SSR fingerprinting of 203 sweetpotato (Ipomoea batatas (L.) Lam.)varieties
2018-01-04MENGYushaZHAONingLlHuiZHAlHongHEShaozhenLlUQingchang
MENG Yu-sha, ZHAO Ning, Ll Hui, ZHAl Hong, HE Shao-zhen, LlU Qing-chang
Key Laboratory of Sweetpotato Biology and Biotechnolog, Ministry of Agriculture/Beijing Key Laboratory of Crop Genetic Improvement/Laboratory of Crop Heterosis and Utilization of Ministry of Education, China Agricultural University, Beijing 100193, P.R.China
RESEARCH ARTICLE
SSR fingerprinting of 203 sweetpotato (Ipomoea batatas (L.) Lam.)varieties
MENG Yu-sha, ZHAO Ning, Ll Hui, ZHAl Hong, HE Shao-zhen, LlU Qing-chang
Key Laboratory of Sweetpotato Biology and Biotechnolog, Ministry of Agriculture/Beijing Key Laboratory of Crop Genetic Improvement/Laboratory of Crop Heterosis and Utilization of Ministry of Education, China Agricultural University, Beijing 100193, P.R.China
Simple sequence repeat (SSR) markers have been shown to be a powerful tool for varieties identi fication in plants. However, SSR fingerprinting of sweetpotato varieties has been a little reported. In this study, a total of 1 294 SSR primer pairs,including 1 215 genomic-SSR and 79 expressed sequence tag (EST)-SSR primer pairs, were screened with sweetpotato varieties Zhengshu 20 and Luoxushu 8 and their 2 F1individuals randomly sampled, and 273 and 38 of them generated polymorphic bands, respectively. Four genomic-SSR and 3 EST-SSR primer pairs, which showed good polymorphism,were selected to amplify 203 sweetpotato varieties and gave a total of 172 bands, 85 (49.42%) of which were polymorphic.All of the 203 sweetpotato varieties showed unique fingerprint patterns, indicating the utility of SSR markers in variety identi fication of this crop. Polymorphism information content (PIC) ranged from 0.5824 to 0.9322 with an average of 0.8176.SSR-based genetic distances varied from 0.0118 to 0.6353 with an average of 0.3100 among these varieties. Thus, these sweetpotato varieties exhibited high levels of genetic similarity and had distinct fingerprint pro files. The SSR fingerprints of the 203 sweetpotato varieties have been successfully constructed. The highly polymorphic SSR primer pairs developed in this study have the potential to be used as core primer pairs for variety identi fication, genetic diversity assessment and linkage map construction in sweetpotato and other plants.
EST-SSR, fingerprinting, genetic distance, genomic-SSR, sweetpotato
1. lntroduction
Sweetpotato, Ipomoea batatas (L.) Lam., is the seventh most important food crop worldwide and its annual production reaches 104.46 million tons globally (FAO 2014). In many developing countries, sweetpotato is a staple food crop because it produces large quantities of energy per day(Hagenimana and Low 2000; Srinivas et al. 2009; Li et al.2014). Sweetpotato is a vegetatively propagated species,and each variety is a clone. Phenotypic markers are usually used to provide descriptors for identifying sweetpotato varieties, but they are unreliable due to their paucity and vulnerability to environmental in fluence (Prakash et al. 1996).
Molecular marker techniques have been proved to be powerful tools for identifying plant species, varieties, clones,individuals and even plant products (Karihaloo 2015).Prakash et al. (1996) used DNA ampli fication fingerprinting(DAF) to investigate genetic relationships of sweetpotato varieties and developed fingerprint pro files of the 30 USA varieties with 7 octamer primers. Guo et al. (2003) and Wang H Y et al. (2009) constructed random ampli fied polymorphic DNA (RAPD) fingerprints of the 10 and 30 sweetpotato varieties with one RAPD primer, respectively. In the study of Liu et al. (2012), the 98 sweetpotato varieties were distinguished with one ampli fied fragment length polymorphism (AFLP)primer combination. Simple sequence repeat (SSR) is short tandem nucleotide repeats and is known for its high polymorphism level (Hayden and Sharp 2001; Chen H L et al. 2016;Dos Santos et al. 2016). SSR markers are easy to detect,highly informative, stable, and comparatively inexpensive to use in research, and have been shown to be a powerful tool for varieties identi fication in several plant species such as potato (Moisan-Thiery et al. 2005), cabbage (Louarn et al.2007), tomato (Caramante et al. 2011), rose (Akond et al.2012), mango (Kumar et al. 2013), rice (Jiang et al. 2010;Lu et al. 2014) and oil camellia (Chen Y N et al. 2016). To date, SSR markers have been used to assess the genetic diversity of accessions and construct genetic linkage maps in sweetpotato (Hwang et al. 2002; Zhao et al. 2013; Hundayehu et al. 2014; Koussao et al. 2014; Rodriguez-Bonilla et al. 2014; Yada et al. 2015; Yang et al. 2015; Ngailo et al.2016). However, the SSR fingerprinting of sweetpotato varieties has been a little reported. Luo et al. (2014) constructed the DNA fingerprinting of 52 sweetpotato accessions with 2 expressed sequence tag (EST)-SSR primers.
In this study, 4 genomic-SSR and 3 EST-SSR primer pairs were selected from 1 215 genomic-SSR and 79 EST-SSR primer pairs, respectively, to construct the fingerprints of the 203 sweetpotato varieties. All of the varieties showed unique fingerprint patterns, which indicates the utility of SSR markers in variety identi fication of sweetpotato.
2. Materials and methods
2.1. Plant materials
Sweetpotato varieties Zhengshu 20 and Luoxushu 8 and their 2 F1individuals randomly sampled, which were provided by the Crop Research Institute, Shandong Academy of Agricultural Sciences, China, were used for screening polymorphic SSR primer pairs. The 2 varieties have distinct differences in yield, starch content and diseases resistance.For SSR fingerprinting, the 203 sweetpotato varieties were collected from China and Japan (Appendix A).
2.2. DNA extraction
Genomic DNA from each variety was extracted from young leaves with the cetyltrimethylammonium bromide (CTAB)method (Saghai-Maroof et al. 1984). The DNA quality was quantitated on a 1% (w/v) agarose gel, and the DNA concentration of each sample was quanti fied under UV illumination.
2.3. SSR primer designing and screening
A total of 1 215 genomic SSR primer pairs with di-, tri- and tetra-nucleotide motifs were developed from whole genome sequencing of sweetpotato cv. Xushu 18. These genomic SSR primer pairs were provided by Dr. Sachiko Isobe, the Kazusa DNA Research Institute, Japan. Meanwhile, a total of 6 130 EST sequences were downloaded from the EST database of NCBI (http://www.ncbi.nlm.nih.gov/nucest). After removing redundant sequences, the remaining sequences were used to search EST-SSR sequences by software AutoSSR (Wang C B et al. 2009), and the number of repeats containing di-, tri-, tetra-, penta-, and hexanucleotide motifs was 9, 6, 5, 4 and 3, respectively. The selected EST-SSR sequences longer than 150 bp in general were used to design SSR primers by Primer Premier 5.0 according to the following standard parameters: target amplicon lengths of 100–900 bp, annealing temperatures of 55–65°C, GC contents of 40–60% and primer sizes of 18–28 bp. A total of 79 EST-SSR primer pairs were designed. All of the genomic-SSR and EST-SSR primer pairs were synthesized at BGI-Tech (Shenzhen, China).
A total of 1 294 SSR primer pairs were screened with Zhengshu 20 and Luoxushu 8 and their 2 F1individuals randomly sampled to detect their polymorphism. PCR ampli fication was performed in a 20-μL reaction solution consisting of 3 μL (50 ng μL–1) DNA template, 2 μL 10×PCR buffer, 0.8 μL (10 mmol L–1) dNTPs, 0.2 μL (5 U μL–1)EasyTaq®DNA polymerase (TransGen Biotech, Beijing,China), 1 μL (10 μmol L–1) of each SSR primer (BGI-Tech,Shenzhen, China) and 12 μL deionized distilled water.PCR conditions for ampli fication were as follows: 95°C for 5 min followed by 35 cycles at 95°C for 1 min, 56.8–59°C(depending on the SSR primers, Table 1) for 30 s, 72°C for 1 min and a final extension at 72°C for 10 min. The PCR products were denatured and analyzed by Bio-Rad (USA)electrophoresis on 6% denaturing polyacrylamide gel in 1×TBE buffer and visualized by silver staining.
2.4. SSR fingerprinting
Genomic-SSR and EST-SSR primer pairs showing good polymorphism were selected to amplify the 203 sweetpotato varieties for constructing their fingerprint pro files as described above. Polymorphic bands were visually scored as binary data with presence as “1” and absence as “0” by GeneRulerTM100 bp DNA Ladder.
Characteristics of the SSR primer pairs for constructingsweetpotato fingerprints were evaluated with the 203 sweetpotato varieties in terms of effective number of alleles (Ne*),Nei’s gene diversity (H*), Shannon’s information index (I*)and polymorphism information content (PIC) using POPGENE version 1.32 (Yeh and Boyle 1997) and PIC-Calc version 0.6 (Nagy et al. 2012), respectively.

Table 1 Characteristics of the seven simple sequence repeat (SSR) primer pairs for constructing sweetpotato fingerprints1)
2.5. Genetic diversity analysis
A genetic similarity matrix based on the “proportion of shared alleles” among the 203 sweetpotato varieties was generated using NTSYS-pc version 2.2 statistical package (Rohlf et al.1992). An unweighted pair group method with arithmetic mean (UPGMA) tree based on the shared allele distances was constructed using molecular evolutionary genetics analysis (MEGA) version 4.0 software (Tamura et al. 2007)for evaluating genetic relationships among the sweetpotato varieties. Principal component analysis (PCA) was used to indicate the distribution of individual varieties in scatter diagram and 2-dimensional PCA graph was drawn with R statistical package (Team 2012).
3. Results
3.1. Polymorphism of SSR primer pairs
Of 1 215 genomic SSR primer pairs, 273 showed polymorphism among Zhengshu 20 and Luoxushu 8 and their 2 F1individuals randomly sampled. They generated 2-44 bands with an average of 8.5 bands per primer pair. And 38 of 79 EST-SSR primer pairs were polymorphic, which gave 6-27 bands with an average of 14.8 bands. Especially,it was found that 4 genomic-SSR and 3 EST-SSR primer pairs ampli fied more polymorphic bands. These 7 primers pairs were named SPGS1, SPGS2, SPGS3, SPGS4,SPES1, SPES2 and SPES3, respectively (Appendix A),and were used to construct SSR fingerprints of the 203 sweetpotato varieties.
3.2. Construction of SSR fingerprints
Four genomic-SSR and 3 EST-SSR primer pairs were used to amplify the 203 sweetpotato varieties and generated a total of 172 bands, 85 (49.42%) of which were polymorphic(Fig. 1). For each primer pair, the number of bands ranged from 15 to 41 with an average of 21 and the number of polymorphic bands varied from 5 to 27 with an average of 12 (Table 1). Ne*ranged from 1.2532 to 1.4172 with an average of 1.3223, H*varied from 0.1599 to 0.2409 with an average of 0.1974, the I*was from 0.2540 to 0.3573 with an average of 0.3046, and PIC from 0.5824 to 0.9322 with an average of 0.8167 (Table 1). These results indicated that these 7 SSR primer pairs were suitable for constructing SSR fingerprint pro files of the 203 sweetpotato varieties.
Each SSR primer pair could distinguish 11 to 187 sweetpotato varieties tested in this study (Appendix B).All of the 203 sweetpotato varieties could be completely distinguished and showed unique fingerprint patterns based on the 85 clear polymorphic bands ampli fied with the 7 primer pairs, indicating the utility of SSR markers in variety identi fication of sweetpotato (Appendix A). The SSR fingerprinting of each variety was converted into digital fingerprint by following computer language (1, presence; 0,absence). Each sweetpotato variety was recorded to form a binary matrix which could be processed by a computer easily (Appendix A). According to the formula P=(1/2)n,the 7 SSR primer pairs detected 85 polymorphic alleles,the probability appearing the same fingerprint in the tested varieties was P=(1/2)85=2.5849×10–26. Thus, the developed SSR fingerprinting can be used for identifying these 203 sweetpotato varieties.

Fig. 1 Fingerprinting of sweetpotato varieties constructed with SPES3 primer pairs.
3.3. Analysis of genetic diversity
Based on SSR data from the 7 primer pairs, the genetic distances among the 203 sweetpotato varieties ranged from 0.0118 to 0.6353 with an average of 0.3100 (Appendix C).Using UPGMA analysis, a dendrogram was constructed based on genetic distance matrix to evaluate the genetic relationship among the 203 sweetpotato varieties (Fig. 2). The dendrogram revealed that sweetpotato varieties from the same regions were not well clustered in the same groups.For example, the 39 varieties from Sichuan of China were scattered, only a few of them gathered together. PCA results also showed that the 203 sweetpotato varieties were not clustered together according to their regions (Fig. 3). The first 2 principal components accounted for 7.26 and 4.84%of the molecular variance, respectively (Fig. 3). These results indicated that the 203 sweetpotato varieties tested had high levels of genetic similarity and distinct fingerprint pro files, which further showed the reliability of SSR markers for variety identi fication in sweetpotato.
4. Discussion
SSR markers have been widely used for genetic diversity assessment and identi fication and purity analysis of crop varieties because of its advantages of co-dominance,higher reliability, reproducibility and higher polymorphism compared with other DNA molecular markers (Powell et al.1996; Jones et al. 1997; Karihaloo et al. 2015). Though SSR markers have been used for analyzing the genetic diversity and developing linkage maps in sweetpotato (Hwang et al.2002; Zhao et al. 2013; Hundayehu et al. 2014; Koussao et al. 2014; Rodriguez-Bonilla et al. 2014; Yada et al. 2015;Yang et al. 2015; Ngailo et al. 2016), the SSR fingerprinting of sweetpotato varieties has been a little reported. Luo et al.(2014) constructed the DNA fingerprinting of 52 sweetpotato accessions with 2 EST-SSR primers.
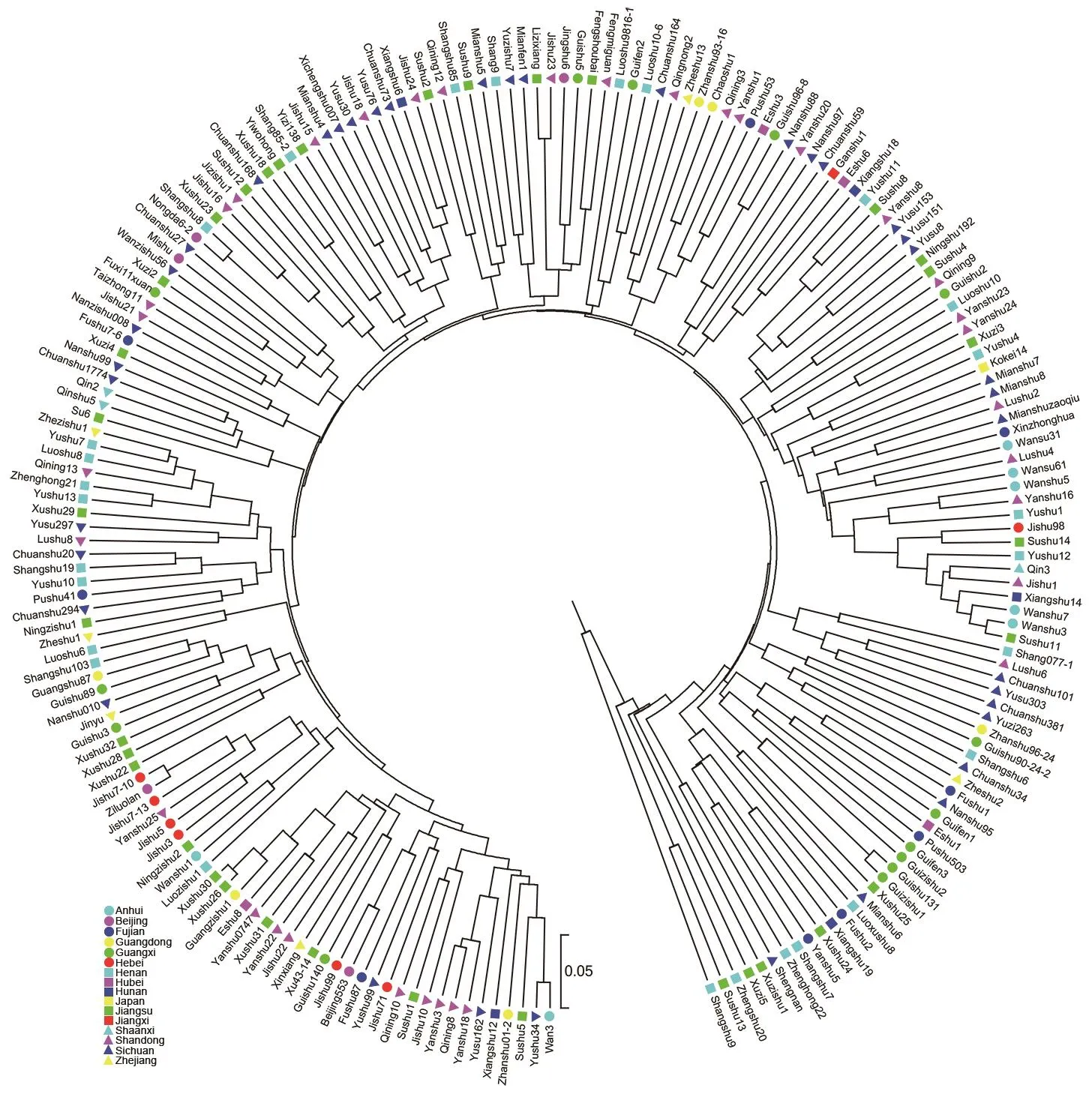
Fig. 2 Unweighted pair group method with arithmetic mean (UPGMA) dendrogram of the 203 sweetpotato varieties in Japan and 15 regions of China (represented by 16 different shapes and colors) based on simple sequence repeat (SSR) markers.
In the present study, a total of 1 294 SSR primer pairs,including 1 215 genomic-SSR and 79 EST-SSR primer pairs, were screened, and the 4 genomic-SSR and 3 ESTSSR primer pairs were found to show high polymorphism(Table 1). PIC value ranged from 0.5824 to 0.9322 with an average of 0.8167 (Table 1), which implied that the SSR markers were very informative and of high discriminating ability according to Botstein’s PIC guideline (Botstein et al.1980). The PIC values obtained in the present study were in agreement with those reported by Koussao et al. (2014),Rodriguez-Bonilla et al. (2014) and Ngailo et al. (2016) who analyzed the genetic diversity of sweetpotato accessions using SSR markers. The high levels of polymorphism of sweetpotato SSR primer pairs observed could be due to outcrossing polyploid with a large number of chromosomes(2n=6x=90), large genome size (about 2.4 Gb) and high heterozygosis of this crop (Hwang et al. 2002; Zhao et al.2013; Li et al. 2015; Ngailo et al. 2016).
To date, DAF, RAPD, AFLP and EST-SSR markers have been used to construct the fingerprints of some sweetpotato varieties (Prakash et al. 1996; Guo et al. 2003; Wang H Y et al. 2009; Liu et al. 2012; Luo et al. 2014). The present results revealed that the 4 genomic-SSR and 3 EST-SSR primer pairs could completely distinguish each one of the 203 sweetpotato varieties tested, with the very tiny probability appearing the same fingerprint in the tested varieties,which showed successful construction of the SSR finger-prints of the 203 sweetpotato varieties. These 7 SSR primer pairs were also used for the first time. Therefore, these highly polymorphic SSR primer pairs can be applied as core primer pairs for variety identi fication, genetic diversity assessment and linkage map construction in sweetpotato and other plants.
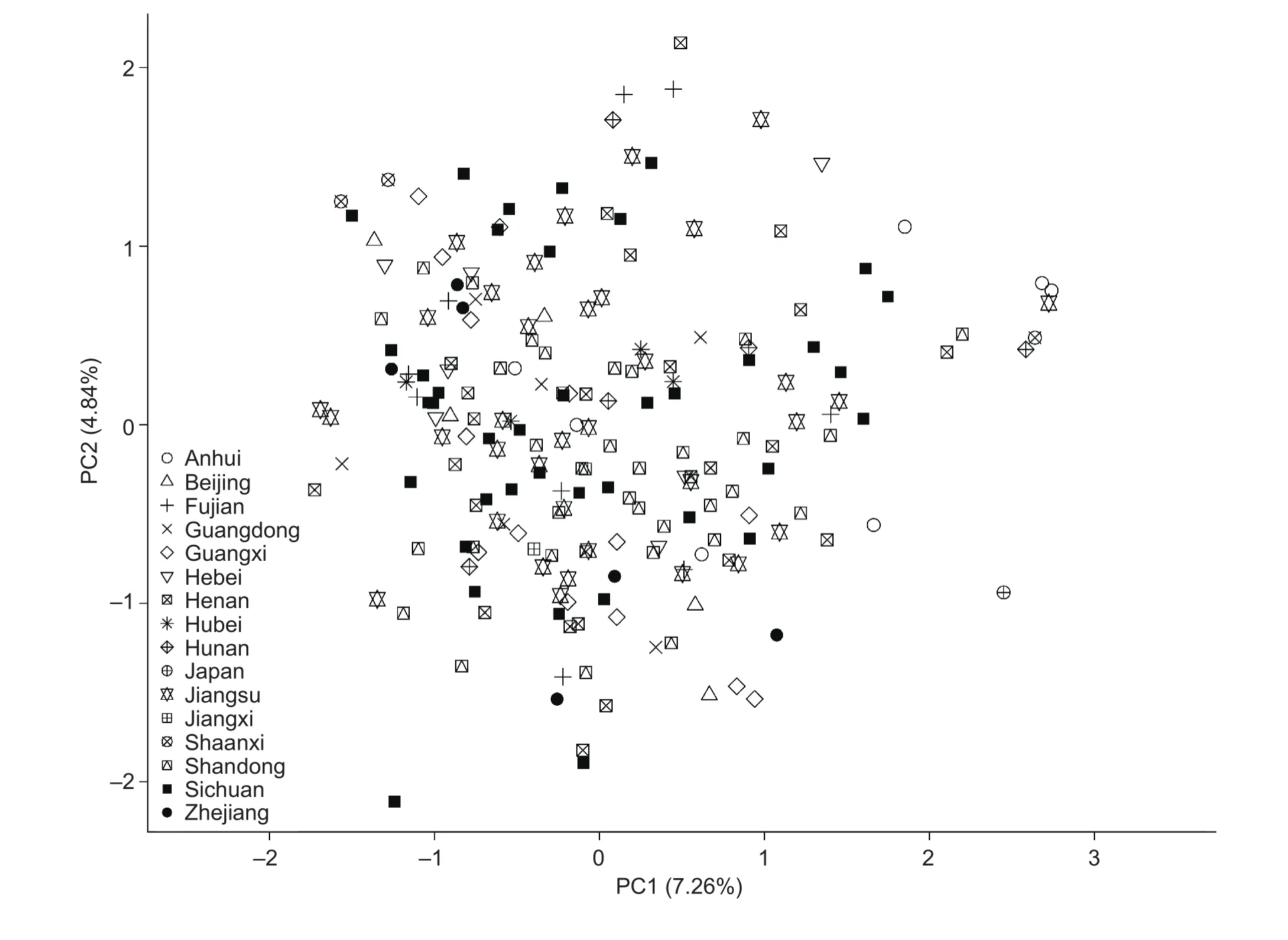
Fig. 3 Two-dimensional principal component analysis (PCA) graph of the 203 sweetpotato varieties from Japan and 15 regions of China (represented by 16 different shapes). The rate of contribution of PC1 and PC2 were 7.26 and 4.84%, respectively.
In the present study, SSR-based genetic distance ranged from 0.0118 to 0.6353 with an average of 0.3100 among the 203 sweetpotato varieties (Appendix C). PCA results also showed that most of the varieties were quite clustered(Fig. 3). These results revealed that these sweetpotato varieties had high levels of genetic similarity. Actually, 202 of the 203 sweetpotato varieties (except Kokei 14 of Japan)in this study were released since the 1970s in China, most of which have the genetic background of Okinawa 100 from Japan and Nancy Hall from the USA (Lu et al. 1998;Li et al. 2008; Liu et al. 2012). Though these sweetpotato varieties have narrow genetic background and high levels of genetic similarity, each one of them shows the unique fingerprint pro file with the 7 SSR primer pairs, which further indicates the utility of SSR markers for variety identi fication in sweetpotato.
5. Conclusion
We have successfully developed the fingerprints of the 203 sweetpotato varieties using 4 genomic-SSR and 3 ESTSSR primer pairs. These sweetpotato varieties exhibit high levels of genetic similarity and each one of them shows the unique SSR fingerprint pro file, which indicate the utility of SSR markers for variety identi fication in sweetpotato. These highly polymorphic SSR primer pairs have the potential to be used as core primer pairs for variety identi fication, genetic diversity assessment and linkage map construction in sweetpotato and other plants.
Acknowledgements
We thank Dr. Sachiko Isobe, the Kazusa DNA Research Institute, Japan, for providing genomic SSR primer pairs and Prof. Tang Jun, Xuzhou Sweetpotato Research Center,China, for providing sweetpotato materials. This work was supported by the earmarked fund for the China Agriculture Research System (CARS-11), the National Natural Science Foundation of China (31461143017) and the Science and Technology Planning Project of Guangdong Province, China(2015B020202008).
Appendixassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Akond M, Jin S M, Wang X W. 2012. Molecular characterization of selected wild species and miniature roses based on SSR markers. Scientia Horticulturae,147, 89–97.
Botstein D, White R L, Skolnick M, Davis R W. 1980.Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics,32, 314–331.
Caramante M, Corrado G, Monti L M, Rao R. 2011. Simple sequence repeats are able to trace tomato cultivars in tomato food chains. Food Control,22, 549–554.
Chen H L, Chen X, Tian J, Yang Y, Liu Z X, Hao X Y, Wang L X, Wang S H, Liang J, Zhang L Y, Yin F X, Chen X Z.2016. Development of gene-based SSR markers in rice bean (Vigna umbellata L.) based on transcriptome data.PLOS ONE,11, e0151040.
Chen Y N, Dai X G, Hou J, Guan H W, Wang Y X, Li Y, Yin T M.2016. DNA fingerprinting of oil camellia cultivars with SSR markers. Tree Genetics & Genomes,12, 1–8.
FAO (Food and Agriculture Organization). 2014. Online statistical database: food balance. FAOSTAT. [2016-12-05].http://faostat3.fao.org/compare/E
Guo J P, Pan D R, Xu L P, Yuan Z N. 2003. Establishment of DNA fingerprint and its application to variety identi fication in sweet potato. Journal of Fujian Agriculture and Forestry University,32, 19–22. (in Chinese)
Hagenimana V, Low J. 2000. Potential of orange- fleshed sweet potatoes for raising vitamin A intake in Africa. Food and Nutrition Bulletin,21, 414–418.
Hayden M J, Sharp P J. 2001. Targeted development of informative microsatellite (SSR) markers. Nucleic Acids Research,29, 1003–1010.
Hundayehu M C, du Toit E, Laurie S M, Steyn M, Greyling R,Myeza N. 2014. Effect of long-term in vitro subculturing on quality degeneration of sweetpotato varieties: Morphoanatomic assessment and simple sequence repeat (SSR)analysis. In: Low J, Nyongesa M, Quinn S, Parker M, eds.,Potato and Sweetpotato in Africa: Transforming the Value Chains for Food and Nutrition Security. CABI Publishing,England. pp. 311-321.
Hwang S Y, Tseng Y T, Lo H F. 2002. Application of simple sequence repeats in determining the genetic relationships of cultivars used in sweet potato polycross breeding in Taiwan. Scientia Horticulturae,93, 215–224.
Jiang S K, Huang C, Zhang X J, Wang J Y, Chen W F, Xu Z J. 2010. Development of a highly informative microsatellite(SSR) marker framework for rice (Oryza sativa L.)genotyping. Agricultural Sciences in China,9, 1697–1704.
Jones C J, Edwards K J, Castaglione S, Win field M O, Sala F, van de Wiel C, Bredemeijer G, Vosman B, Matthes M,Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E,Malcevschi A. 1997. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Molecular Breeding,3, 381–390.
Karihaloo J L. 2015. DNA fingerprinting techniques for plant identi fication. In: Plant Biology and Biotechnology. Springer,India. pp. 205–221.
Koussao S, Gracen V, Asante I, Danquah E Y, Ouedraogo J T, Baptiste T J, Vianney T M. 2014. Diversity analysis of sweetpotato (Ipomoea batatas (L.) Lam.) germplasm from Burkina Faso using morphological and simple sequence repeats markers. African Journal of Biotechnology,13,729–742.
Kumar M, Ponnuswami V, Nagarajan P, Jeyakumar P, Senthil N. 2013. Molecular characterization of ten mango cultivars using simple sequences repeat (SSR) markers. African Journal of Biotechnology,12, 6568–6573.
Li H, Zhao N, Yu X X, Liu Y X, Zhai H, He S Z, Li Q, Ma D F,Liu Q C. 2014. Identi fication of QTLs for storage root yield in sweetpotato. Scientia Horticulturae,170, 182–188.
Li Q, Liu Q C, Zhai H, Ma D F, Wang X, Li X Q, Wang Y P. 2008.Genetic diversity in main parents of sweetpotato in China as revealed by ISSR marker. Acta Agronomica Sinica,34,972–977. (in Chinese)
Li R J, Zhai H, Kang C, Liu D G, He S Z, Liu Q C. 2015. De novo transcriptome sequencing of the orange- fleshed sweetpotato and analysis of differentially expressed genes related to carotenoid biosynthesis. International Journal of Genomics,2015, 843802.
Liu D G, Zhao N, Zhai H, Yu X X, Jie Q, Wang L J, He S Z,Liu Q C. 2012. AFLP fingerprinting and genetic diversity of main sweetpotato varieties in China. Journal of Integrative Agriculture,11, 1424–1433.
Louarn S, Torp A M, Holme I B, Andersen S B, Jensen B D.2007. Database derived microsatellite markers (SSRs)for cultivar differentiation in Brassica oleracea. Genetic Resources and Crop Evolution,54, 1717–1725.
Lu S Y, Liu Q C, Li W J. 1998. Sweetpotato Breeding. China Agriculture Press, Beijing, China. pp. 7–17. (in Chinese)
Lu X Z, Ni J L, Li L, Wang X F, Ma H, Zhang X J, Yang J B. 2014.Construction of rice variety identity using SSR fingerprint and commodity information. Acta Agronomica Sinica,40,823–829. (in Chinese)
Luo Z X, Fang B P, Li R, Wang Z Y, Huang L F, Chen J Y, Zhang X J, Li Y J, Chen X L, Huang S H. 2014. Construction of DNA fingerprint database based on EST-SSR markers for sweet potato germplasm. Journal of Plant Genetic Resource,15,810–814. (in Chinese)
Moisan-Thiery M, Marhadour S, Kerlan M C, Dessenne N,Perramant M, Gokelaere T, Hingrat L. 2005. Potato cultivar identi fication using simple sequence repeats markers(SSR). Potato Research,48, 191–200.
Nagy S, Poczai P, Cernák I, Gorji A M, Hegedűs G,Taller J. 2012. PICcalc: An online program to calculate polymorphic information content for molecular genetic studies. Biochemical Genetics,50, 670–672.
Ngailo S, Shimelis H, Sibiya J, Amelework B, Mtunda K. 2016.Genetic diversity assessment of Tanzanian sweetpotato genotypes using simple sequence repeat markers. South African Journal of Botany,102, 40–45.
Powell W, Machray G C, Provan J. 1996. Polymorphism revealed by simple sequence repeats. Trends in Plant Science,1, 215–222.
Prakash C S, He G H, Jarret R L. 1996. DNA marker-based study of genetic relatedness in United States sweetpotato cultivars. Journal of the American Society for Horticultural Science,121, 1059–1062.
Rodriguez-Bonilla L, Cuevas H E, Montero-Rojas M, Bird-Pico F, Luciano-Rosario D, Siritunga D. 2014. Assessment of genetic diversity of sweetpotato in PuertoRico. PLOS ONE,9, e116184.
Rohlf F J. 1992. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System. Setauket, New York.
Saghai-Maroof M A, Soliman K M, Jorgensen R A, Allard R W. 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the United States of America,81, 8014–8018.
Dos Santos L F, Fregapani R M, Falcao L L, Togawa R C, do Carmo Costa M M, Lopes U V, Gramacho K P, Alves R M,Micheli F, Marcellino L H. 2016. First microsatellite markers developed from cupuassu ESTs: Application in diversity analysis and cross-species transferability to Cacao. PLOS ONE,11, e0151074.
Srinivas T. 2009. Economics of sweetpotato production and marketing. In: Loebenstein G, Thottappilly G, eds., The Sweetpotato. Springer, New York. pp. 235–267.
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0.Molecular Biology and Evolution,24, 1596–1599.
Team R C. 2013. R: A Language and Environment for Statistical Computing. Computing Team, RDCVienna, Austria.
Wang C B, Guo W Z, Zhang T Z, Li Y E, Liu H M. 2009.AutoSSR: An improved automatic software for SSR analysis from large-scale EST sequences. Cotton Science,21,243–247. (in Chinese)
Wang H Y, Zhai H, Wang Y P, He S Z, Liu Q C. 2009.RAPD fingerprints and genetic variations of the 30 main sweetpotato varieties in China. Molecular Plant Breeding,5, 879–884.
Yada B, Brown-Guedira G, Alajo A, Ssemakula G N, Mwanga R O M, Yencho G C. 2015. Simple sequence repeat marker analysis of genetic diversity among progeny of a biparental mapping population of sweetpotato. Hortscience,50,1143–1147.
Yang X S, Su W J, Wang L J, Lei J, Chen S S, Liu Q C. 2015.Molecular diversity and genetic structure of 380 sweetpotato accessions as revealed by SSR markers. Journal of Integrative Agriculture,14, 633–641.
Yeh F C, Boyle T J B. 1997. Sample genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany,129, 157.
Zhao N, Yu X X, Jie Q, Li H, Li H, Hu J, Zhai H, He S Z, Liu Q C. 2013. A genetic linkage map based on AFLP and SSR markers and mapping of QTL for dry-matter content in sweetpotato. Molecular Breeding,32, 807–820.
1 December, 2016 Accepted 7 March, 2017
MENG Yu-sha, E-mail: mengyusha200@163.com; Correspondence LIU Qing-chang, Tel: +86-10-62733710, E-mail: liuqc@cau.edu.cn
© 2018 CAAS. Publishing services by Elsevier B.V. All rights reserved.
10.1016/S2095-3119(17)61687-3
Managing editor SHI Hong-liang
杂志排行
Journal of Integrative Agriculture的其它文章
- Climate change and agriculture: Impacts and adaptive responses in Iran
- Development of elite restoring lines by integrating blast resistance and low amylose content using MAS
- Evaluation of stability and yield potential of upland rice genotypes in North and Northeast Thailand
- ldenti fication of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao
- New clues concerning pigment biosynthesis in green colored fiber provided by proteomics-based analysis
- Constitutive expression of feedback-insensitive cystathionine γ-synthase increases methionine levels in soybean leaves and seeds
