition,V.mangachapoitrees begin fructifying after 15 years(Chen 1964).The older mature trees are rarely seen after the historical large-scale deforestation and land conversion.The remnant trees cannot provide effective or suf fi cient gene fl ow.Because of the outcrossing and selfincompatible mating system ofV.mangachapoi,the gene fl ow barriers probably occurred,which may result in increasing genetic differentiation among remnant populations by genetic drift(Li and Jin 2008).Therefore,population isolation,which is driven and aggravated by historical human disturbances,might be the cause of signi fi cantgenetic reduction and differentiation ofV.mangachapoi.
Implications for conservation
Current remnant populations have potential to re fl ect historical forest composition and changes.However,the longterm effects of deforestation may be only detected in their progeny,because of time-lag effects between disturbances and expression of traits in plants(Hamrick et al.1992).Even worse,compared with the impact of deforestation on animals,the biological consequence of deforestation on plants are not easily detectable and may not appear in a short-term period,especially at genetic level.Thus,it often may not be obvious enough to attract policymakers’attention.
In addition,tropical deforestation is an expanding global phenomenon(Gaino et al.2010;Zhai et al.2012),resulting in erosion of genetic diversity(Aguilar et al.2008).Although the government has implemented great conservation efforts,recent studies show that tropical plantations,especially rubber and pulp plantation,are still threatening Hainan’s remaining natural tropical forests(Zhai et al.2012,2014)with the reemergence of forest shrinkage on Hainan Island (annual forest density change rate=-0.94%)(Shi et al.2011).
To date,the natural forest in Hainan,even more than 65%nature reserves,is still facing an increasing of fragmentation(Wang et al.2013).Therefore,detailed assessment of these long-term historical disturbances on genetic diversity is extremely important for the urgent conservation management of trees,especially for endangered species(e.g.V.mangachapoi).
We recommend effective conservation pathways to bene fi t to the survival ofV.mangachapoipopulations on Hainan Island,based on our fi ndings and recommended conservation methods(Schemske et al.1994):
1. In situ conservation is urged as a priority becauseV.mangachapoishould grow completely free from disturbance,such as slashing in the population DF1 and TS1.
2. The low disturbance populations with higher diversity are the key protected units.
3. Establishment of corridors,which connected the central population from those key populations ofV.mangachapoi,would be an effective strategy to facilitate the gene communication and remove or decrease the in fl uences of geographical isolation.
4. A new reserve should be established for the population WC1,which harbors high genetic diversity and suffers from high geographical isolation and serious anthropogenic activities.
5. Effective restoration of remnant populations with seedlings from populations that possess higher genetic diversity needs to be implemented.
Seeds ofV.mangachapoican keep their vitality within a few days after ripening(Sist et al.2003).There is a clear and urgent need for seeds to be collected and reared to seedling stage,then replanted into the natural populations.
Conclusions
The genetic variation at the species level ofV.mangachapoiis relatively high,with a considerable proportion of genetic variation among populations.However,the anthropogenic disturbances decreased genetic diversity and caused genetic differentiation among populations ofV.mangachapoi,especially highly disturbed populations.
AcknowledgementsWe thank all forestry departments in Hainan,China,for their kind cooperation with distribution of surveys and sample collections.We kindly thank Chai-ShianKua(Xishuangbanna Tropical Botanical Garden,Chinese Academy of Sciences,China)for her helpful comments on the early version of this manuscript.We are grateful to the anonymous reviewers and the journal editors for their comments that have helped to improve this manuscript.
Compliance with ethical standards
Con fl ict of interestThe authors declare that they have no con fl ict of interest.
Aguilar R,Quesada M,Ashworth L,Herrerias-Diego Y,Lobo J(2008)Genetic consequences of habitat fragmentation in plant populations:susceptible signals in plant traits and methodological approaches.Mol Ecol 17:5177–5188
Ashton P(1998)Vatica mangachapoi.IUCN red list of threatened species.Version 2010.2.(http://www.iucnredlist.org/):International Union for Conservation of Nature and Natural Resources(IUCN)
Bittencourt JV,Sebbenn AM(2007)Patterns of pollen and seed dispersal in a small,fragmented population of the windpollinated treeAraucaria angustifoliain southern Brazil.Heredity 99:580–591
Breinholt JW,Van Buren R,Kopp OR,Stephen CL (2009)Population genetic structure of an endangered Utah endemic,Astragalus ampullarioides(Fabaceae).Am J Bot 96:661–667
Bunyavejchewin S,LaFrankie JV,Baker PJ,Kanzaki M,Ashton PS,Yamakura T(2003)Spatial distribution patterns of the dominant canopy dipterocarp species in a seasonal dry evergreen forest in western Thailand.For Ecol Manag 175:87–101
Chen HY(1964)Flora of Hainan,vol 1.Science Press,Beijing,pp 516–517(in Chinese)
Dai ZC,Zhong QX,Si CC,Lin Y,Wang KR,Zhang B,Du DL(2008)A review of study on endangered mechanism and conservation ecology of endangeredVatica mangachapoi.J Hainan Normal University(Natural Science)21:82–86(in Chinese with English abstract)
Dai ZC,Si CC,Zhai DL,Huang P,Qi SS,Zhong QX,Hu X,Li HM,Du DL (2013)Human impacts on genetic diversity and differentiation in six natural populations ofMadhuca hainanensis,an endemic and endangered timber species in China.Biochem Syst Ecol 50:212–219
Dai ZC,Qi SS,Miao SL,Liu YT,Tian YF,Zhai DL,Huang P,Du DL(2015)Isolation of NBS-LRR RGAs from invasiveWedelia trilobataand the calculation of evolutionary rates to understand bioinvasion from a molecular evolution perspective.Biochem Syst Ecol 61:19–27
Ding Y,Zang RG,Jiang Y-X(2006)Effect of hillslope gradient on vegetation recovery on abandoned land of shifting cultivation in Hainan Island,South China.J Integr Plant Biol 48:642–653
Fatemi M,Gross CL(2009)Life on the edge—high levels of genetic diversity in a cliff population ofBertya ingramiiare attributed toB.rosmarinifolia(Euphorbiaceae).Biol Conserv 142:1461–1468
Feyissa T,Nybom H,Bartish I,Welander M(2007)Analysis of genetic diversity in the endangered tropical tree speciesHagenia abyssinicausing ISSR markers.Genet Resour Crop Evol 54:947–958
Gaino APSC,Silva AM,Moraes MA,Alves PF,Moraes MLT,Freitas MLM,Sebbenn AM (2010)Understanding the effects of isolation on seed and pollen fl ow,spatial genetic structure and effective population size of the dioecious tropical tree speciesMyracrodruon urundeuva.Conserv Genet 11:1631–1643
Hamrick JL,Godt MJW,Sherman-Broyles SL(1992)Factors in fl uencing levels of genetic diversity in woody plant species.New For 6:95–124
Herrera JM,Garcia D(2010)Effects of forest fragmentation on seed dispersal and seedling establishment inOrnithochoroustrees.Conserv Biol 24:1089–1098
Jump AS,Marchant R,Penuelas J(2009)Environmental change and the option value of genetic diversity.Trends Plant Sci 14:51–58
Lewontin RC(1972)The apportionment of human diversity.Evol Biol 6:381–398
Li JM,Jin ZX(2008)Genetic structure of endangeredEmmenopterys henryiOliv.based on ISSR polymorphism and implications for its conservation.Genetica 133:227–234
Lopez-Pujol J,Zhang FM,Ge S(2006)Plant biodiversity in China:richly varied,endangered,and in need of conservation.Biodivers Conserv 15:3983–4026
Luo S,He Y,Ning G,Zhang J,Ma G,Bao M(2011)Genetic diversity and genetic structure of different populations of the endangered speciesDavidia involucratain China detected by inter-simple sequence repeat analysis.Trees Struct Funct 25:1063–1071
Manohara TN,Linto EL,Renuka C(2010)Diversity and conservation of palms in Andaman and Nicobar archipelago.Biodivers Conserv 19:3655–3666
Matesanz S,Gimeno TE,de la Cruz M,Escudero A,Valladares F(2011)Competition may explain the fi ne-scale spatial patterns and genetic structure of two co-occurring plant congeners.J Ecol 99:838–848
Mayer AL,Kauppi PE,Angelstam PK,Zhang Y,Tikka PM(2005)Importing timber, exporting ecological impact. Science 308:359–360
McDermott JM,McDonald BA(1993)Gene fl ow in plant pathosystems.Annu Rev Phytopathol 31:353–373
Nei M(1972)Genetic distance between populations.Am Nat 106:283–292
Nei M(1973)Analysis of gene diversity in subdivided populations.Proc Natl Acad Sci USA 70:3321–3323
Nybom H(2004)Comparison of different nuclear DNA markers for estimating intraspeci fi c genetic diversity in plants.Mol Ecol 13:1143–1155
Peakall ROD,Smouse PE(2006)Genalex 6:genetic analysis in Excel.Population genetic software for teaching and research.Mol Ecol Notes 6:288–295
Rucinska A,Puchalski J(2011)Comparative molecular studies on the genetic diversity of an ex situ garden collection and its source population of the critically endangered polish endemic plantCochlearia polonicaE.Frohlich.Biodivers Conserv 20:401–413
SAS Institute Inc(2004)SAS/STAT user’s guide.Version 9.1.SAS Institute Inc.,Cary
Schemske DW,Husband BC,Ruckelshaus MH,Goodwillie C,Parker IM,Bishop JG(1994)Evaluating approaches to the conservation of care and endangered plants.Ecology 75:584–606
Shi L,Zhao S,Tang Z,Fang J(2011)The changes in china’s forests:an analysis using the forest identity.PLoS ONE 6:e20778
Sist P,Fimbel R,Sheil D,Nasi R,Chevallier MH(2003)Towards sustainable management of mixed dipterocarp forests of Southeast Asia:moving beyond minimum diameter cutting limits.Environ Conserv 30:364–374
State Forestry Administration PR,China(1999)List of National key protected wild plants(1st Part).State Forestry Administration,Beijing(in Chinese)
Stewart CN Jr,Via L(1993)A rapid CTAB DNA isolation technique useful for RAPD fi ngerprinting and other PCR applications.Biotechniques 14:748–750
Wang W,Pechacek P,Zhang M,Xiao N,Zhu J,Li J(2013)Effectiveness of nature reserve system for conserving tropical forests:a statistical evaluation of Hainan Island,China.PLoS ONE 8:e57561
Yeh F,Boyle T,Rongcai Yang,Ye Z,Xian JM(1999)POPGENE 32,Microsoft windows-based freeware for population genetic analysis,version 1.32.University of Alberta,Edmonton
Zhai DL,Cannon CH,Slik JWF,Zhang CP,Dai ZC(2012)Rubber and pulp plantations represent a double threat to Hainan’s natural tropical forests.J Environ Manag 96:64–73
Zhai DL,Xu JC,Dai ZC,Cannon CH,Grumbine RE(2014)Increasing tree cover while losing diverse natural forests in tropical Hainan,China.Reg Environ Change 14:611–621
Zhang YQ,Uusivuori J,Kuuluvainen J(2000)Econometric analysis of the causes of forest land use changes in Hainan,China.Can J For Res 30:1913–1921
猜你喜欢
杂志排行
Journal of Forestry Research的其它文章
- Measurement of lumber moisture content based on PCA and GSSVM
- Characterization of mean stem density, fi bre length and lignin from two Acacia species and their hybrid
- Theoretical modeling of the effects of temperature and moisture content on the acoustic velocity of Pinus resinosa wood
- The properties of fl ax fi ber reinforced wood fl our/high density polyethylene composites
- Risks involved in fecal DNA-based genotyping of microsatellite loci in the Amur tiger Panthera tigris altaica:a pilot study
- Genecological zones and selection criteria for natural forest populations for conservation:the case of Boswellia papyrifera in Ethiopia
