2个玉米光敏色素C基因的转录丰度对多种光质处理的响应
2017-07-24牛骧郭林杨宗举孙蕾李红丹游光霞徐宏孟凡华佘跃辉杨建平
牛骧,郭林,杨宗举,3,孙蕾,3,李红丹,3,游光霞,徐宏,孟凡华,佘跃辉,杨建平
(1四川农业大学农学院,成都 611130;2中国农业科学院作物科学研究所,北京 100081;3中国农业科学院研究生院,北京100081;4河南省潢川县农业科学研究所,河南潢川 465150)
2个玉米光敏色素C基因的转录丰度对多种光质处理的响应
牛骧1,2,郭林2,杨宗举2,3,孙蕾2,3,李红丹2,3,游光霞2,徐宏4,孟凡华2,佘跃辉1,杨建平2
(1四川农业大学农学院,成都 611130;2中国农业科学院作物科学研究所,北京 100081;3中国农业科学院研究生院,北京100081;4河南省潢川县农业科学研究所,河南潢川 465150)
【目的】通过NCBI数据库获得2个玉米PHYC及相关数据,并进行相关生物信息学分析。利用实时荧光定量PCR(qRT-PCR)分析2个玉米PHYC在玉米各器官的转录丰度,以及其转录丰度对多种光质处理、黑暗到各种光质转换和光周期处理(长日照和短日照)的响应,为研究玉米PHYC在玉米幼苗去黄化与开花期的调控机制奠定基础。【方法】采用玉米B73自交系为研究材料,通过RT-PCR分别对ZmPHYC1和ZmPHYC2的全长ORF序列进行克隆;借助相关软件对其进行生物信息学分析,利用qRT-PCR分析这两个基因在玉米各器官中的转录丰度,及其转录丰度对各种光照处理的响应。【结果】ZmPHYC1和ZmPHYC2的全长ORF均为3 408 bp,编码1 135个氨基酸基序,分子量分别为126.14和126.07 kD。生物信息学分析表明,玉米phyC蛋白可以分为6个功能区段:节奏周期蛋白—Ah核转运接受蛋白—专一蛋白区段(Per-Arnt-Sim,PAS)、cGMP受激磷酸二酯酶区段(GAF)、色素区段(PHY)和PAS相关区段(PRD,包含2个PAS区段)、组氨酸激酶A区段和组氨酸激酶ATP酶区段,但是ZmphyC2在PRD区段仅有一个PAS区段。氨基酸水平的系统发育树分析表明,ZmphyC1和ZmphyC2与禾本科物种phyC有很高的一致性,且与甘蔗和高粱phyC的亲缘关系较近。qRT-PCR分析表明,ZmPHYC1和ZmPHYC2的表达在根和叶中的转录丰度均较高,同时对持续蓝光和白光响应强烈;在黑暗到各种光质转换处理中,这两个PHYC的表达模式相似。在黑暗转到远红光、红光、蓝光和白光的0.5 h,ZmPHYC1和ZmPHYC2的转录表达均急剧上升,随后迅速下降到自身起始黑暗时的水平以下,并上下波动。这两个基因对长日照和短日照的光周期处理也能积极响应,在长日照条件下,2个ZmPHYC出现了极其相似的表达模式,均在光照和黑暗阶段各出现1个峰值;在短日照条件下,这两个基因的表达模式差异较大,ZmPHYC1的峰值出现在进入黑暗后6 h,而ZmPHYC2的峰值出现在进入光照阶段2 h。【结论】玉米phyC蛋白可以分成6个功能区段,但是ZmphyC2在PRD区段仅具有一个PAS相关区段。2个玉米PHYC转录丰度具有组织特异性。在各种光质处理中,ZmPHYC1和ZmPHYC2的表达模式相近,可能二者存在功能冗余,在转录水平上前者的丰度高于后者,推测ZmPHYC1在玉米中起更重要的作用,并且可能二者在功能上存在分工。ZmPHYC1和ZmPHYC2对各种光质和光周期处理均有较强的响应,推测二者在调控玉米光形态建成和开花中具有重要作用。
玉米;ZmPHYC;光信号转导;表达模式;光质处理
0 引言
【研究意义】植物中的光敏色素具有两个方面的主要作用[1],一是通过不同信号途径调节光应答基因的表达,影响植物光形态建成[2-3];二是通过感知周围环境的光信息,参与生物钟调控开花时间[4-5]。玉米是重要的粮食、饲料和工业原料[6],研究玉米光敏色素基因及其光信号途径[7],为了解它们在玉米幼苗去黄化与开花中的调控机制奠定基础。【前人研究进展】光与植物的生长发育密不可分。一方面,光给植物提供不可或缺的能量;另一方面,植物也能通过感知周围环境中光照的变化而做出相应的生理反应,从而调节自身的生长发育以适应环境,优化生存和繁衍[8]。模式植物拟南芥幼苗在光暗不同生长条件下其形态差异极其显著。在黑暗条件下,拟南芥幼苗呈现较长的下胚轴、子叶闭合呈钩状、其前质体发育为黄化体,该形态特征称之为暗形态建成(黄化反应);在光照条件下,拟南芥幼苗呈现较短的下胚轴、子叶打开并伸展,并且其前质体发育为成熟的叶绿体,此时的形态特征称之为光形态建成(去黄化反应)[7,9]。植物是通过光受体来感知外界环境光条件变化的[1]。光受体主要有4种类型:光敏色素、隐花色素、向光素以及UV-B的受体[10]。光敏色素是一类接受远红光和红光的色素蛋白[11],通常形成二聚体,其N端形成的疏水区以共价键的方式结合线性四吡咯生色团,负责感受光的变化;C端为2个光敏色素分子形成二聚体所必需,负责信号传递及核定位[12-13]。光敏色素调节植物从萌发到成熟的整个生长发育过程,也参与昼夜节律生物钟[14]。拟南芥中有5个不同的光敏色素基因,它们在调节植物生长发育的过程中的作用既有协同,又存在拮抗[11,15]。依据其蛋白在红光或白光下的稳定性可以分为两类,phyA是唯一属于光不稳定类型,而phyB、phyC、phyD和phyE均为光稳定型[16-17]。在光下生长的植物中,phyB的含量最多,phyC、phyD和phyE相对较低,但它们与phyB在功能存在冗余[18-19]。对拟南芥phyC的研究表明,phyC参与植物生长发育的调节,尽管它含量较低,却参与了红光特异的幼苗去黄化。与野生型相比,phyC突变体幼苗对持续红光的敏感性降低,表现出下胚轴变长和子叶变小。通过对双突变体phyB/phyC与单突变体phyB和phyC在红光条件下表型的对比发现,phyB/phyC黄化反应与phyB单突变体没有明显差异,可见phyC在phyB缺失背景下很难呈现加性效应,这表明幼苗对红光的反应主要由phyB调控[8,20]。在红光条件下,phyA/phyC双突变体的下胚轴比phyC单突变体更长,表明phyA和phyC在抑制红光条件的下胚轴伸长具有加性效应。拟南芥phyC也能够在蓝光条件下抑制下胚轴伸长,却不参与远红光条件下的幼苗去黄化,而水稻phyC却参与远红光反应[8,20-21]。水稻phyA/phyB/phyC三重突变体能够引起极强烈的黄化反应,并且胚芽鞘和叶片加长[21]。拟南芥PHYA、PHYB和PHYC的过量表达(35S启动子驱动)均增强幼苗的去黄化能力[22-23]。拟南芥phyC在最初的叶片开张中发挥的作用与phyA和phyB有异[24]。转入光敏感型烟草中的拟南芥PHYC并不影响下胚轴伸长,但促进红光依赖的幼苗子叶开张和成株叶面积增加[25]。光敏色素同时参与调控植物生物钟,影响植物开花。研究发现PHYB是一个调控开花的重要因子。对拟南芥、玉米、水稻等的研究发现,PHYB的缺失均能引起早花现象[7,26-27]。其他研究还表明phyC也能够感知光周期的长短,进而调节植物开花。在长日条件下,长日照植物拟南芥phyA/phyC双突变体比phyA突变体开花延迟,表明phyC在phyA突变体背景下能够促进开花;在短日条件下,拟南芥phyB/phyC双突变体的开花期却与phyB类似,表明在开花期phyB上位于phyC[8]。水稻是短日照植物,水稻phyC突变体能够在长日照条件下提前一周开花,phyA/phyC双突变体的开花期更加提前,说明抑制水稻PHYC的活性能够促进其在长日照条件下的开花[21,28]。小麦phyC能够激活PPD1的表达,进而小麦在长日照条件下开花期提前[29]。另外,大麦和珍珠栗等研究也同样发现phyC与开花期紧密相关[30-32]。【本研究切入点】PHYC在植物光形态建成与开花期调控中具有重要作用,但目前关于玉米ZmPHYC的研究鲜有报道。【拟解决的关键问题】本研究利用NCBI数据库检索得到ZmPHYC1和ZmPHYC2序列及其相关数据,利用生物信息学分析ZmphyC1和ZmphyC2的功能结构域,利用qRT-PCR的方法分析ZmPHYC1和ZmPHYC2在不同器官以及响应不同光质与光周期处理的表达模式,以期为研究ZmPHYC1和ZmPHYC2响应光对玉米光形态建成和开花期调控的作用提供依据,为玉米基因工程育种奠定基础。
1 材料与方法
1.1 试验材料
采用玉米B73自交系为研究材料(由李新海博士惠赠、由杨建平实验室繁育并保存)。大肠杆菌DH5α菌株、凝胶回收试剂盒和质粒提取试剂盒购自天根生化科技(北京)有限公司,克隆载体pEASY®- Blunt Simple Cloning Kit购自北京全式金生物技术有限公司。PrimeSTAR®HS酶、Recombinant DNaseⅠ(RNasefree)、SYBR®Premix Ex Taq Ⅱ购自宝生物工程(大连)有限公司,Revert Aid First Strand cDNA Synthesis Kit购自赛默飞世尔科技公司,限制性内切酶和T4 DNA连接酶购自NEB(北京)公司,Trizol购自美国英骏生命技术有限公司,其他试剂均为国产分析纯。
1.2 材料准备
基因克隆样品的准备:玉米种子在室温下浸泡于烧杯中24 h,然后平放于铺有三层湿润滤纸的培养皿中,置于28℃恒温培养箱中萌发1 d,挑选萌动良好的种子定植于装有营养土的塑料小盆中,移入22℃白光培养箱中生长13 d。叶片取样后速冻于液氮,保存于-80℃备用。
qRT-PCR分析样品的准备:(1)器官特异性分析样品处理:将玉米B73自交系的种子在北京地区4月下旬播种于花盆中,待自然条件下生长60 d后分别取成株的根、茎、叶、叶枕、叶鞘、花丝、花柄、雄花、苞叶和幼穗,取样后速冻于液氮,保存于-80℃备用。(2)持续光质处理:种子的萌发前处理同上述基因克隆样品的准备,再将定植过萌动良好B73种子的塑料小盆分别置于22℃的黑暗(Dk)、远红光(FR,1.9 µmol·m-2·s-1)、红光(R,22.3 µmol·m-2·s-1)、蓝光(B,13 µmol·m-2·s-1)和白光(WL,17 µmol·m-2·s-1)培养箱中生长13 d,分别取样。(3)黑暗转换各种光质:将22℃黑暗中生长13 d的幼苗分别转入如(2)的远红光、红光、蓝光和白光条件下0.25、0.5、1、2、4、8、12和24 h分别取样。(4)长日照和短日照处理:将定植过萌动良好B73种子的塑料小盆分别置于长日照(LD,16 h-光照/8 h-黑暗)或短日照(SD,8 h-光照/16 h-黑暗)条件下生长13 d,22℃,每隔2 h取样,WL和Dk互相转换时提前5 min取样。以上(2)—(4)均将幼苗的地上部分全部取样,速冻于液氮后-80℃保存备用。
1.3 总RNA提取及cDNA合成
用Trizol(Invitrogen, USA)法提取玉米幼苗的总RNA,用岛津紫外可见分光光度计(UV-2550)检测RNA浓度与纯度,1%的琼脂糖凝胶电泳检测其完整性。经DNaseⅠ处理后,利用RT-PCR试剂盒(Revert Aid Frist Strand cDNA Synthesis Kit,Thermo Scientific,USA)按照商家说明合成cDNA第一链。
1.4 ZmPHYC1和ZmPHYC2全长ORF克隆
依据NCBI(http://www.ncbi.nlm.nih.gov/)中的ZmPHYC1(AY234829)、ZmPHYC2(AY234830)的序列设计引物(表1),以克隆基因制备样品的cDNA为模板,分别进行PCR扩增。PCR程序为94℃ 5 min;98℃ 10 s,58℃ 15 s,72℃ 4 min,25个循环;72℃ 10 min。PCR产物经凝胶回收试剂盒回收后连接到pEASY®-Blunt Simple Cloning Vector(北京全式金生物技术有限公司)中,再转化大肠杆菌感受态细胞DH5α,对进行PCR和酶切鉴定正确的白斑克隆交北京奥克鼎盛生物科技有限公司测序,测序正确即为含有目的基因的克隆。
1.5 ZmphyC1和ZmphyC2蛋白序列分析
使用DNAMAN(Version 8)、ExPASy和NCBI分别对ZmphyC1和ZmphyC2氨基酸序列相似性,蛋白质的分子量、等电点及其蛋白质结构域进行分析。利用DNAMAN(Version 8)对植物同源phyC蛋白进行氨基酸多序列比对,并构建系统发育树。
1.6 Real-time PCR分析
根据目的基因序列,利用Primer Premier 5.0软件设计荧光定量PCR引物,以玉米Tubulin(ZmTubulin)为内参基因(表2)。PCR程序为95℃ 30 s;95℃ 5 s,60℃ 30 s,40个循环,95℃ 5 s,60℃ 1 min,95℃绘制融解曲线。每个样品设3个重复,在LightCycler480ⅡSystem(Roche,瑞士)上进行试验。采用2-ΔΔCT的方法计算结果[33]。

表1 克隆基因所用引物Table 1 Primers for cloning genes

表2 qRT-PCR引物Table 2 qRT-PCR primers
2 结果
2.1 2个ZmPHYC的克隆及序列分析
根据NCBI中ZmPHYC1和ZmPHYC2的mRNA对应cDNA序列设计引物,经PCR扩增得到二者完整的ORF片段(图1),均包含3 408个核苷酸,利用NCBI网站(https://blast.ncbi.nlm.nih.gov/)推测编码1 135个氨基酸残基的多肽,利用ExPASy网站(http://web.expasy.org/protparam/)预测二者分子量分别为126.14 kD和126.07 kD,等电点理论值分别为5.89和5.93。
利用NCBI网站和DNAMAN(Version 8)软件对ZmphyC1和ZmphyC2进行蛋白质结构域分析(图2-A),并在氨基酸水平进行了多序列比对(图2-B)。分析表明ZmphyC1可以分成6个功能区段:节奏周期蛋白—Ah核转运接受蛋白—专一蛋白区段(Per-Arnt-Sim,PAS)、cGMP受激磷酸二酯酶区段(GAF)、色素区段(PHY)和PAS相关区段(PRD,包含2个PAS区段)、组氨酸激酶A区段、组氨酸激酶ATP酶区段。与ZmphyC1相比,ZmphyC2缺失一个PAS保守结构域,包括一个假定激活位点、一个血红素结合位点、一个磷酸化作用位点以及一个二聚体结合位点。
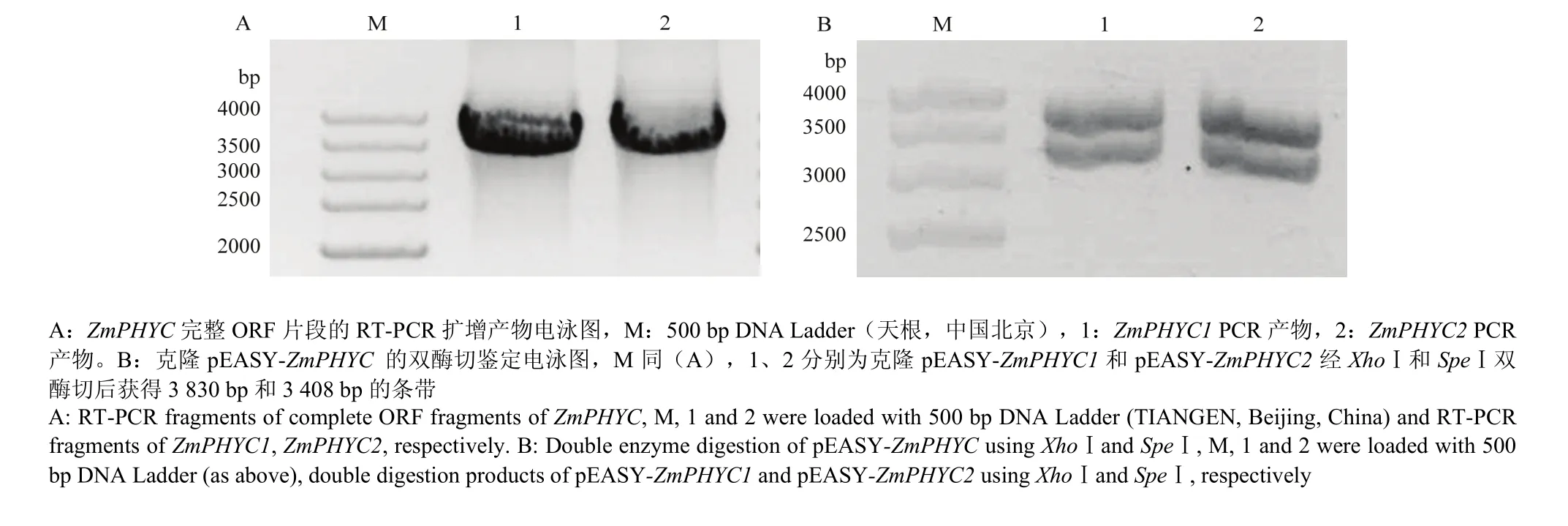
图1 2个ZmPHYC完整ORF的RT-PCR产物和pEASY-ZmPHYC双酶切电泳鉴定Fig. 1 Electrophoretic identification of RT-PCR fragments of complete ORF of both ZmPHYC genes and double digestion of pEASY-ZmPHYC using XhoⅠand SpeⅠ
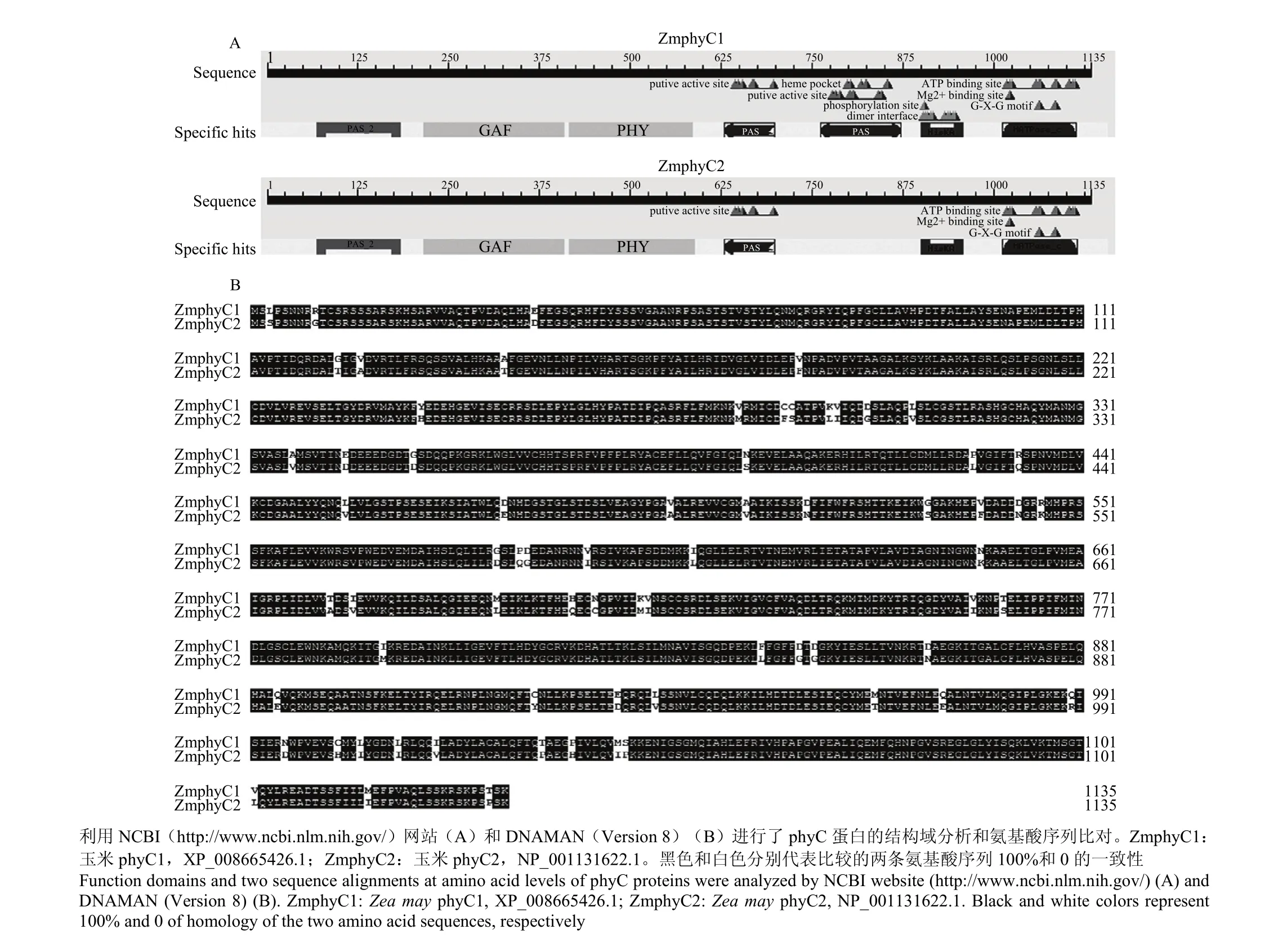
图2 2个ZmphyC蛋白的结构域分析和氨基酸序列比对Fig. 2 Function domains and multiple sequence alignment of both ZmphyC proteins
序列比对结果表明ZmphyC1和ZmphyC2相互间氨基酸水平上的相似性为93.92%;它们与禾本科的大麦、小麦、水稻、高粱和甘蔗phyC蛋白的相似性逐渐升高,从92.53%到97.18%,而与拟南芥的相似性只有83.04%。基于氨基酸序列的系统发育树分析表明,2个ZmphyC蛋白与甘蔗和高粱的亲缘关系较近,而与拟南芥的亲缘关系相对较远(图3)。
2.2 2个ZmPHYC转录表达的组织特异性分析
2个ZmPHYC在不同器官中的相对表达水平分析表明,它们在各个器官中均表达,但其丰度差异较大(图4)。ZmPHYC1的相对表达丰度以花柄最低,故设为对照。ZmPHYC1在各器官中的表达丰度均高于ZmPHYC2(图4),并且2个ZmPHYC在根中的表达丰度较高,分别是对照的68.3和4.9倍;其次是叶,分别是对照的45.9倍和5.5倍。其余器官中ZmPHYC1和ZmPHYC2的转录丰度值范围分别是对照的1—6倍和0.1—0.6倍。
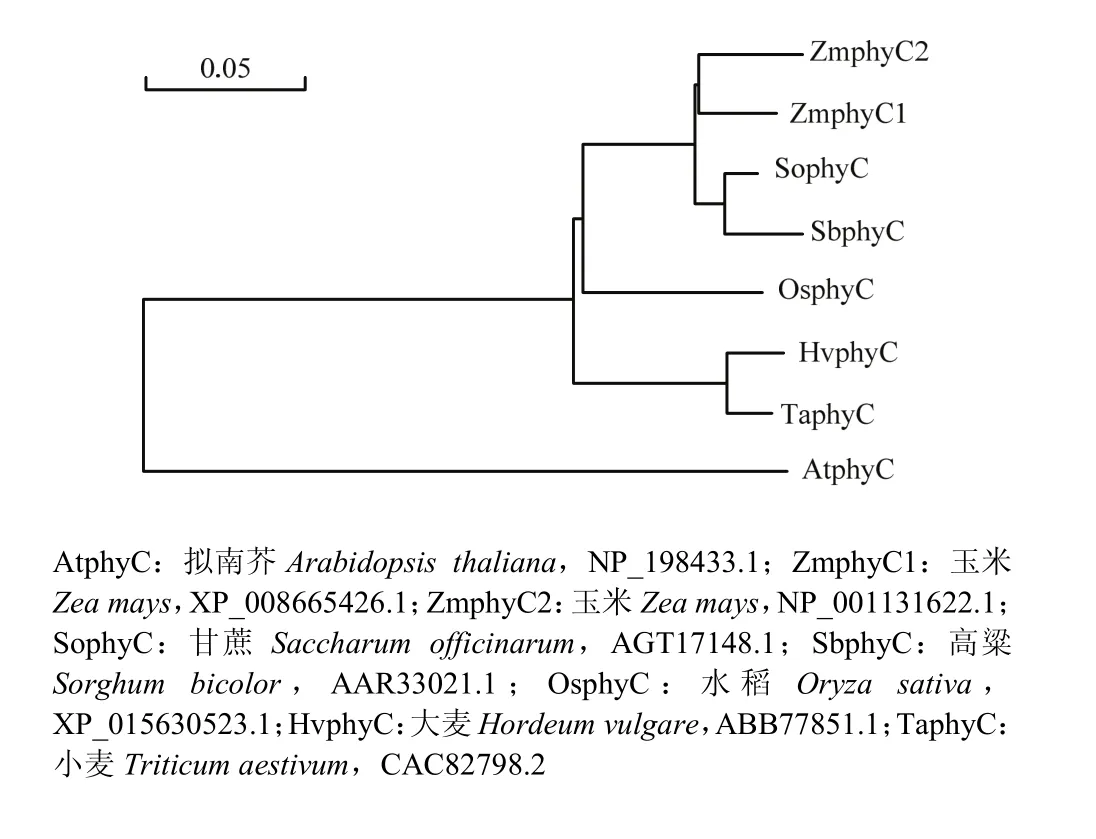
图3 2个ZmphyC蛋白与常见植物phyC蛋白的氨基酸水平系统发育分析Fig. 3 Phylogenetic analysis of phyC proteins of maize and other common plants at amino acid level

图4 2个ZmPHYC在不同器官中的相对表达水平Fig. 4 Relative expression levels of both ZmPHYC genes in different organs
2.3 2个ZmPHYC转录表达对持续光质的响应

图5 2个ZmPHYC在不同持续光质下的相对表达水平Fig. 5 Relative expression levels of both ZmPHYC genes under different continuous light quality conditions
为了研究2个ZmPHYC对各种光质的响应,采用qRT-PCR方法分析它们在黑暗(Dk)和持续远红光(FRc)、红光(Rc)、蓝光(Bc)和白光(WLc)下表达丰度(图5)。ZmPHYC1在黑暗条件下的表达丰度较低,作为对照并设为1。ZmPHYC1和ZmPHYC2对不同光质均有响应,尤其在蓝光和白光条件下的响应均较为强烈。ZmPHYC1在蓝光条件下表达丰度最高,为对照的4.6倍;ZmPHYC2在白光条件下表达丰度最高,为对照的4.4倍。在远红光下,ZmPHYC1和ZmPHYC2的表达丰度相当,为对照的1.6倍。在红光条件下,ZmPHYC1和ZmPHYC2的表达丰度略有差异,分别是对照的1.2和1.7倍。
2.4 2个ZmPHYC转录表达对不同光质转换的响应
将黑暗中生长13 d的B73幼苗转入到远红光、红光、蓝光和白光下0.25、0.5、1、2、4、8、12和24 h,来进一步研究2个ZmPHYC表达对黑暗到不同光质转换的响应(图6)。同样将ZmPHYC1在黑暗条件下的表达丰度作为对照,并设为1。qRT-PCR结果可以看出,这两个ZmPHYC的转录丰度在黑暗到不同光质转换中变化趋势很接近。在黑暗到任何光质转换1 h以内,2个ZmPHYC的转录丰度均变化强烈,尤其是转入远红光中,此时ZmPHYC1和ZmPHYC2的转录丰度分别上升至对照的320和71倍,随后在1 h时降至对照的50%左右(图6-C)。在黑暗到任何光质转换1 h后至24 h期间,2个ZmPHYC的转录丰度绝大多数情况下均在自身黑暗相对表达丰度以下的范围内起伏。值得指出的是,ZmPHYC1和 ZmPHYC2的转录丰度在转入远红光1 h至12 h期间分别为对照的2.7—6.6倍和2.4—4.7倍。
2.5 2个ZmPHYC转录表达对光周期的响应
为了更全面地了解短日照植物玉米对光周期的反应能力,进一步检测了2个ZmPHYC转录表达对光周期(长日照和短日照处理)的响应。将黑暗转光照临界期的2个ZmPHYC各自的转录丰度作对照。长日照条件下,2个ZmPHYC转录表达的趋势一致(图7-A、图7-B),它们在进入光照阶段10 h时和进入黑暗阶段4 h时均有一个峰值;ZmPHYC1转录丰度2个峰值分别是自身黑暗时的7.9和5.5倍;ZmPHYC2的2个峰值分别是自身黑暗时的2.2和1.2倍。短日照条件下,ZmPHYC1与ZmPHYC2的转录模式的最大区别是前者的峰值发生在进入黑暗阶段6 h时(自身黑暗时的12.7倍),而后者是进入光照阶段2 h时(自身黑暗时的7.4倍)(图7-C、图7-D)。2个ZmPHYC短日照条件下的黑暗阶段均有4个波动峰。
3 讨论
玉米在远古时代经历了一个四倍体化的过程,其染色体组经历基因组和片段复制、染色体融合及易位等,这个过程造成玉米中光敏色素A、B和C基因均包括2个拷贝[34-35]。2个ZmPHYC来源的差异,可能造成二者在蛋白功能、结构和转录表达的差异。利用生物信息学的手段对ZmPHYC1和ZmPHYC2编码蛋白的保守结构域进行分析,结果表明,2个ZmphyC蛋白具有其他一些物种的phyC蛋白保守结构域,但ZmphyC2蛋白相比ZmphyC1蛋白缺少部分功能结构域,包括一个假定激活位点、一个血红素结合位点、一个磷酸化作用位点以及一个二聚体结合位点(图2)。ZmphyC1的激活位点可以结合某些具有激活作用的物质,起着激活蛋白的作用;血红素不仅参与植物许多氧化还原酶辅基的合成,同时也在蛋白质的稳定性中发挥作用。对光敏色素来说,将亚铁血红素转变为胆绿素是合成活性光敏色素的前提[36-38];蛋白质磷酸位点是蛋白质磷酸化的作用部位,蛋白质磷酸化可以参与其他重要酶促反应作用,也能介导蛋白活性,还可以发挥各种独特的生理效应[39];二聚体结合位点发生蛋白互作[40];这些功能位点的缺失可能是造成ZmphyC2蛋白功能有别于ZmphyC1。
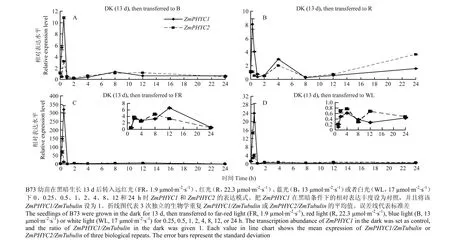
图6 2个ZmPHYC在黑暗不同光质转换下的相对表达水平Fig. 6 Relative expression levels of both ZmPHYC genes during transitions from the dark to different light conditions
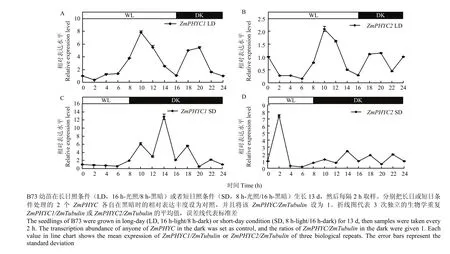
图7 2个ZmPHYC响应光周期(长日照和短日照)的相对表达水平Fig. 7 Relative expression levels of both ZmPHYC genes in response to photoperiod (long-day and short-day)
2个ZmPHYC在玉米各种器官转录表达丰度差异明显,表明它们的表达调控具有器官特异性(图4);并且ZmPHYC1的表达丰度均远高于ZmPHYC2,这与ZmPHYB1和ZmPHYB2的情形类似[41]。已有的研究表明,光敏色素参与红光调控下根的伸长和向光性[42-44]。2个ZmPHYC在根中的表达丰度均很高,其次是它们在与叶相关的器官中表达丰度也较高(图4),暗示它们可能参与根的向地性和叶的负向地性,因此,ZmPHYC在玉米极性伸长上的作用值得进一步研究。另外,光敏色素也能够影响植物气孔发育,并且促进气孔发育基因的表达[45-46]。因此,光敏色素可能通过影响植物的光合作用和蒸腾作用来调节植物生长发育。
2个ZmPHYC在光下的转录丰度均高于黑暗(图5),尤其在蓝光和白光的条件下转录丰度均较高,其中ZmPHYC1在蓝光下的表达丰度最高,ZmPHYC2在白光下最高(图5)。尽管光敏色素作为红光和远红光的受体,2个ZmPHYC在红光和远红光下的转录丰度均较低,特别是ZmPHYC1在红光下的表达量相比其他光质下最低(图5)。在黑暗到各种光质转换初期,2个ZmPHYC的转录丰度均剧烈上升后骤然下降,随后普遍维持在一个低于自身黑暗时丰度的区间内上下波动(图6),这与黑暗到各种光质转换下2个ZmPHYA的研究结果相似[47],推测它们在光信号早期起作用。尤其是黑暗到远红光的转换,可能是PHYC与PHYA在进化上相近有关[48]。这两个基因参与远红光信号途径,可能类似于水稻OsPHYC[21]。这些结果表明2个ZmPHYC不但参与了红光和白光途径,也参与了蓝光和远红光途径。
模式植物拟南芥的研究表明,高等植物开花的调控是由光信号转导途径与生物钟途径相关基因如GI(GIGANTEA)、CO(CONSTANS)和FT(FLOWERING LOCUS T)共同完成的[49-51]。对于玉米来说,光是影响其生长发育的重要因素,减少光照时间会造成早花现象[52]。本研究表明2个ZmPHYC均强烈地响应长日照和短日照处理(图7),它们与玉米开花调控基因之间的关系需进一步验证。2个ZmPHYC对各种光质及长日照、短日照响应,推测它们参与玉米光形态建成及开花调控。由于多数情况下,ZmPHYC1转录丰度均超过ZmPHYC2,可能ZmPHYC1在响应幼苗去黄化和光周期过程中起主要作用。综上所述,光信号转导途径与开花期紧密相关,通过修饰玉米光信号转导途径,可以有效改良植物的开花期性状。因此,2个ZmPHYC在玉米改良中的作用值得进一步探讨。
4 结论
ZmphyC1蛋白可以分成6个功能区段,但是ZmphyC2在PRD区段仅有一个PAS相关区段。在各种光质处理中,ZmPHYC1和ZmPHYC2的表达模式相近,可能二者存在功能冗余,在转录水平上前者的丰度高于后者,推测前者在玉米起更重要的作用。ZmPHYC1和ZmPHYC2对各种光质和光周期处理均有较强的响应,推测二者在调控玉米光形态建成和开花中具有重要作用。
[1] ÉVA K, FERENC N. Phytochrome controlled signalling cascades in higher plants. Physiologia Plantarum, 2003, 117(3): 305-313.
[2] WANG H, DENG X W. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends in Plant Science, 2003, 8(4): 172-178.
[3] GAO Y, JIANG W, DAI Y, XIAO N, ZHANG C Q, LI H, LU Y, WU M Q, TAO X Y, DENG D X, CHEN J M. A maize phytochromeinteracting factor 3 improves drought and salt stress tolerance in rice. Plant Molecular Biology, 2015, 87(4/5): 413-428.
[4] SHIN J, ANWER M U, DAVIS S J. Phytochrome-Interacting Factors (PIFs) as bridges between environmental signals and the circadian clock: Diurnal regulation of growth and development. Molecular Plant, 2013, 6(3): 283-300.
[5] KOLMOS E, HERRERO E, BUJDOSO N, MILLAR A J, TOTH R, GYULA P, NAGY F, DAVIS S J. A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. The Plant Cell, 2011, 23(9): 3230-3246.
[6] 王莉, 胡胜德. 玉米用途之争: 粮食消费还是能源消费. 农业经济, 2008(11): 8-9.
WANG L, HU S D. Corn use of the dispute: Food consumption or energy consumption. Agricultural Economy, 2008(11): 8-9. (in Chinese)
[7] 詹克慧, 李志勇, 侯佩, 习雨琳, 肖阳, 孟凡华, 杨建平. 利用修饰光敏色素信号途径进行作物改良的可行性. 中国农业科学, 2012, 45(16): 3249-3255.
ZHAN K H, LI Z Y, HOU P, XI Y L, XIAO Y, MENG F H, YANG J P. A new strategy for crop improvement through modification of phytochrome signaling pathways. Scientia Agricultura Sinica, 2012, 45(16): 3249-3255. (in Chinese)
[8] MONTE E, ALONSO J M, ECKER J R, ZHANG Y L, LI X, YOUNGJ, PHILLIPS S A, QUAIL P H. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. The Plant Cell, 2003, 15(9): 1962-1980.
[9] FANKHAUSER C, CASAL J J. Phenotypic characterization of a photomorphogenic mutant. The Plant Journal, 2004, 39(5): 747-760.
[10] BATSCHAUER A. Photoreceptors of higher plants. Planta, 1998, 206(4): 479-492.
[11] LI J G, LI G, WANG H Y, DENG X W. Phytochrome signaling mechanisms. The Arabidopsis Book, 2011, 9: e0148.
[12] BAE G, CHOI G. Decoding of light signals by plant phytochromes and their interacting proteins. Annual Review of Plant Biology, 2008, 59: 281-311.
[13] SAKAMOTO K, NAGATANI A. Nuclear localization activity of phytochrome B. The Plant Journal, 1996, 10(5): 859-868.
[14] 廖祥儒, 史海水, 尚丹, 韩国辉. 植物中的光敏色素. 生物技术通讯, 2004, 15(1): 95-97.
LIAO X R, SHI H S, SHANG D, HAN G H. Photochromes in plants. Letters in Biotechnology, 2004, 15(1): 95-97. (in Chinese)
[15] REED J W, NAGATANI A, ELICH T D, FAGAN M, CHORY J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology, 1994, 104(4): 1139-1149.
[16] SHARROCK R A, CLACK T. Heterodimerization of type II phytochromes in Arabidopsis. Proceedings of the National Academy of Sciences of the USA, 2004, 101(31): 11500-11505.
[17] SHARROCK R A, CLACK T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiology, 2002, 130(1): 442-456.
[18] CLACK T, MATHEWS S, SHARROCK R A. The phytochrome apoprotein family in Arabidopsis, is encoded by five genes: The sequences and expression of PHYD, and PHYE. Plant Molecular Biology, 1994, 25(3): 413-427.
[19] QUAIL P H, BOYLAN M T, PARKS B M, SHORT T W, XU Y, WAGNER D. Phytochromes: Photosensory perception and signal transduction. Science, 1995, 268(5211): 675-680.
[20] FRANKLIN K A, DAVIS S J, STODDART W M, VIERSTRA R D, WHITELAM G C. Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. The Plant Cell, 2003, 15(9): 1981-1989.
[21] TAKANO M, INAGAKI N, XIE X Z, YUZURIHARA N, HIHARA F, ISHIZUKA T, YANO M, NISHIMURA M, MIYAO A, HIROCHIKA H, SHINOMURA T. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. The Plant Cell, 2005, 17(12): 3311-3325.
[22] ROBSON P R H, SMITH H. Fundamental and biotechnological applications of phytochrome transgenes. Plant Cell & Environment, 1997, 20(6): 831-839.
[23] FRANKLIN K A, WHITELAM G C. Light signals, phytochromes and cross-talk with other environmental cues. Journal of Experimental Botany, 2004, 55(395): 271-276.
[24] QIN M M, KUHN R, MORAN S, QUAIL P H. Overexpressed phytochrome C has similar photosensory specificity to phytochrome B but a distinctive capacity to enhance primary leaf expansion. The Plant Journal, 1997, 12(5): 1163-1172.
[25] HALLIDAY K J, THOMAS B, WHITELAM G C. Expression of heterologous phytochromes A, B or C in transgenic tobacco plants alters vegetative development and flowering time. The Plant Journal, 1997, 12(5): 1079-1090.
[26] SAWERS R J H, LINLEY P J, FARMER P R, HANLEY N P, COSTICH D E, TERRY M J, BRUTNELL T P. Elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiology, 2002, 130(1): 155-163.
[27] IZAWA T, OIKAWA T, TOKUTOMI S, OKUNO K, SHIMAMOTO K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). The Plant Journal, 2000, 22(5): 391-399.
[28] TAKANO M, HIROCHIKA H, MIYAO A. Control of plant flowering time by regulation of phytochrome c expression: US 7566815 B2[P]. 2009.
[29] CHEN A, LI C X, HU W, LAU M Y, LIN H Q, ROCKWELL N C, MARTIN S S, JERNSTEDT J A, CLARK L J, DUBCOVSKY J. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proceedings of the National Academy of Sciences of the USA, 2014, 111(28): 10037-10044.
[30] SAÏDOU A A, CLOTAULT J, COUDERC M, MARIAC C, DEVOS K M, THUILLET A C, AMOUKOU I A, VIGOUROUX Y. Association mapping, patterns of linkage disequilibrium and selection in the vicinity of the PHYTOCHROME C gene in pearl millet. Theoretical and Applied Genetics, 2014, 127(1): 19-32.
[31] SAÏDOU A A, MARIAC C, LUONG V, PHAM J L, BEZANCON G, VIGOUROUX Y. Association studies identify natural variation at PHYC linked to flowering time and morphological variation in pearl millet. Genetics, 2009, 182(182): 899-910.
[32] NISHIDA H, ISHIHARA D, ISHII M, KANEKO T, KAWAHIGASHI H, AKASHI Y, SAISHO D, TANAKA K, HANDA H, TAKEDA K, KATO K. Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiology, 2013, 163(2): 804-814.
[33] RAJEEVAN M S, RANAMUKHAARACHCHI D G, VERNON S D, UNGER E R. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods, 2001, 25(4): 443-451.
[34] GAUT B S, DOEBLEY J F. DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences of the USA, 1997, 94(13): 6809-6814.
[35] WILSON W A, HARRINGTON S E, WOODMAN W L, LEE M, SORRELLS M E, MCCOUCH S R. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics, 1999, 153(1): 453-473.
[36] MURAMOTO T, TSURUI N, TERRY M J, YOKOTA A, KOHCHI T. Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiology, 1958, 130(4): 1958-1966.
[37] CORNEJO J, WILLOWS R D, BEALE S I. Phytobilin biosynthesis: Cloning and expression of a gene encoding soluble ferredoxindependent heme oxygenase from Synechocystis sp. PCC 6803. The Plant Journal, 1998, 15(1): 99-107.
[38] WELLER J L, TERRY M J, RAMEAU C, REID J B, KENDRICK R E. The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα. The Plant Cell, 1996, 8(1): 55-67.
[39] 梁前进, 王鹏程, 白燕荣. 蛋白质磷酸化修饰研究进展. 科技导报, 2012, 30(31): 73-79.
LIANG Q J, WANG P C, BAI Y R. Summarization on the progress in protein phosphorylation. Science & Technology Review, 2012, 30(31): 73-79. (in Chinese)
[40] EDGERTON M D, JONES A M. Localization of protein-protein interactions between subunits of phytochrome. The Plant Cell, 1992, 4(2): 161-171.
[41] SHEEHAN M J, KENNEDY L M, COSTICH D E, BRUTNELL T P. Subfunctionalization of PHYB1, and PHYB2, in the control of seedling and mature plant traits in maize. Plant Journal for Cell & Molecular Biology, 2007, 49(2): 338-353.
[42] KISS J Z, MULLEN J L, CORRELL M J, HANGARTER R P. Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiology, 2003, 131(3): 1411-1417.
[43] JOHNSON E M, PAO L I, FELDMAN L J. Regulation of phytochrome message abundance in root caps of maize. Plant Physiology, 1991, 95(S1): 544-550.
[44] CORRELL M J, KISS J Z. The roles of phytochromes in elongation and gravitropism of roots. Plant & Cell Physiology, 2005, 46(2): 317-323.
[45] CASSON S A, FRANKLIN K A, GRAY J E, GRIERSON C S, WHITELAM G C, HETHERINGTON A M. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Current Biology, 2009, 19(3): 229-234.
[46] BOCCALANDRO H E, RUGNONE M L, MORENO J E, PLOSCHUK E L, SERNA L, YANOVSKY M J, CASAL J J. Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis. Plant Physiology, 2009, 150(2): 1083-1092.
[47] 杨宗举, 闫蕾, 宋梅芳, 苏亮, 孟凡华, 李红丹, 白建荣, 郭林, 杨建平. 玉米光敏色素A1与A2在各种光处理下的转录表达特性.作物学报, 2016, 42(10): 1462-1470.
YANG Z J, YAN L, SONG M F, SU L, MENG F H, LI H D, BAI J R, GUO L, YANG J P. Transcription characteristics of ZmPHYA1 and ZmPHYA2 under different light treatments in maize. Acta Agronomica Sinica, 2016, 42(10): 1462-1470. (in Chinese)
[48] MATHEWS S. Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Molecular Ecology, 2006, 15(12): 3483-3503.
[49] SUÁREZ-LÓPEZ P, WHEATLEY K, ROBSON F, ONOUCHI H, VALVERDE F, COUPLAND G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature, 2001, 410(6832): 1116-1120.
[50] PIÑEIRO M, GEORGE C. The control of flowering time and floral identity in Arabidopsis. Plant Physiology, 1998, 117(1): 1-8.
[51] SAMACH A, ONOUCHI H, GOLD S E, DITTA G S, SCHWARZSOMMER Z, YANOFSKY M F, COUPLAND G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science, 2000, 288(5471): 1613-1616.
[52] MARKELZ N H, COSTICH D E, BRUTNELL T P. Photomorphogenic responses in maize seedling development. Plant Physiology, 2003, 133(4): 1578-1591.
(责任编辑 李莉)
Transcription Abundances of Two Phytochrome C in Response to Different Light Treatments in Zea mays
NIU Xiang1,2, GUO Lin2, YANG ZongJu2,3, SUN Lei2,3, LI HongDan2,3, YOU GuangXia2, XU Hong4, MENG FanHua2, SHE YueHui1, YANG JianPing2
(1College of Agronomy, Sichuan Agricultural University, Chengdu 611130;2Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing 100081;3Graduate School, Chinese Academy of Agricultural Sciences, Beijing 100081;4Huangchuan Institute of Agricultural Sciences of Henan Province, Huangchuan 465150, Henan)
Zea mays; ZmPHYC; light signal transduction; expression pattern; light treatment
2016-12-16;接受日期:2017-02-15
国家转基因生物新品种培育科技重大专项(2016ZX08010002-003-002)、国家重点研究发展计划(2016YFD0101002)、北京市自然科学基金(重点)(6151002)、中国农业科学院科技创新工程
联系方式:牛骧,Tel:010-82105851;E-mail:cnau323_niux@163.com。通信作者杨建平,Tel/Fax:010-82105859;E-mail:yangjianping02@caas.cn。通信作者佘跃辉,Tel:13880283512;E-mail:syuehui@sina.com
Abstract:【Objective】To study the functions of phytochrome C genes in seedling de-etiolation and flowering regulation in maize (Zea mays L.), two phytochrome C genes of maize (ZmPHYC1 and ZmPHYC2) were selected from the NCBI database and analyzed by bioinformatic methods. The transcription abundances of two ZmPHYC genes was analyzed in different tissues and in response to light qualities, transitions from the dark to different light conditions, photoperiod treatment (long day and short day) by quantitative RT-PCR (qRT-PCR).【Method】B73 inbred line was used in this study, the full length ORFs of two ZmPHYC genes were cloned by RT-PCR. The proper clones were sequenced and analyzed by bioinformatics software. The transcription abundances of two ZmPHYC genes in different tissues and in response to light treatments was detected using qRT-PCR.【Result】Both the full length ORFs of ZmPHYC1 and ZmPHYC2 contained 3408 nucleotides and encoded two polypeptides with 1135 amino acid motifs, and their molecular weight was 126.14 kD and 126.07 kD, respectively. Two ZmphyC proteins were able to be further divided into six domains: Per (period circadian protein)-Arnt (Ah receptor nuclear translocator protein)-Sim (single-minded protein) (PAS), cGMP-stimulated phosphodiesterase (GAF), phytochrome (PHY), PAS-related domain (PRD) containing two PAS, His Kinase A domain (HisKA), Histidine kinase-like ATPases (HATPase_c), while ZmphyC2 lacked a PAS in PRD domain. Phylogenetic analysis indicated that the two ZmphyC proteins belonged to the same branch with phyC proteins from other graminaceous species, especially with the phyC proteins from sugarcane and sorghum. qRT-PCR assays showed that both ZmPHYC1 and ZmPHYC2 belonged to tissue-specific genes and highly expressed in roots and leaves. The transcription abundances of the both genes were very high under blue and white light conditions. Both ZmPHYC1 and ZmPHYC2 displayed similar expression patterns during transitions from the dark to different light conditions. The transcription abundances of the both genes went dramatically up at 0.5 h after transitions from the dark to far-red, red, blue, or white light. And then they quickly dropped and waved below their own levels in the dark. Both ZmPHYC1 and ZmPHYC2 were also able to respond to long-day or short-day treatments. During long-day treatment, they likely had one peak in either light or dark period. During short-day treatment, they showed different expression patterns, the peak of ZmPHYC1 happened at 6 h after conversion to dark period, while ZmPHYC2 occurred at 2 h after conversion to light period.【Conclusion】ZmphyC1 protein kept six domains, while ZmphyC2 lacks a PAS in PRD domain. The transcription abundances of the both PHYC genes were tissue-specific in maize. Similar expression patterns of ZmPHYC1 and ZmPHYC2 genes in response to different light treatments suggest that they both might keep redundant functions. Since the transcription abundances of ZmphyC1 were higher than these of ZmphyC2, ZmphyC1 might have more important role in seedling responding to light than ZmphyC2, which may be due to the existence of different functions in maize. Both ZmPHYC1 and ZmPHYC2 respond to light and photoperiod treatments, suggesting that they are involved in seedling de-etiolation and flowering time control in maize. Thus their roles in crop improvement are worthy of more exploration in the future.
