酰胺酶的挖掘及在有机合成中的应用进展
2018-01-23吴哲明金建强郑仁朝
吴哲明,金建强,郑仁朝
(1. 浙江工业大学 生物工程学院,浙江 杭州 310014;2. 浙江省生物有机合成技术研究重点实验室, 浙江 杭州 310014)
酰胺酶(amidase,EC 3.5.1.X),又称酰胺水解酶(amidohydrolase),是一类催化酰胺化合物水解生成相应羧酸和氨的重要水解酶。该酶促反应的本质是催化酰基从供体(酰胺)转移至受体(如水),因此,当体系中存在比水亲核性更高的羟胺时,则生成相应的氧肟酸(图1)[1-3]。作为腈转化酶家族的重要成员,酰胺酶的底物谱很广,能够水解各种天然及人工合成的脂肪族、芳香族及杂环类酰胺。酰胺酶所具有的高立体选择性、广底物谱等特性使其在动力学拆分外消旋酰胺制备手性羧酸、手性酰胺衍生物及光学纯氨基酸等方面具有独特优势,正日益受到研究者的重视(表1)[4-7]。
酰胺酶最早由Kelly等[20]和Jakoby等[21]分别在Pseudomonasaeruginosa和P.fluorescens中发现,其来源十分广泛,存在于各类原核、真核微生物和动植物中[22-23]。其中,细菌是酰胺酶的主要来源,如红球菌属Rhodococcus[24-25]、假单孢菌属Pseudomonas[26]、嗜硫菌属Sulfolobus[27-28]、芽孢杆菌属Bacillus[29]、代尔夫特菌属Delftia[30-31]、克雷伯氏菌属Klebsiella[32]和苍白杆菌属Ochrobactrum[33]等。
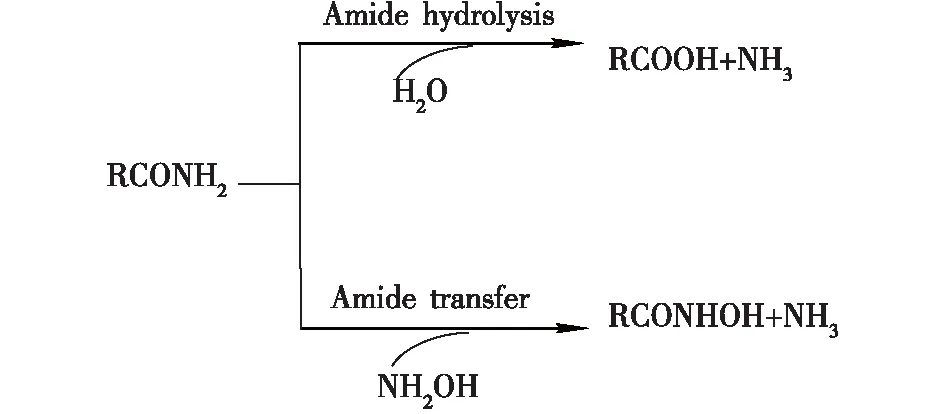
图1 酰胺酶催化的水解和酰基转移反应Fig.1 The hydrolysis and acyl transfer reaction catalyzed by amidase

表1 酰胺酶催化的外消旋酰胺的立体选择性水解
1 酰胺酶的分类及结构
酰胺酶的种类很多,不同来源的酰胺酶结构和性质差异显著,目前仍未有一个系统的分类方法。根据不同的标准,酰胺酶有多种分类方式。根据底物特异性差异,可分为广谱类酰胺酶、α-氨基酰胺酶、脂肪族酰胺酶和芳香族酰胺酶等;根据酰胺酶基因上下游是否存在腈水合酶基因编码,可以分为与腈水合酶耦联的酰胺酶和非耦联的酰胺酶。近年来,基于氨基酸序列的酰胺酶分类方法受到普遍认可。Chebrou等[34]通过氨基酸序列比对分析,提出将酰胺酶分为腈水解酶超家族(nitrilase superfamily)和酰胺酶标签(amidase signature,AS)家族两大类。
腈水解酶家族酰胺酶是一类含有保守亲核半胱氨酸的巯基酶。该家族酰胺酶之间序列同源性高,与腈水解酶具有序列相似性,并含有保守的Glu、Cys和Lys催化三联体负责共价键催化(图2)。目前,已知的腈水解酶家族酰胺酶底物谱较窄,催化短链脂肪族酰胺的水解。酶蛋白通常以同源四聚体、六聚体的形式存在,单体结构一般表现为α-β-β-α式的夹心折叠。Novo等[35]以蠕虫腈水解酶融合蛋白晶体结构为模版,假定催化三联体Glu-Lys-Cys在所有腈水解酶家族成员中均为保守,通过对比模拟,建立了P.aeruginosa酰胺酶3D结构模型,证明了催化三联体Glu-Lys-Cys负责共价键的催化,其中Cys为亲核体。Kimani等[36]对GeobacilluspallidusRAPc8酰胺酶晶体结构研究表明,其单体具有典型的腈水解酶超家族α-β-β-α折叠,同时也证实了在腈水解酶超家族成员中Glu-Lys-Cys催化三联体的保守性。
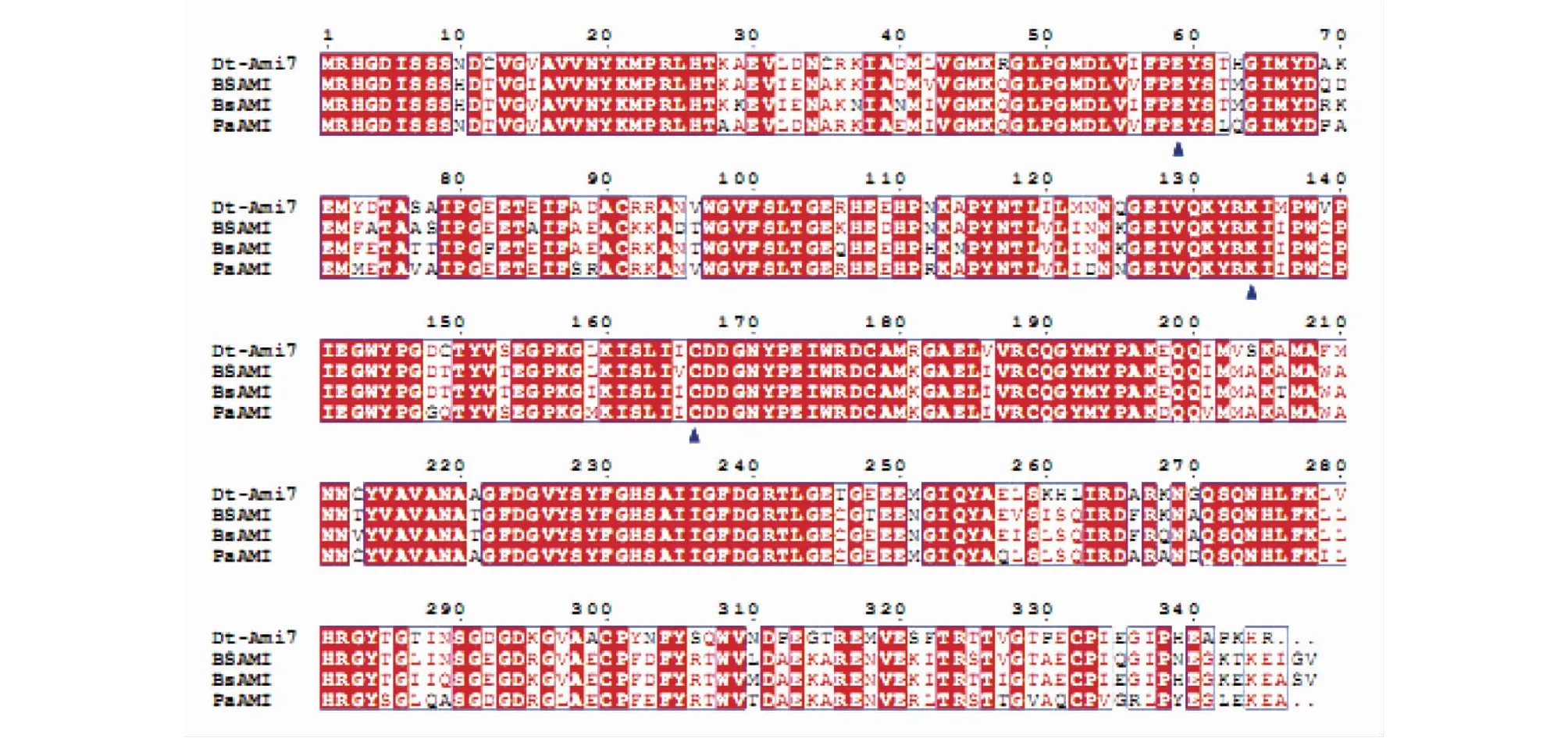
Dt-Ami7—Delftia tsuruhatensis ZJB-05174酰胺酶(KP943495);BS AMI—B. stearothermophilus酰胺酶(Q9RQ17);Bs AMI—Bacillus sp. BR449酰胺酶 (AF257487);Pa AMI—P. aeruginosa酰胺酶(AAA25697)图2 腈水解酶家族酰胺酶序列比对Fig.2 Sequence alignment of nitrilase superfamily amidases
有别于腈水解酶家族酰胺酶,酰胺酶标签家族酰胺的一级结构中含有一段高度保守的GGSS区域,且具有含约130个富含甘氨酸、丝氨酸和丙氨酸等氨基酸组成的高度保守序列(AS序列),其催化三联体为Ser、Ser和Lys(图3),一般呈同源二聚体或同源八聚体。酰胺酶标签家族酰胺酶的底物谱通常较广,能够水解脂肪族、芳香族和杂环酰胺[24,37]。目前已报道结构的标签家族酰胺酶较多,如来源于Stenotrophomonasmaltophilia的PAM是酰胺酶标签家族中第一个已解析三级结构的酰胺酶,具有清晰的α-β夹心折叠结构,中心的β-折叠片核心被α-螺旋包围,该折叠在标签酰胺酶家族成员中保守[38]。Ohtaki等[39]发现Rhodococcussp. N-771酰胺酶晶体结构含有3个结构域:N端α螺旋结构域,小结构域和大结构域。大结构域含有一个α/β结构,由一个疏水核心(18个β折叠片)和8个位于β折叠片两侧的α螺旋构成,其包含了一段酰胺酶标签序列。小结构域有5个α螺旋,位于大结构域顶端。Ser-cisSer-Lys催化三联体位于大结构域,而小结构域的一些疏水性氨基酸残基则参与底物的识别。Lee等[40]报道了芳基酰基酰胺酶(AAA)的晶体结构,发现其属于α/β水解酶家族,催化三联体为Ser187、Ser163与Lys84。该酶外部结构为α螺旋,内部则由β折叠结构组成,并且拥有一个独特的由两个loop环和一个α螺旋构成的底物结合口袋。

Dt-Ami 2、Dt-Ami 6—D. tsuruhatensis ZJB-05174酰胺酶(KP943493、KP943494);Ca. AMI—R. rhodochrous Jl 酰胺酶(BAA03744);PAM—S. maltophilia酰胺酶 (CAC93616);Re. AMI—R. erythropolis MP50酰胺酶(AY026386)图3 腈水解酶家族酰胺酶序列比对Fig.3 Sequence alignment of amidase signature family amidases
2 酰胺酶的催化机制
目前,酰胺酶的催化反应机制尚不十分明晰。Maestracci等[41]以Rhodococcussp. R312酰胺酶为研究对象,以乙酰胺和羟胺为双底物,发现并证明其介导的酰基转移反应符合双底物乒乓反应机制,即底物先与酰胺酶结合形成酰基-酶复合物,进而将底物的酰基转移至其受体羟胺,生成相应的氧肟酸。Kobayashi等[42]认为底物酰胺的羰基受到亲核进攻时,与酶形成一个四面体中间体,中间体因氨的形成并解离而快速转变为酰基-酶复合物。在水分子加入后,复合物发生水解生成相应的酸(图4)。
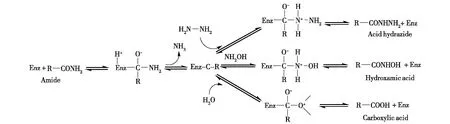
图4 酰胺酶水解反应和酰基转移反应作用机制Fig.4 Mechanism of amide hydrolysis reactionand acyl transfer reaction from amide to hydrazine and hydroxylamine
随着酰胺酶结构的不断解析,基于催化三联体的酰胺酶催化机制被研究者广泛报道。脂肪酸酰胺水解酶(FAAH)是一个典型的标签家族酰胺酶,其催化机制被学者广泛研究[43-45]。Mileni等[46]推测FAAH的反应机制主要为以下四步:1)亲核体Ser241被极化激活并进攻底物酰胺键,形成酶-底物复合体;2)酰胺底物C—N键断裂,氨基部分脱离,形成酰基-酶中间体;3)活化的水分子进攻酰基-酶中间体并形成新的四面体中间体;4)复合体分解产生羧酸,而酰胺酶则获得再生(图5)。

图5 酰胺酶FAAH假设的催化机制Fig.5 Proposed reaction mechanism of FAAH
Labahn等[38]研究标签家族酰胺酶PAM催化机制主要分为4步(图6):1)cisSer202侧链羟基的质子转移至碱催化剂Lys的氨基上,增强了其对底物酰胺羰基氧的质子化能力;Ser226亲核进攻酰胺羰基碳原子,同时cisSer202质子化Ser226的羰基氧;随后,失去质子的cisSer202夺取Ser226的质子,形成酶-底物复合体;2)复合体上的氨基夺取cisSer202的质子,而失去质子的cisSer202转而从带正电的Lys123捕获质子,恢复稳定;3)质子化的氨基形成氨脱离复合体,而Lys123重新夺回cisSer202上的质子;失去质子后的cisSer202转而捕获底物上的羟基质子,两者恢复最初状态;同时,底物的羰基也重新产生,形成酶-酰基中间体;4)水分子进攻酶-酰基中间体使其分解,形成产物羧酸。Ser226获得水分子的质子,重新恢复稳定。

图6 酰胺酶PAM假设的催化机制Fig.6 Proposed reaction mechanism of PAM
Lee等[40]报道了标签家族酰胺酶AAA的晶体结构,并解释了其催化机制。该酶以Ser163、Ser187与Lys84作为催化三联体,其中Lys84作为广义碱催化剂接受来自于Ser163通过氢键传递的质子,激活Ser163(图7)。具体催化步骤如下:1)在碱性环境下的Lys84处于去质子化状态,其通过氢键网络介导cisSer163的Oγ与亲核体Ser187的OγH通过氢键发生极化并使之激活;2)Ser187的OγH与底物(对乙酰氨基酚)的羰基碳通过共价键形成四面体中间体,同时Ser187的Oγ脱去质子;3)形成酰基-酶复合体,随后在1分子水的作用下发生脱酰反应;4)酶与产物分离,酰胺酶获得再生。

图7 酰胺酶AAA假设的催化机制Fig.7 Proposed reaction mechanism of AAA
3 酰胺酶的筛选与挖掘
传统的酰胺酶筛选是从土壤或菌种库中筛选具有酰胺酶活力的微生物,但效率低,耗时长,而且无法区分酰胺酶是否具有立体选择性。笔者所在团队的Zheng等[47]在首次证明酰胺酶催化的酰基转移和水解反应立体选择性一致的基础上,利用酰基转移反应产物氧肟酸与铁离子在酸性条件下螯合显色的特性,建立了高通量立体选择性酰胺酶筛选模型(图8)。该方法与已有的基于底物官能团的酰胺酶筛选模型[48]相比,其更显著的优势在于对底物的普适性,是目前报道的普适性最强的酰胺酶筛选模型。

图8 基于酰基转移反应的比色法筛选立体选择性酰胺酶Fig.8 Selective colorimetric screen for enantioselective amidase-producing microorganisms
近20年来,虽然已从自然环境中筛选获得大量产酰胺酶的微生物(表2),但由于野生菌中酰胺酶的表达水平通常较低,且可能同一微生物中存在几种不同立体选择性的酰胺酶,酶活、对映选择性低等问题成为其大规模应用的瓶颈。因此,研究人员把目光投向了如何快速、高效获得高立体选择性、高催化活力的酰胺酶。近年来,得益于DNA测序技术的进步,急剧增加的生物信息为新酶的开发带来了前所未有的机遇,基因挖掘已成为快速开发新酶的有力手段。截至2017年10月,根据基因组测序计划统计网站GOLD(www.genomesonline.org)的统计数据,目前已完成全基因组测序的生物多达282 640种,其中细菌完成测序250 327种,真核生物20 956种,这为酰胺酶的挖掘提供了宝库(图9)。笔者根据酰胺酶保守序列,通过基因挖掘从DelftiatsuruhatensisZJB-05174、ParvibaculumlavamentivoransZJB14001[63]以及BurkholderiaphytofirmansZJB-15079[64]全基因组中分别获得了若干重组酰胺酶。同时,Ruan等[65]利用基因组同源比对和HiTAIL-PCR对未知区域的高效扩增,从B.epidermidisZJB-07021基因组中克隆获得2个酰胺酶基因,并实现了异源表达。

表2 不同来源酰胺酶的性质
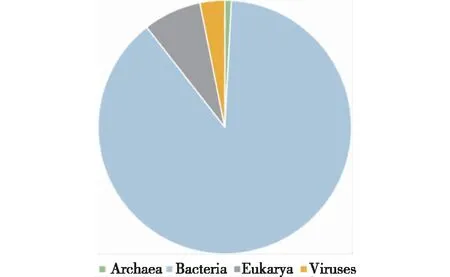
图9 完成全基因组测序生物组成Fig.9 Sequenced genome projects in GOLD (www.genomesonline.org)
4 酰胺酶的应用
手性羧酸和酰胺衍生物是重要的旋光性模块化合物,可用于大量手性生物活性分子的合成,包括羟基酸、氨基酸、甲基氨基酸、伯胺和伪核苷化合物等,在精细化工、医药、农用化学品及功能材料等方面有广泛应用。酰胺酶底物谱广、催化活性高以及对映选择性严格等特点使其在制备复杂结构手性羧酸及酰胺衍生物具有无可比拟的优势。笔者所在课题组多年来致力于酰胺酶生物催化工业应用开发,建立了一系列以酰胺酶为催化剂的(手性)羧酸及酰胺的生物合成新工艺。
4.1 (S)-2,2-二甲基环丙甲酰胺的合成
(S)-2,2-二甲基环丙甲酰胺是抗重症感染首选药物亚胺培南/西司他丁钠的关键手性中间体,开发其高效合成技术对控制西司他丁钠成本具有重要意义。目前工业上应用的化学法合成工艺步骤冗长,反应条件苛刻,工艺总收率低,且需大量有毒有害试剂,环境负担大。
笔者所在团队的Zheng等[31]通过建立普适性高通量立体选择性酰胺酶筛选模型,获得了能够R型立体选择性水解2,2-二甲基环丙烷甲酰胺的酰胺酶产生菌D.tsuruhatensisZJB-05174,对映体选择率E>100(图10)。在添加5%的乙腈作为共溶剂后,(S)-2,2-二甲基环丙甲酰胺的收率为43.6%,光学纯度达到99%以上。
4.2 1-氰基环己基乙酸的合成
加巴喷丁(Gabapentin)是一种新型抗癫痫药物,具有疗效好、安全性高和耐受性好等特点[66-67]。1-氰基环己基乙酸(1-CCHAA)是合成加巴喷丁的关键中间体。笔者等[68]通过基因挖掘,筛选获得了可高效水解1-氰基环己基乙酰胺(1-CCHAM)合成1-CCHAA 的酰胺酶Pa-Ami,比酶活达297.6 U/mg。以含Pa-Ami的重组菌为催化剂水解1-CCHAM(图11),底物质量浓度为80 g/L,湿菌体质量浓度为1 g/L时,反应20 min,转化率可达100%。
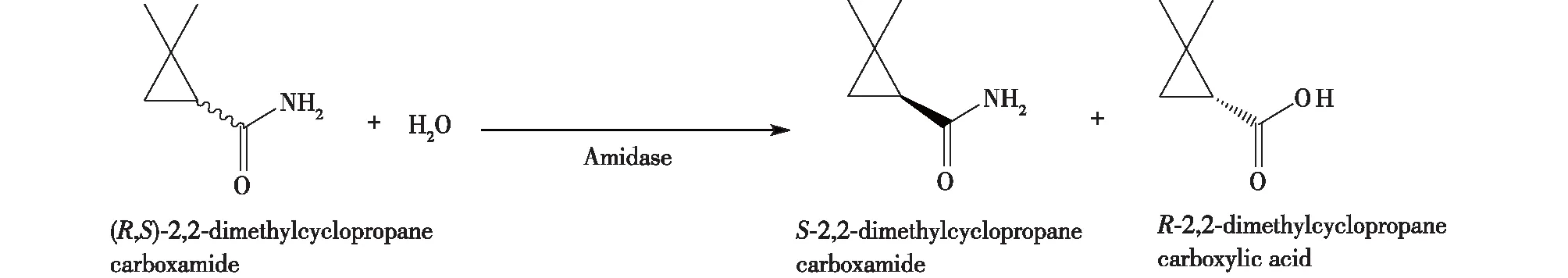
图10 R,S-2,2-二甲基环丙烷甲酰胺的酶法拆分Fig.10 Enzymatic resolution of (R,S)-2,2-dimethylcyclopropane carboxamide

图11 酰胺酶法制备加巴喷丁路线Fig.11 Enzymatic route for synthesis of gabapentin by amidase
4.3 R -3,3,3-三氟-2-羟基-2-甲基丙酸的合成
3,3,3-三氟-2-羟基-2-甲基丙酸(TFHMA)是一种手性医药中间体,其S型或R型异构体均可用于系列药物的合成[69-70],如S型异构体可用于制备新型ATP敏感性钾(KATP)通道开放剂[71];R型异构体可合成丙酮酸脱氢酶激酶(PDK)抑制剂等[72]。瑞士Lonza公司的Shaw等[73]利用产酸克雷伯氏菌KlebsiellaoxytocaR-酰胺酶拆分外消旋3,3,3-三氟-2-羟基-2-甲基丙酰胺(TFHMM)(图12),制备R-3,3,3-三氟-2-羟基-2-甲基丙酸和S-3,3,3-三氟-2-羟基-2-甲基丙酰胺,底物质量浓度100 g/L,产率接近50%,产物e.e.>98%。笔者等[64]利用来源于B.phytofirmansZJB-15079的重组酰胺酶工程菌拆分3,3,3-三氟-2-羟基-2-甲基丙酰胺,在底物质量浓度200 g/L、湿菌体质量浓度5 g/L、80 ℃下反应10 min,R-3,3,3-三氟-2-羟基-2-甲基丙酸质量浓度可达86.2 g/L,e.e.>95%,E值达86。

图12 R,S-3,3,3-三氟-2-羟基-2-甲基丙酰胺的酰胺酶法拆分Fig.12 Enzymatic resolution of (R,S)-3,3,3-trifluoro-2-hydroxy-2-methylpropanamide
4.4 2-氯烟酸的合成
2-氯烟酸是一种重要精细化工中间体,广泛用于杀菌剂、杀虫剂、抗生素和心血管疾病药物的合成[74-76]。金建强[77]通过大量筛选,获得了可高活性水解2-氯烟酰胺合成2-氯烟酸的重组酰胺酶工程菌(图13),并考察了其对系列氯代烟酰胺的反应动力学。全细胞(湿菌体质量浓度5 g/L)催化2-氯烟酰胺水解底物浓度可达400 mmol/L,转化率达94%。

图13 酰胺酶水解合成2-氯烟酸Fig.13 Enzymatic route for synthesis of 2-chloronicotinic acid by amidase
5 结论
不对称生物合成方兴未艾,酰胺酶正崛起为不对称生物催化的重要工具酶。作为腈转化酶家族中的重要一员,酰胺酶所具有的高立体选择性、广底物谱等特性使其在动力学拆分外消旋酰胺制备复杂结构手性羧酸及酰胺衍生物中占据无可比拟的优势。随着酰胺酶空间结构的解析,其反应催化机制也正逐渐被揭示。然而,由于对酰胺酶底物特异性和立体选择性规律研究的缺乏,酰胺酶的开发仍只能局限于以“底物”为探针筛选“催化剂”的传统研究范畴,无法主动进行生物催化剂的选择和理性改造。通过探究酰胺酶的“结构和功能”的关系,在掌握酶的结构信息的情况下对酰胺酶进行分子改造,可进一步促进酰胺酶的工业化应用。
[1] NEL A J,TUFFIN I M,SEWELL B T,et al.Unique aliphatic amidase from a psychrotrophic and haloalkaliphilicNesterenkoniaisolate[J].Appl Environ Microbiol,2011,77(11):3696-3702.
[2] SHEN W,CHEN H,JIA K,et al.Cloning and characterization of a novel amidase fromParacoccussp. M-1,showing aryl acylamidase and acyl transferase activities[J].Appl Microbiol Biotechnol,2012,94(4):1007-1018.
[3] ZHANG J,YIN J G,HANG B J,et al.Cloning of a novel arylamidase gene fromParacoccussp. strain FLN-7 that hydrolyzes amide pesticides[J].Appl Environ Microbiol,2012,78(14):4848-4855.
[5] LORENZ P,ECK J.Screening for novel industrial biocatalysts[J].Eng Life Sci,2010,4(6):501-504.
[6] MA D Y,WANG D X,PAN J,et al.Nitrile biotransformations for the synthesis of highly enantioenrichedβ-hydroxy andβ-amino acid and amide derivatives:a general and simple but powerful and efficient benzyl protection strategy to increase enantioselectivity of the amidase[J].J Org Chem,2008,73(11):4087-4091.
[7] JAEGER K E,DIJKSTRA B W,REETZ M T.Bacterial biocatalysts:molecular biology,three-dimensional structures,and biotechnological applications of lipases[J].Annu Rev Microbiol,1999,53(1):315-351.
[8] HIRRLINGER B,STOLZ A,KNACKMUSS H J.Purification and properties of an amidase fromRhodococcuserythropolisMP50 which enantioselectively hydrolyzes 2-arylpropionamides[J].J Bacteriol,1996,178(12):3501-3507.
[9] DORAN J P,DUGGAN P,MASTERSON M,et al.Expression and purification of a recombinant enantioselective amidase[J].Protein Exp Purif,2005,40(1):190-196.
[10] BIANCHI D,BATTISTEL E,CESTI P,et al.Substrate specificity and stereoselectivity of hydrolytic enzymes fromBrevibacteriumimperialeB222[J].Appl Microbiol Biotechnol,1993,40(1):53-56.
[11] NOJIRI M,TAOKA N,YASOHARA Y.Characterization of an enantioselective amidase fromCupriavidussp.KNK-J915 (FERM BP-10739) useful for enzymatic resolution of racemic 3-piperidine carboxamide[J].J Mol Catal B:Enzymatic,2014,109:136-142.
[12] SNELL D,COLBY J.Enantioselective hydrolysis of racemic ibuprofen amide toS-(+)-ibuprofen byRhodococcusAJ270[J].Enzyme Microb Technol,1999,24(4):160-163.
[13] EICHHORN E,RODUIT J P,SHAW N,et al.Preparation of (S)-piperazine-2-carboxylic acid,(R)-piperazine-2-carboxylic acid,and (S)-piperidine-2-carboxylic acid by kinetic resolution of the corresponding racemic carboxamides with stereoselective amidases in whole bacterial cells[J].Tetrahedron Asymmetry,1998,29(1):2533-2536.
[14] WEGMAN M A,HEINEMANN U,RANTWIJK F V,et al.Hydrolysis of D,L -phenylglycine nitrile by new bacterial cultures[J].J Mol Catal B:Enzymatic,2001,11(4):249-253.
[15] KOMEDA H,ASANO Y.Gene cloning,nucleotide sequencing,and purification and characterization of the D-stereospecific amino-acid amidase fromOchrobactrumanthropiSV3[J].Eur J Biochem,2000,267(7):2028-2035.
[16] BAEK D H,KWON S J,HONG S P,et al.Characterization of a thermostable D-stereospecific alanine amidase fromBrevibacillusborstelensisBCS-1[J].Appl Environ Microbiol,2003,69(2):980-986.
[17] VAN DEN TWEEL W J J,VAN DOOREN T J G M,DE JONGE P H,et al.OchrobactrumanthropiNCIMB 40321:a new biocatalyst with broad-spectrum L-specific amidase activity[J].Appl Microbiol Biotechnol,1993,39(3):296-300.
[18] KRIEG L,KULA M R.Screening for amidases:isolation and characterization of a novel D-amidase fromVariovoraxparadoxus[J].Adv Synth Catal,2002,344(9):965-973.
[19] KOMEDA H,ASANO Y.A novel D-stereoselective amino acid amidase fromBrevibacteriumiodinum:gene cloning,expression and characterization[J].Enzyme Microb Technol,2008,43(3):276-283.
[20] KELLY M,KORNBERG H L.Purification and properties of acyltransferases fromPseudomonasaeruginosa[J].Biochem J,1964,93(3):557-566.
[21] JAKOBY W B,FREDERICKS J.Reactions catalyzed by amidases[J].J Biol Chem,1964,239(6):1978-1982.
[22] SHARMA M,SHARMA N N,BHALLA T C.Amidases:versatile enzymes in nature[J].Rev Environ Sci Biotechnol,2009,8(4):343-366.
[23] FOURNAND D,ARNAUD A.Aliphatic and enantioselective amidases:from hydrolysis to acyl transfer activity[J].J Appl Microbiol,2001,91(3):381-393.
[24] PERTSOVICH S I,GURANDA D T,PODCHERNYAEV D A,et al.Aliphatic amidase fromRhodococcusrhodochrousM8 is related to the nitrilase/cyanide hydratase family[J].Biochemistry,2005,70(11):1280-1287.
[25] JIN L Q,LI Y F,LIU Z Q,et al.Characterization of a newly isolated strainRhodococcuserythropolisZJB-09149 transforming 2-chloro-3-cyanopyridine to 2-chloronicotinic acid[J].New Biotechnol,2011,28(6):610-615.
[26] NOVO C,FARNAUD S,TATA R,et al.Support for a three-dimensional structure predicting a Cys-Glu-Lys catalytic triad forPseudomonasaeruginosaamidase comes from site-directed mutagenesis and mutations altering substrate specificity[J].Biochem J,2002,365(3):731-738.
[27] CILIA E,FABBRI A,URIANI M,et al.The signature amidase fromSulfolobussolfataricusbelongs to the CX3C subgroup of enzymes cleaving both amides and nitriles:Ser195 and Cys145 are predicted to be the active site nucleophiles[J].FEBS J,2010,272(18):4716-4724.
[28] GIORDANO C,AMMENDOLA S.Characterization of mutants ofSulfolobussolfataricussignature amidase able to hydrolyseR-ketoprofen amide[J].Protein Pept Lett,2008,15(6):617-623.
[29] AGARWAL S,CHOUDHURY B.Presence of multiple acyltranferases with diverse substrate specificity inBacillussmithiistrain IITR6b2 and characterization of unique acyltransferase with nicotinamide[J].J Mol Catal B:Enzymatic,2014,107:64-72.
[30] HONGPATTARAKERE T,KOMEDA H,ASANO Y.Purification,characterization,gene cloning and nucleotide sequencing of D-stereospecific amino acid amidase from soil bacterium:Delftiaacidovorans[J].J Ind Microbiol Biotechnol,2005,32(11/12):567-576.
[31] ZHENG R C,WANG Y S,ZHENG Y G,et al.Kinetic resolution of (R,S)-2,2-dimethylcyclopropanecarboxamide byDelftiatsuruhatensisZJB-05174:role of organic cosolvent in reaction medium[J].Catal Commun,2012,18(1):68-71.
[32] GUO F M,WU J P,YANG L R,et al.Soluble and functional expression of a recombinant enantioselective amidase fromKlebsiellaoxytocaKCTC 1686 inEscherichiacoliand its biochemical characterization[J].Process Biochem,2015,50(8):1264-1271.
[33] KOMEDA H,ISHIKAWA N,ASANO Y.Enhancement of the thermostability and catalytic activity ofd-stereospecific amino-acid amidase fromOchrobactrumanthropiSV3 by directed evolution[J].J Mol Catal B:Enzymatic,2003,21(4):283-290.
[34] CHEBROU H,BIGEY F,ARNAUD A,et al.Study of the amidase signature group[J].Biochim Biophys Acta,1996,1298(2):285-293.
[35] NOVO C,FARNAUD S,TATA R,et al.Support for a three-dimensional structure predicting a Cys-Glu-Lys catalytic triad forPseudomonasaeruginosaamidase comes from site-directed mutagenesis and mutations altering substrate specificity[J].Biochem J,2002,365(3):731-738.
[36] KIMANI S W,AGARKAR V B,COWAN D A,et al.Structure of an aliphatic amidase fromGeobacilluspallidusRAPc8[J].Acta Crystallogr,2007,63:1048-1058.
[37] WU Z M,ZHENG R C,ZHENG Y G.Exploitation and characterization of three versatile amidase super family members fromDelftiatsuruhatensisZJB-05174[J].Enzyme Microb Technol,2016,86:93-102.
[38] LABAHN J,NEUMANN S,BÜLDT G,et al.An alternative mechanism for amidase signature enzymes[J].J Mol Biol,2002,322(5):1053-1064.
[39] OHTAKI A,MURATA K,SATO Y,et al.Structure and characterization of amidase fromRhodococcussp.N-771:insight into the molecular mechanism of substrate recognition[J].Biochim Biophys Acta,2010,1804(1):184-192.
[40] LEE S,PARK E H,KO H J,et al.Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family[J].Biochem Biophys Res Commun,2015,467(2):268-274.
[41] MAESTRACCI M,THIERY A,ARNAUD A,et al.A study of the mechanism of the reactions catalyzed by the amidaseBrevibacteriumsp.R312[J].Agric Biol Chem,2006,50(9):2237-2241.
[42] KOBAYASHI M,SHIMIZU S.Identification of active sites in amidase:evolutionary relationship between amide bond- and peptide bond-cleaving enzymes[J].PANS,1997,94(22):11986-11991.
[43] MCKINNEY M K,CRAVATT B F.Structure and function of fatty acid amide hydrolase[J].Annu Rev Biochem,2005,74(74):411-432.
[44] LABAR G,MICHAUX C.Fatty acid amide hydrolase:from characterization to therapeutics[J].Chem Biodivers,2007,38(47):1882-1902.
[45] PALERMO G,ROTHLISBERGER U,CAVALLI A,et al.Computational insights into function and inhibition of fatty acid amide hydrolase[J].Eur J Med Chem,2015,91:15-26.
[46] MILENI M,KAMTEKAR S,WOOD D C,et al.Crystal structure of fatty acid amide hydrolase bound to the carbamate inhibitor URB597:discovery of a deacylating water molecule and insight into enzyme inactivation[J].J Mol Biol,2010,400(4):743-754.
[47] ZHENG R C,ZHENG Y G,SHEN Y C.A screening system for active and enantioselective amidase based on its acyl transfer activity[J].Appl Microbiol Biotechnol,2007,74(1):256-262.
[48] DUCHATEAU A L,HILLEMANS-CROMBACH M G,VAN DUIJNHOVEN D A,et al.A colorimetric method for determination of amino amidase activity[J].Anal Biochem,2004,330(2):362-364.
[49] WU Z M,ZHENG R C,ZHENG Y G.Exploitation and characterization of three versatile amidase super family members fromDelftiatsuruhatensisZJB-05174[J].Enzyme Microb Technol,2016,86:93-102.
[50] ZHENG R C,WANG Y S,LIU Z Q,et al.Isolation and characterization ofDelftiatsuruhatensisZJB-05174,capable ofR-enantioselective degradation of 2,2-dimethylcyclopropanecarboxamide[J].RES Microbiol,2007,158(3):258-264.
[51] CHEONG T K,ORIEL P J.Cloning of a wide-spectrum amidase fromBacillusstearothermophilusBR388 inEscherichiacoliand marked enhancement of amidase expression using directed evolution[J].Enzyme Microb Technol,2000,26(2/3/4):152-158.
[52] KOMEDA H,HARIYAMA N,ASANO Y.L-Stereoselective amino acid amidase with broad substrate specificity fromBrevundimonasdiminuta:characterization of a new member of the leucine aminopeptidase family[J].Appl Microbiol Biotechnol,2006,70(4):412-421.
[53] MAYAUX J F,CEREBELAUD E,SOUBRIER F,et al.Purification,cloning,and primary structure of an enantiomer-selective amidase fromBrevibacteriumsp.strain R312:structural evidence for genetic coupling with nitrile hydratase[J].J Bacteriol,1990,172(12):6764-6773.
[54] RUAN L T,ZHENG R C,ZHENG Y G.A novel amidase fromBrevibacteriumepidermidisZJB-07021:gene cloning,refolding and application in butyrylhydroxamic acid synthesis[J].J Ind Microbiol Biotechnol,2016,43(8):1071-1083.
[55] YAMAMOTO K,OTSUBO K,MATSUO A,et al.Production ofR-(2)Ketoprofen from an amide compound byComamonasacidovoransKPO-2771-4[J].Appl Environ Microbiol,1996,62(1):152-155.
[56] MAKHONGELA H S,GLOWACKA A E,AGARKAR V B,et al.A novel thermostable nitrilase superfamily amidase fromGeobacilluspallidusshowing acyl transfer activity[J].Appl Microbiol Biotechnol,2007,75(4):801-811.
[57] NAWAZ M S,KHAN A D,SIITONEN P H,et al.Physical,biochemical,and immunological characterization of a thermostable amidase fromKlebsiellapneumoniaeNCTR 1[J].J Bacteriol,1996,178(8):2397-2401.
[58] CISKANIK L M,WILCZEK J M,FALLON R D.Purification and characterization of an enantioselective amidase fromPseudomonaschlororaphisB23[J].Appl Environ Microbiol,1995,61(3):998-1003.
[59] KOMEDA H,HARADA H,WASHIKA S,et al.A novelR-stereoselective amidase fromPseudomonassp. MCI3434 acting on piperazine-2-tert-butylcarboxamide[J].Eur J Biochem,2004,271(8):1580-1590.
[60] EGOROVA K,TRAUTHWEIN H,VERSECK S,et al.Purification and properties of an enantioselective and thermoactive amidase from the thermophilic actinomycetePseudonocardiathermophila[J].Appl Microbiol Biotechnol,2004,65(1):38-45.
[61] SCOTTO D A A,AMMENDOLA S,SCANDURRA R,et al.Molecular and biochemical characterization of the recombinant amidase from hyperthermophilic archaeonSulfolobussolfataricus[J].Extremophiles,2001,5(3):183-192.
[62] SUZUKI Y,OHTA H.Identification of a thermostable and enantioselective amidase from the thermoacidophilic archaeonSulfolobustokodaiistrain 7[J].Protein Exp Purif,2006,45(2):368-373.
[63] WU Z M,ZHENG R C,ZHENG Y G.Identification and characterization of a novel amidase signature family amidase fromParvibaculumlavamentivoransZJB14001[J].Protein Exp Purif,2017,129:60-68.
[64] WU Z M,ZHENG R C,TANG X L,et al.Identification and characterization of a thermostable and cobalt-dependent amidase fromBurkholderiaphytofirmansZJB-15079 for efficient synthesis of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid[J].Appl Microbiol Biotechnol,2017,101(5):1953-1964.
[65] RUAN L T,ZHENG R C,ZHENG Y G.Mining and characterization of two amidase signature family amidases fromBrevibacteriumepidermidisZJB-07021 by an efficient genome mining approach[J].Protein Exp Purif,2016,126:16-25.
[66] 陈宝泉,刘肖英,李彩文.新型抗癫痫药加巴喷丁[J].中国新药杂志,2007,16(11):900-902.
[67] ZHU D,MUKHERJEE C,BIEHL E R,et al.Nitrilase-catalyzed selective hydrolysis of dinitriles and green access to the cyanocarboxylic acids of pharmaceutical importance[J].Adv Synth Catal,2007:349(10):1667-1670.
[68] WU Z M,ZHENG R C,DING X,et al.Enzymatic production of key intermediate of gabapentin by recombinant amidase fromPantoeasp.with high ratio of substrate to biocatalyst[J].Process Biochem,2016,51(5):607-613.
[69] MENZEL K,MACHROUHI F,BODENSTEIN M,et al.Process development of a potent Bradykinin 1 antagonist[J].Org Process Res Dev,2015,13(3):519-524.
[70] PARKER J S,BOWER J F,MURRAY P M,et al.Kepner-Tregoe decision analysis as a tool to aid route selection:part 3.application to a back-up series of compounds in the PDK project[J].Org Process Res Dev,2008,12(6):1060-1071.
[71] OHNMACHT C J,RUSSELL K,EMPFIELD J R,et al.N-aryl-3,3,3-trifluoro-2-hydroxy-2-methylpropanamides:KATP potassium channel openers:modifications on the western region[J].J Med Chem,1996,39(23):4592-4601.
[72] AICHER T D,ANDERSON R C,GAO J,et al.Secondary amides of (R)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase[J].J Med Chem,2000,43(2):236-249.
[73] SHAW N M,NAUGHTON A,ROBINS K,et al.Selection,purification,characterisation,and cloning of a novel heat-stable stereo-specific amidase fromKlebsiellaoxytoca,and its application in the synthesis of enantiomerically pure (R)- and (S)-3,3,3-trifluoro-2-hydroxy-2-methylpropionic acids and (S)-3,3,3-trifluoro-2-hydroxy-2-methylpropionamide[J].Org Process Res Dev,2002,6(4):497-504.
[74] 徐加利,王金信.烟嘧磺隆的研究与开发进展[J].农药科学与管理,2007,28(6):151-154.
[75] 胡斐.超高效磺酰脲类除草剂烟嘧磺隆的合成[D].杭州:浙江工业大学,2014.
[76] 陈升.抗抑郁药米氮平及其中间体的合成工艺研究[D].南京:东南大学,2007.
[77] 金建强.泛生菌酰胺酶的改造及催化合成2-氯烟酸研究[D].杭州:浙江工业大学,2017.
