Genetically modifying transcription factors to promote CNS axon regeneration
2017-06-05SaloniT.Mehta,JohnL.Bixby,VanceP.Lemmon
Genetically modifying transcription factors to promote CNS axon regeneration
Following injury to the central nervous system (CNS), severed axons fail to regenerate and re-form functional connections. Accordingly, CNS neurons appear to lack the intrinsic ability to modify their gene expression patterns to activate a robust regenerative response, perhaps as a result of limited plasticity in the adult brain (Davis, 2013). On the other hand, neurons of the peripheral nervous system (PNS) have been shown to modulate gene expression in response to injury, promoting axon regrowth that results in restoration of function. Transcriptomic analyses of differential gene expression between PNS and CNS neurons following injury suggest the involvement of numerous genes in successful PNS regeneration (Smith et al., 2011; Chandran et al., 2016). These studies provide insight into the identities of transcription factors (TFs) that, by controlling large suites of regeneration associated genes, could aid in promoting CNS regeneration.
Previous research by the Lemmon-Bixby group using microarrays, subtractive hybridization and RNA-seq found the TF “signal transducer and activator of transcription 3” (Stat3) to be enriched and likely activated in dorsal root ganglion (DRG) neurons compared to CNS neurons (Smith et al., 2011). Stat3 had previously been implicated,viaphosphorylation and translocation to the nucleus, in the successful regeneration of the peripheral branch of DRG neurons following injury. Furthermore, blocking Stat3 impeded the regeneration of the DRG central branch that follows a conditioning lesion to the peripheral branch (Tedeschi, 2011). Overexpression of a constitutively active form of Stat3 promoted regeneration in the optic nerve (Pernet et al., 2013). These findings suggested that overexpression of Stat3 in the CNS could play a role in promoting axon growth and regeneration.
Various studies have found that altering gene expression –viaoverexpression or knockdown – in the CNS can lead to regeneration. While overexpression of genes such as CREB, Klf7, Jun, and Sox11, and knockdown of PTEN, SOCS3, and Klf4, have led to promising regeneration, functional recovery remains elusive. VP16 is a viral activation domain responsible for activation of immediate early genes in herpes simplex virus. It functions by recruiting co-factors, histone acetyltransferases, and chromatin remodeling proteins, allowing TFs to initiate transcription more effectively (Hirai et al., 2010). In the cases of Klf7 and CREB, fusion of VP16 to these TFs helped promote growth and regeneration following TF overexpression (Gao et al., 2004; Blackmore et al., 2012). This indicated that increasing TF activity with methods such as VP16 fusion can promote increased regeneration, motivating our Stat3 experiments. Our studies focused on constitutive activation of Stat3viamutation, as well as fusion of constitutively activated (CA) Stat3 with the VP16 transactivation domain, in order to promote CNS axon regeneration. Our major finding was that overexpression of “hyperactivated” Stat3 – VP16-Stat3CA –substantially promoted neurite outgrowth in cortical neuronsin vitroas well as axon regeneration in retinal ganglion cellsin vivo, and did both to a greater extent that Stat3CA alone (Mehta et al., 2016).
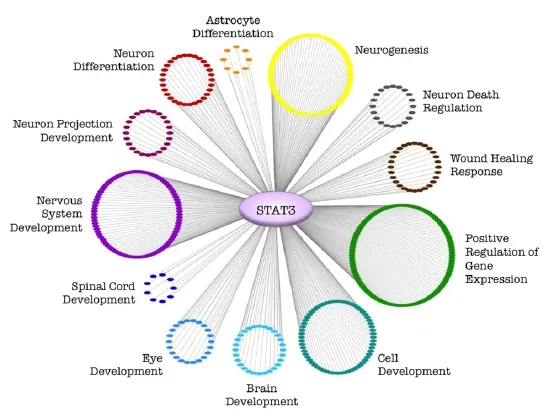
Figure 1 Clustering of Stat3 downstream genes by GO term analysis suggests pathways promoting successful regenerationviaStat3.
In addition to fusing wild type Stat3 (Stat3WT) to a VP16 activation domain, we tested this activation domain in the context of Stat3CA. In order for Stat3 to become active and translocate to the nucleus, it must be phosphorylated and homodimerize. The Stat3CA variant, however, has two cysteines introduced into Stat3’s SH2 binding domain, allowing Stat3CA to homodimerize constitutively, bypassing phosphorylation by upstream activators such as CNTF/Janus kinases. Thus, Stat3CA fused to VP16 would be “hyperactivated”, translocating to the nucleus without upstream activation, and able to regulate downstreamgenes more effectively by chromatin modification. Following this logic, the hyperactivated form of Stat3 – VP16-Stat3CA –should not only promote neurite outgrowth and axon regeneration (see above), but also strongly increase expression levels of regeneration-associated genes. Indeed, qPCR analysis following VP16-Stat3CA overexpression showed an increase in Stat3-associated transcriptional activity, as indicated by an increase in expression of the downstream regeneration-associated genes ATF3 and Sprr1a (as well as Stat3 itself), compared to an inactive (mCherry) control and to overexpression of Stat3CA without VP16 fusion (Mehta et al., 2016).
To test whether overexpression of VP16-Stat3CA could promote CNS axon regenerationin vivo, we used the optic nerve crush model, which has been widely used to study regenerationin vivo, and has led to the identification of PTEN knockdown and CNTF treatment as robust promoters of axon regeneration. Since a previous study indicated that overexpression of Stat3CA leads to optic nerve regeneration following a crush injury (Pernet et al., 2013), our experiment compared regeneration following treatment with GFP (control), Stat3CA, and VP16-Stat3CA. We confirmed that overexpression of Stat3CA led to significant regeneration (with axons growing up to 200 μm), but found that overexpression of VP16-Stat3CA promoted axon regeneration up to 1 mm past the injury site at 2 weeks following the crush (Mehta et al., 2016).
Our results, along with those of other studies, indicate that manipulation of Stat3 has strong effects on axon regeneration in the CNS. An analysis of Stat3 downstream genes followed by gene ontology (GO) term analysis suggests an explanation for the ability of Stat3 overexpression to promote CNS regeneration (Figure 1). Stat3 regulates pathways of neurogenesis, nervous system development, and the wound healing response in addition to other relevant pathways. Moreover, our qPCR results indicate that hypractivated Stat3 (VP16-Stat3CA) increased expression of the regeneration-associated genes ATF3 and Sprr1a more strongly than Stat3CA alone (Mehta et al., 2016). These findings suggest that overexpression of hyperactivated Stat3 activates regeneration-relevant pathways more effectively than Stat3CA without the VP16 addition.
While we showed successful CNS regenerationin vivowith VP16-Stat3CA overexpression, other transcription factors, such as Klf7, CREB, and cJun, are also likely to play major roles in this process (Gao et al., 2004; Blackmore et al., 2012; Lerch et al., 2014). Our studies showed that it was crucial to use VP16 as well as a constitutively active Stat3 to promote robust axon regeneration. Thus it is worth considering that simple overexpression of a TF may not lead to successful regeneration. Furthermore, another study showed that while overexpression of the TF Sox11 did promote axon regeneration of the corticospinal tract after a pyramidotomy, it led to worse behavioral recovery compared to animals without Sox11 overexpression (Wang et al., 2015). A combinatorial approach to TF overexpression showed that while a combination of Stat3, Smad1, Jun, and ATF3 promoted axon regeneration of the central branch of DRG neurons, it was not more effective than overexpression of Jun alone (Fagoe et al., 2015). Interestingly, we previously showed that overexpression of certain combinations of 2 transcription factors can increase neurite outgrowthin vitrocompared to overexpression of a single transcription factor (Lerch et al., 2014). Thus, it is possible that the right combination of hyperactivated transcription factors could lead to more substantial CNS axon regeneration as well as functional recovery. Taken together, these studies show that, when it comes to axon regeneration, one must consider the activation state of the TF(s), their ability to work together, and the ultimate effects on behavior.
This work was supported by NIH HD057632 to VPL and JLB. VPL holds the Walter G. Ross Distinguished Chair in Developmental Neuroscience.
Saloni T. Mehta, John L. Bixby, Vance P. Lemmon*
Miami Project to Cure Paralysis, University of Miami Miller School of Medicine, Miami, FL, USA (Mehta ST, Bixby JL, Lemmon VP) Center for Computational Science, University of Miami Miller School of Medicine, Miami, FL, USA (Bixby JL, Lemmon VP) Neurological Surgery, University of Miami Miller School of Medicine, Miami, FL, USA (Bixby JL, Lemmon VP) Molecular & Cellular Pharmacology, University of Miami Miller School of Medicine, Miami, FL, USA (Bixby JL)
*Correspondence to:Vance P. Lemmon, Ph.D., vlemmon@med.miami.edu.
Accepted:2017-04-17
orcid:0000-0003-3550-7576 (Vance P. Lemmon)
How to cite this article:Mehta ST, Bixby JL, Lemmon VP (2017) Genetically modifying transcription factors to promote CNS axon regeneration. Neural Regen Res 12(5):737-738.
Open access statement:This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Open peer reviewer:Barbara Haenzi.
Additional file:Open peer review report 1.
Blackmore MG, Wang Z, Lerch JK, Motti D, Zhang YP, Shields CB, Lee JK, Goldberg JL, Lemmon VP, Bixby JL (2012) Krüppel-like factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc Natl Acad Sci U S A 109:7517-7522.
Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, Blesch A, Michaelevski I, Davis-Turak J, Gao F, Langfelder P, Horvath S, He Z, Benowitz L, Fainzilber M, Tuszynski M, et al. (2016) A systems-level analysis of the peripheral nerve intrinsic axonal growth program. Neuron 89:956-970.
Davis GW (2013) Homeostatic signaling and the stabilization of neural function. Neuron 80:718-728.
Fagoe ND, Attwell CL, Kouwenhoven D, Verhaagen J, Mason MRJ (2015) Overexpression of ATF3 or the combination of ATF3, c-Jun, STAT3 and Smad1 promotes regeneration of the central axon branch of sensory neurons but without synergistic effects. Hum Mol Genet 24:6788-6800.
Gao Y, Deng K, Hou J, Bryson JB, Barco A, Nikulina E, Spencer T, Mellado W, Kandel ER, Filbin MT (2004) Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron 44:609-621.
Hirai H, Tani T, Kikyo N (2010) Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int J Dev Biol 54:1589-1596.
Lerch JK, Martínez-Ondaro YR, Bixby JL, Lemmon VP (2014) cJun promotes CNS axon growth. Mol Cell Neurosci 59:97-105.
Mehta ST, Luo X, Park KK, Bixby JL, Lemmon VP (2016) Hyperactivated Stat3 boosts axon regeneration in the CNS. Exp Neurol 280:115-120.
Pernet V, Joly S, Jordi N, Dalkara D, Guzik-Kornacka A, Flannery JG, Schwab ME (2013) Misguidance and modulation of axonal regeneration by Stat3 and Rho/ROCK signaling in the transparent optic nerve. Cell Death Dis 4:e734.
Smith RP, Lerch-Haner JK, Pardinas JR, Buchser WJ, Bixby JL, Lemmon VP (2011) Transcriptional profiling of intrinsic PNS factors in the postnatal mouse. Mol Cell Neurosci 46:32-44.
Tedeschi A (2011) Tuning the orchestra: transcriptional pathways controlling axon regeneration. Front Mol Neurosci 4:60.
Wang Z, Reynolds A, Kirry A, Nienhaus C, Blackmore MG (2015) Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J Neurosci 35:3139-3145.
10.4103/1673-5374.206637
杂志排行
中国神经再生研究(英文版)的其它文章
- Cerebral mechanism of puncturing at He-Mu point combination for functional dyspepsia: study protocol for a randomized controlled parallel trial
- Therapeutic opportunities and challenges of induced pluripotent stem cells-derived motor neurons for treatment of amyotrophic lateral sclerosis and motor neuron disease
- Inhibition and enhancement of neural regeneration by chondroitin sulfate proteoglycans
- Collapsin response mediator protein-2 plays a major protective role in acute axonal degeneration
- Hypoxia inducible factor-1 alpha stabilization for regenerative therapy in traumatic brain injury
- Minocycline targets multiple secondary injury mechanisms in traumatic spinal cord injury
