Preparation and evaluation of mixed-mode resins with tryptophan analogues as functional ligands for human serum albumin separation☆
2017-05-29QiciWuQileiZhangShiwenXuChengyongGeShanjingYaoDongqiangLin
Qici Wu,Qilei Zhang,Shiwen Xu,Chengyong Ge,Shanjing Yao,Dongqiang Lin*
Key Laboratory of Biomass Chemical Engineering of Ministry of Education,College of Chemical and Biological Engineering,Zhejiang University,Hangzhou 310027,China
1.Introduction
Mixed-mode chromatography(MMC)isa relatively new technology for protein separation[1].Specially-designed MMC ligands can interact with target proteinsviamultiple modes including hydrophobic interaction,electrostatic force and hydrogen bonding to improve binding selectivity[2].Generally,MMC has advantages ofhigh capacity,good selectivity,facile elution and salttolerance.Burtonetal.[1]prepared MMC resins with hydrophobic amine and/or alkylcarboxylic acid ligands,and 95%-100%recovery of chymosin activity was achieved with one step purification at low or high ionic strength.Johanssonet al.[3]reported that aromatic cation-exchange ligands were optimalfor protein capture at high-salt conditions.One of the most important features of MMC is that its functional ligands can be designed individually for target proteins[4-6],and the principles and approaches to design MMC ligands were reviewed by Zhaoet al.[4].MMC ligands usually have aliphatic or aromatic groups as the hydrophobic moiety and amino,carboxylorsulfonic groupsasthe ionic moiety.Heterocyclic compounds can be unique for specific aromaticity/hydrophobicity and electrostatic interactions[4,7-9].
Human serum albumin(HSA)is the most abundant protein in human plasma(40-50 g·L-1)[10],which has important applications in clinical practice[11].It is also utilized as excipient and stabilizer in pharmaceutic products and supplement for cell culture[12,13].HSA molecule comprises three homologous domains that include two subdomains with common structural motifs[14-16].Two principal binding regions(Site I and Site II)on the surface of HSA were identified[14].Site II in the subdomains IIIA is a hydrophobic pocket surrounded by positively charged amino acids[17].The binding cavity of Site II is known as“indole-binding site”which is specific to indole-containing compounds such as tryptophan[18-20].Mcmenamyetal.[18,21,22]investigated the binding behaviors between HSA and indole analogues.The specificity requirements for tryptophan-HSA binding can be satisfied by virtue of indole-protein bond,carboxyl-protein bond,and the α-hydrogen of tryptophan.Electrostatic interaction and van der Waals attraction were found as the main forces in tryptophan-HSA binding.Several indole-based resins were reported for protein separation.For example,a commercial resin MX-TRP-650m with tryptophan as the functional ligand was developed by TOSOH Corporation[23].Zhaoet al.[24]developed a 5-aminoidole ligand for bovine serum albumin separation.Moreover,our previous work indicated that tryptamine as a derivative of tryptophan showed perfect performance for HSA purification[25].Thus,indole-based tryptophan analogues would be good candidates as mixed-mode ligands for HSA separation.
In this work, five indole-based tryptophan analogues including L-tryptophan(L-TRP),L-tryptophan ethyl ester(L-TEE),N-acetyl-L-tryptophan (L-NAT),tryptamine (TA) and 4-(1H-Indol-2-yl)phenylamine(4-HIPA)were investigated as functional MMC ligands for HSA separation.These tryptophan analogues were coupled onto cross-linked agarose beads to prepare MMC resins and HSA adsorption behaviors were compared under varying conditions.The effects of pH,salt type and concentration were studied and discussed with molecular structure of ligands on adsorption behaviors.In addition,the adsorption of recombinant HSA(rHSA)fromPichia pastorisfermentation broth with the five MMC resins was evaluated.
2.Materials and Methods
2.1.Materials
L-tryptophan,N-acetyl-L-tryptophan and tryptamine were purchased from Aladdin Industrial Inc.(Shanghai,China).L-tryptophan ethyl ester and 4-(1H-Indol-2-yl)phenylamine were purchased from J&K Scientific(Beijing,China).Bestarose 6FF(cross-linked 6%agarose gel)was purchased from Bestchrom Bio-Technology Co.,Ltd.(Shanghai,China).Human serum albumin(HSA,plasma-derived,Mw=66700)was purchased from Sigma(Milwaukee,WI,USA).The culture broth ofP.pastoris(pH 6.0 and conductivity of 25 mS·cm-1)with rHSA concentration of 7.2 mg·ml-1was provided by a local biotechnology company.Other reagents were of analytical reagent grade and used as received.
2.2.Preparation of MMC resins
The mixed-mode resins were prepared according to the methods reported previously[25-28]with some modification.The process had three main procedures including allyl bromide(AB)activation,N-bromosuccinimide(NBS)bromination and ligand coupling(Fig.1).10 g drained agarose beads were mixed with suitable amount of AB(5 ml)and sodium hydroxide(2.5 g)in 10 ml 20%dimethyl sulfoxide(DMSO)solution.The mixture was continuously agitated at 180 r·min-1under 30 °C for 24 h.The allyl-activated agarose beads were washed with ethanol and deionized water,and then mixed with 3.0 mol·L-1excess of NBS in 50%acetone at 180 r·min-1and 30 °C for 3 h.The brominated beads were then washed with deionized water.Finally,tryptophan analogues were coupled onto the brominated agarose beads in 1 mol·L-1carbonate buffer(pH 11.0)at 180 r·min-1and 30 °C for 12 h.The beads were washed sequentially with deionized water,0.1 mol·L-1HCl and 0.1 mol·L-1NaOH.The MMC resins with indole-based tryptophan analogue ligands were obtained and named as L-TRP-B,L-TEE-B,L-NAT-B,TA-B,and 4-HIPA-B,respectively.The ligand densities were determinedviatitration as reported previously[29].
2.3.Batch adsorption equilibrium experiments
The adsorption isotherms of HSA on MMC resins were determined by batch adsorption experiments.20 mmol·L-1sodium acetate buffer(pH 4.0 and 5.0)and 20 mmol·L-1sodium phosphate buffer(pH 6.0,7.0 and 8.0)were used as the liquid phases.The resins were pre-equilibrated with appropriate buffer solutions,and then 0.04 g drained resin was added into 0.8 ml aliquots of HSA solutions(0-10 mg·ml-1)with salt(0-1 mol·L-1)in 2 ml tubes. The mixture was agitated in a thermomixer(1500 r·min-1)at 25 °C for 8 h.The resins were then separated by centrifugation(4000g)and decanting.The protein concentrations in the supernatant were determined at 280 nm with One Drop™Spectrophotometer(Nanjing Wins Technology Co.Ltd.,Nanjing,China).The amount of adsorbed protein was calculatedviamass balance.Langmuir equation(Eq.(1))was used to correlate the adsorption isotherms as,

whereQ*andC*are the equilibrium adsorption capacity(mg·g-1)and the equilibrium protein concentration in the liquid phase(mg·ml-1),respectively.Qmis the saturated adsorption capacity(mg·g-1)andKais the association constant(ml·mg-1).
2.4.Adsorption of rHSA from P.pastoris broth
The adsorption of rHSA fromP.pastorisculture broth with the five MMC resins prepared was studied.The culture broth containing 7.2 mg·ml-1rHSA was heated at 68 °C for 30 min with 10 mmol·L-1sodium caprylate,and used as the loading sample after pH adjustment to 5.0.The resins were equilibrated with the equilibrium buffer(20 mmol·L-1acetate buffer,pH 5.0)and then 0.3 g drained resin was added into 0.6 ml aliquots of feedstock in 2 ml tubes.The mixture was then agitated in a thermomixer(1500 r·min-1)at 25 °C for 10 h.The resins were then separated by centrifugation(4000g)and decanting.The collected supernatants were analyzed by SEC-HPLC with TSK G3000SWXLcolumn(7.8 mm×30.0 cm,TOSOH,Japan)and LC3000 HPLC system(Beijing ChuangXinTongHeng Science and Technology Co.,Ltd.,Beijing,China)according to the method published previously[25].
2.5.Molecular docking
The molecular structure of HSA(PDB code:2BXG)was taken from Protein Data Bank(http://www.pdb.org).Molecular docking of tryptamine onto the surface of HSA was carried out with the LibDock module in Discovery Studio(Accelrys,San Diego,CA,USA)according to the method reported previously[30].HSA molecule was surrounded by 23 interaction region spheres with radii of 2 nm in a way that the whole surface of HSA could be fully overlapped,and default settings for small molecule-protein docking were used.Tryptamine was aligned to polar and apolar receptor interaction sites for each interaction region sphere,which included pruning the list of matches for steric clashes,ranking with atom pairwise score,and clustering.Hydrogen bonding and steric potentials were evaluated during re finement with the BFGS optimization algorithm.Finally,all the docked poses were ranked by the scoring algorithm of LibDockScore.
2.6.Calculation of protein and ligand descriptors
The molecular structure of HSA(PDB code:1AO6)was also obtained from the Protein Data Bank.Water molecules and co-solutes were removed from the structural data by Molecular Operating Environment(MOE,Montreal,Québec,Canada).HSA was then protonated at varying pHs using the Protonate 3D function in MOE and subjected to three rounds of tethered energy minimization using the Amber99 force field.The structures of ligands were assembled with MOE and protonated with Protonate 3D function at various pHs,which were followed by energy minimization using the MMFF94x force field.All 2D,i3D and x3D descriptors available in the MOE software package were calculated for HSA and ligands under different pHs.
3.Results and Discussion
3.1.Structural analysis of ligands
L-tryptophan is one ofthe fewnaturalamino acids thatcan bind onto HSA,especially on the indole-binding Site II in a stereospecific manner[18].L-tryptophan has a hydrophobic indole ring,one amino group and one carboxyl group,which shows the typical characteristics of mixedmode ligands with the combination ofhydrophobic and electrostatic interactions.Therefore,L-tryptophan can be a potential ligand candidate for HSA separation.Four analogues based on L-tryptophan were chosen as alternatives as shown in Fig.1.The properties of the five ligands are compared in Table 1.L-TEE is an ethyl esterified derivative of L-tryptophan with higher hydrophobicity(lgP(o/w)=1.85)than L-tryptophan.L-NAT is an acetylated derivative which also shows higher hydrophobicity(lgP(o/w)=1.53)than L-tryptophan.Tryptamine has the simplest structure with lgPof 1.62.In order to further enhance ligand hydrophobicity,a phenyl ring was added between amino group and indole ring of tryptamine,which is 4-HIPA with the highest lgPof 3.56.

Table 1Properties of mixed-mode ligands used
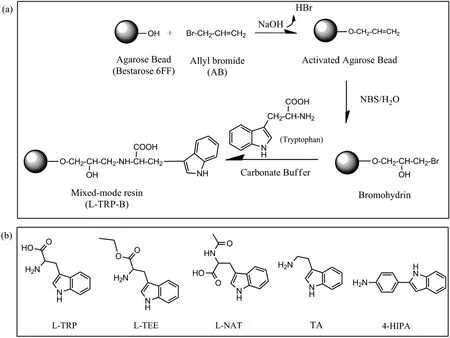
Fig.1.Preparation scheme of mixed-mode resins with tryptophan analogues as the ligands.(a)Typical preparation route ofL-TRP-B with L-tryptophan as the ligand;(b)the structure of the ligands used.
In addition,charged stations are also important for protein bindingviaelectrostatic interactions.L-TRP is a typical amphoteric compound and the pKaof the amino and carboxyl groups on L-TRP is 9.4 and 2.54,respectively.The pKaof--NH2on L-TEE is 6.92,and that value is 15.66 for L-NAT,while the pKaof--COOH on L-NAT is 4.12.The pKaof tryptamine is~10.2 and it is positively charged at the pH range tested(pH 4.0-8.0).On the contrary,4-HIPA is not charged under liquid conditions tested due to low pKa(4.42).Therefore,the five tryptophan analogue ligands in the present work have different combination of hydrophobic and electrostatic properties,and a great diversity of impacts on the binding of HSA would be expected.
3.2.Preparation of MMC resins
The MMC resinswith tryptophan analoguesas the functionalligands were prepared with three procedures,i.e.activation with AB,bromination with NBS and ligand coupling.The first two steps(activation and bromination)were the same as our previous works and the optimized conditions were used[31,32].The activation density of agarose beads was 230 μmol·(g gel)-1.Mixed-mode resins with different ligands were obtained after ligand coupling optimization,and varying ligand densities were found due to the difference of ligand coupling efficiency.The highest ligand density(~180 μmol·(g gel)-1)was obtained for L-NAT-B,and the lowest was 60 μmol·(g gel)-1for L-TEE and 4-HIPA-B.L-TRP-B and TA-B had similar ligand densities at the range of 100-130 μmol·(g gel)-1.
3.3.In fluence of pH on HSA adsorption
pH is a key factor in protein adsorption with MMC resins,which could affectnetcharges ofproteins and ligands and thus change electrostatic interactions between proteins and mixed-mode ligands.The adsorption isotherms of HSA on the five MMC resins prepared were measured at different pHs and the Langmuir equation was used to fit the adsorption isotherms.The correlated saturated adsorption capacity(Qm)and association constant(Ka)were obtained and compared in Fig.2.
Fig.2(a)shows thatthe profilesbetween the five resins were similar and the maximumQmwas found at pH 5.0.Qmof the L-NAT-B and TA-B resins reached to 132.05 and 138.02 mg·g-1at pH 5.0,respectively.For L-TRP-B and L-TEE-B resins,Qmwas at the range of 95-100 mg·g-1at pH 5.0.Qmof 4-HIPA-B was the lowest(43.31 mg·g-1)among the five resins tested.Meanwhile,Qmdecreased for all resins at pH<5.0 or pH>5.0.For L-NAT-B,4-HIPA-B and TA-B resins,Qmstill maintained 53.1%,49.3%and 73.2%atpH 7.0,respectively.In general,the TA-B resin showed high adsorption capacity ata wide range ofpH(pH 5.0-8.0)and strong pH-induced desorption at pH 4.0,which would bene fit HSA separation.Fig.2(b)shows that the association constantKawas the highest at pH 5.0 for all five resins tested.Kaof TA-B and L-NAT-B was over 20 ml·mg-1,which indicates strong binding affinity of these two ligands.AlthoughQmof L-TRP-B and L-TEE-B were higher than that of 4-HIPA-B,Kaof 4-HIPA-B was obviously higher due to stronger hydrophobicity with the additional phenyl ring.Kadecreased obviously for all five resins tested when pH<5.0,which means that the binding affinity reduced significantly due to electrostatic repulsion between positively-charged ligands and HSA.Kaalso declined with the increase of pH due to weak hydrophobic interactions at pH>5.0.
Molecular interactions between proteins and mixed-mode ligands are complicated,which combine hydrophobic,electrostatic interactions and hydrogen bonds.The highestQmwas around the isoelectric pointof the targetprotein(pH 4.7 for HSA)for the five MMC resins tested,which is similar as reported in the literature[33,34].In order to better understand molecular interactions,the molecular descriptors of HSA and ligands about surface and partial charges were calculated with MOE.Some importantdescriptorsare listed in Tables 2 and 3,such as the positive and negative accessible surface area of the protein(ASA+and ASA-),hydrophobic surface area of the protein(ASA_H),sums of formal charge(FCharge),total positive partial charge(Q_PC+)and negative partial charge(Q_PC-),number of H-bond acceptor/donor atoms(a_acc/a_don),number of H-bond donor+acceptor atoms(a_donacc),van der Waals volume(vdw_vol)and lgP(o/w)of ligands.It is obvious that the highest hydrophobic surface area of HSA is at pH 5.0,which means that the maximum hydrophobicity of HSA exists around its isoelectric point.The hydrophobicity(corresponding the value of lgP(o/w))of the ligands nearly kept the same at different pHs.The relative high lgP(o/w)of TA and L-NAT(1.62 and 1.53,respectively)leads to high adsorption capacity for HSA,although TA has positive charges and L-NAT has negative charges at pH 5.0.This result indicates that hydrophobic interaction might dominate the binding of HSA on MMC resins at pH 5.0.4-HIPA has the maximum lgP(o/w)value(3.56)but showed the lowest adsorption capacity at pH 5.0,which might be due to steric hindrance and the excessive hydrophobic phenyl ring could not form effective binding with HSA.Moreover,the low ligand density might also result in low adsorption capacity of 4-HIPA-B.The ligand density ofL-TEE-B was also low,but the high hydrophobicity of L-TEE gave approximate adsorption capacity as L-TRP.
When studied at pH different from pH 5.0,the decrease of ASA_H value indicates less surface hydrophobicity of HSA,which induces the decrease of adsorption capacity.For pH>5.0,hydrophobic interactions still dominate and HSA adsorption was less affected for the ligands with high hydrophobicity,such as 4-HIPA,TA and L-NAT.For L-TRP and L-TEE,the adsorption capacities decreased sharply with the increase of pH from 5.0 to 8.0 due to their low hydrophobicity.When studied at pH 4.0,the significant change of ASA+and ASA-of HSA altered electrostatic interactions between HSA and ligands,which certainly affects HSA adsorption.The increase of Q_PC+of HSA induced electrostatic repulsion between the positively-charged HSAand ligands.The increase of Q_PC+of L-NAT ligand further promoted electrostatic repulsion,which resulted in significant decrease ofQmfrom 132.05 mg·g-1at pH 5.0 to 52.11 mg·g-1at pH 4.0,andKafrom 20.22 ml·mg-1at pH 5.0 to 0.40 ml·mg-1at pH 4.0(see Fig.2).
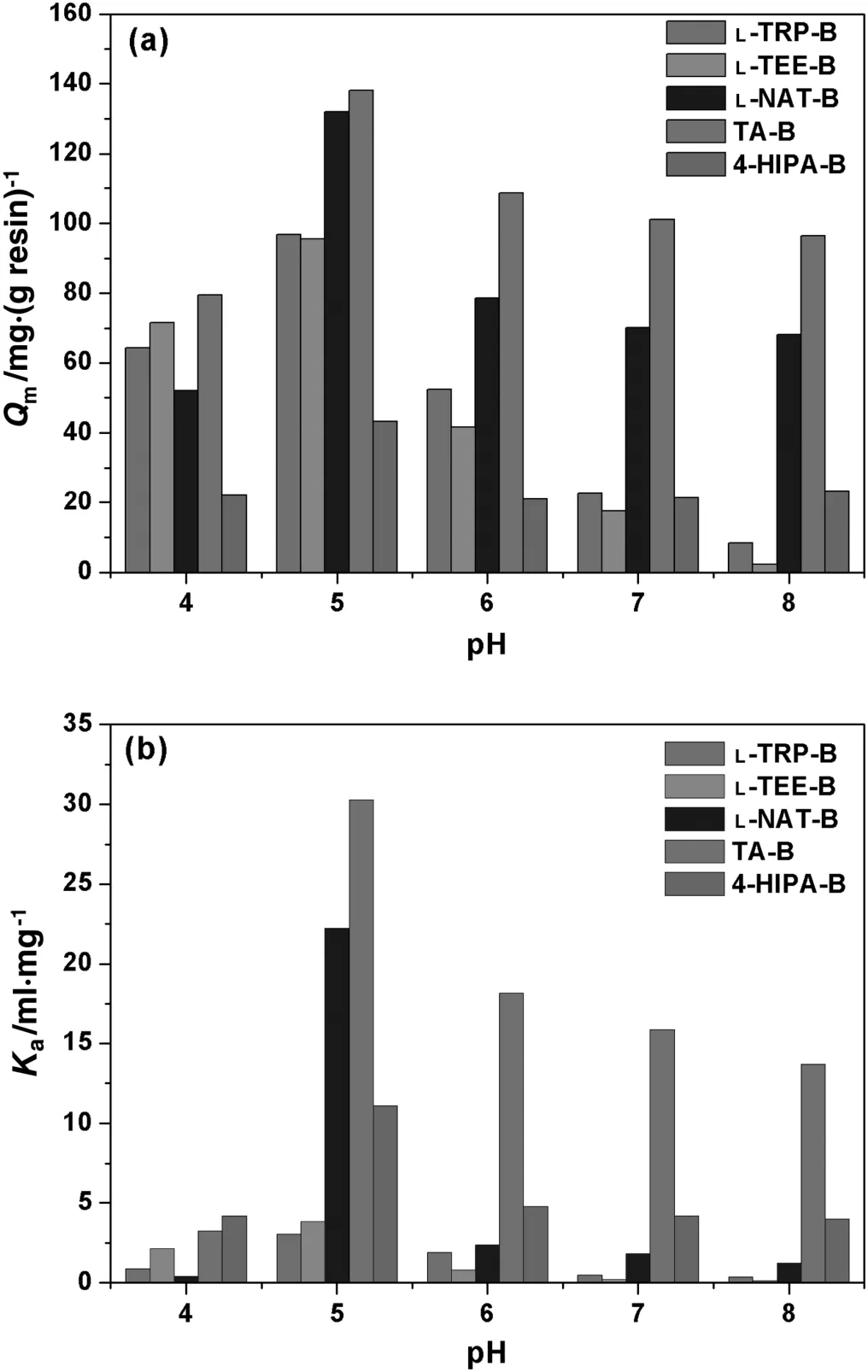
Fig.2.Q m(a)and K a(b)of HSA adsorption with five MMC resins at different pHs.
3.4.In fluence of NaCl addition on HSA adsorption
Salt-tolerant adsorption is another important property of MMC resins.NaClwas used as a neutralsaltto investigate its effecton HSA adsorption at pH 5.0.The adsorption isotherms at varying NaCl concentrations were measured and the correlatedQmandKaare compared in Fig.3.The results show that the five resins have different trends with the increase of NaCl concentration.
Fig.3(a)shows that the addition of NaCl generally reduces HSA adsorption with all of the five resins,and L-NAT-B was the most affected and 4-HIPA-B the least.Qmof L-NAT-B dropped sharply(~84.8%)with the addition of 0.25 mol·L-1NaCl.Qmof L-NAT-B was only~10 mg·g-1at high NaCl concentrations(0.5-1.0 mol·L-1).4-HIPA-B shows a U-shaped adsorption behavior.With the increase of NaClconcentration to 0.125 mol·L-1,Qmof 4-HIPA-B reduced slightly and then increased gradually to 47.50 mg·g-1in 1 mol·L-1NaCl,which is even higherthan thatwithoutsaltaddition(43.31 mg·g-1).In addition,QmofL-TRP-B dropped significantly by about 45.0%when 0.25 mol·L-1NaClwasadded,and thatvalue was 37.5%for L-TEE-B.At high NaCl concentration of 0.5-1.0 mol·L-1,Qmof L-TEE-B was obviously higher than that of L-TRP-B.The adsorption of TA-B showed a typical salt-tolerant property.Qmof TA-B slightly declined in low salt concentration(0.125 mol·L-1NaCl)and then nearly kept stable with the increase of salt concentration(89.57 mg·g-1at 1.0 mol·L-1NaCl).The salttolerant property is consistent with other MMC resins reported[35-37],which bene fits targetprotein capture directly from crude feedstocks with medium-to-high conductivity.Fig.3(b)shows the change ofKawith the increase of NaCl concentration,which has similar trends asQm.L-NAT-B was the most affected and 4-HIPA-B the least.Kaof TA-B kept relatively high among the five resins,which indicates that the TA ligand has strong affinity to HSA at wide ranges of NaCl concentrations.
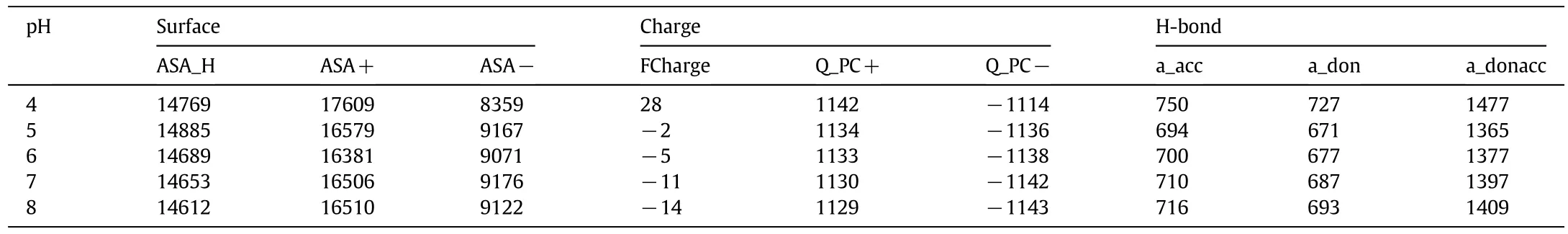
Table 2The descriptors of HSA calculated with MOE at various pHs
These results indicate that hydrophobic interactions dominated HSA adsorption at pH 5.0,while electrostatic attraction also plays an assistant role due to the asymmetrical charge distribution on HSA surface and partial charges on mixed-mode ligands.NaCl is a neutral salt which can shield electrostatic interactions[38].Therefore,the addition ofNaClcan reduce electrostatic attraction between HSAand the ligands,which results in decrease ofQmandKa.The 4-HIPA ligand with the highest hydrophobicity was less affected by NaCl addition and even enhanced hydrophobic binding at high salt concentrations.TA has the lowest total partial charges and the impact of salt addition was also relatively weak.L-TRP-B and L-TEE-B show weak salt tolerance,which means that electrostatic attraction was partly shielded.L-TEE-B had more partial charges and its binding affinity with HSA under high NaCl concentrations was improved.For L-NAT-B,electrostatic interaction may play a crucial role in HSA adsorption,andQmandKareduced significantly with NaCl addition.
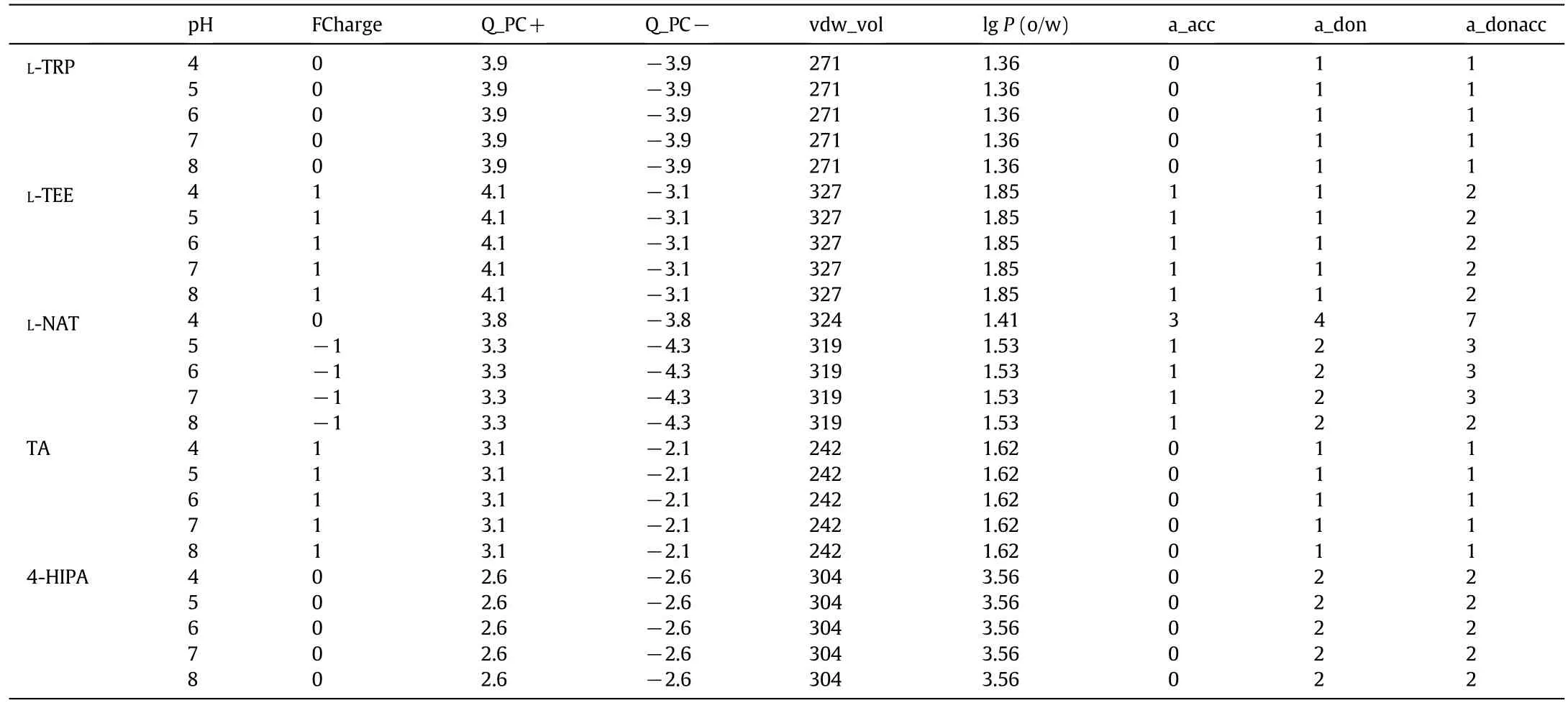
Table 3The descriptors of ligands calculated with MOE at various pHs
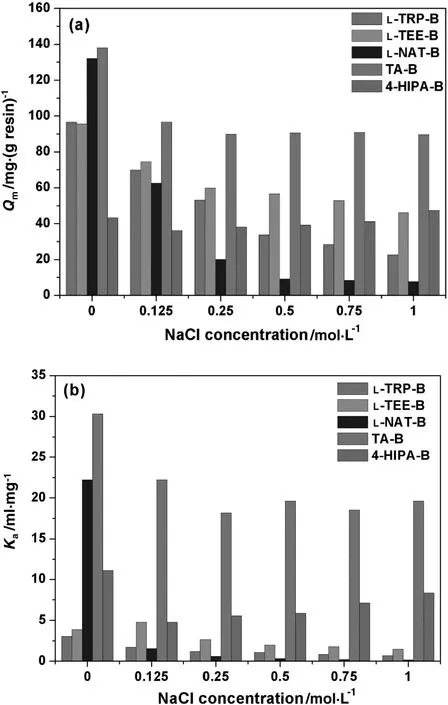
Fig.3.Q m(a)and K a(b)of HSA adsorption with five MMC resins at various NaCl concentrations(pH 5.0).
3.5.In fluence of(NH4)2SO4 addition on HSA adsorption
(NH4)2SO4is a typical kosmotropic salt which can reduce electrostatic interaction atlow concentrationsand improve hydrophobic interactions at high salt concentrations.QmandKaof HSA with the five MMC resins under different(NH4)2SO4concentrations are compared in Fig.4.Typical U-shape trends with(NH4)2SO4addition were found.
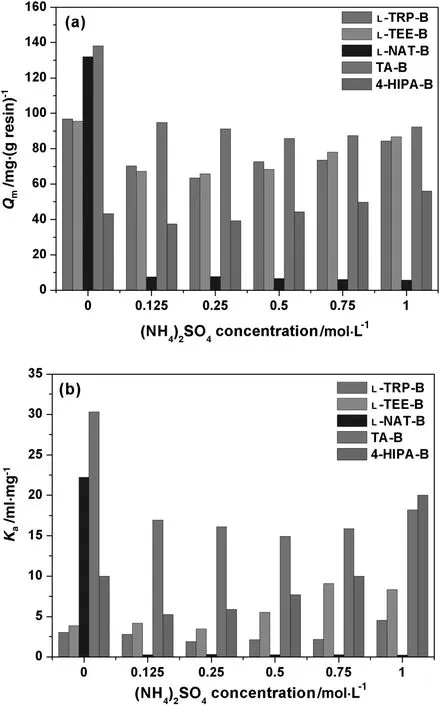
Fig.4.Q m(a)and K a(b)of HSA adsorption with five MMC resins at various(NH4)2SO4 concentrations(pH 5.0).
QmandKaof TA-B decreased by about 31.4%and 44.1%respectively at 0.125 mol·L-1(NH4)2SO4,and then they kept relatively stable with the increase of(NH4)2SO4.The trends of L-TRP-B,L-TEE-B and 4-HIPA-B were similar,but the strongest effects of(NH4)2SO4were found for 4-HIPA-B.Kaat 1 M(NH4)2SO4was obviously higher than thatwithoutsaltaddition,especially for 4-HIPA-B and L-TEE-B.High adsorption reduction was found with L-NAT-B,andQmdropped by more than 90%at the whole range of(NH4)2SO4concentration tested.(NH4)2SO4showed intensive effects than NaCl,which indicates difference on molecular mechanism with salt addition for MMC resins.
(NH4)2SO4has more charges than NaCl,which affects electrostatic interactions at low concentrations.The declining proportions ofQmat 0.125 mol·L-1(NH4)2SO4for L-TRP-B,L-TEE-B and L-NAT-B reins were 27.2%,29.6%and 94.3%,respectively,which were obviously higher than those with NaCl.Meanwhile,(NH4)2SO4could also improve hydrophobic interactions between the MMC ligands and protein molecules at high concentrations.Therefore,the typical U-shape trend with(NH4)2SO4addition was found.TA-B with medium hydrophobicity and partial charges was less affected by(NH4)2SO4addition,and a good salt-tolerant property was found with high adsorption capacity at a wide range of salt concentration.In general,the combination of hydrophobic and electrostatic interactions dominates protein adsorption processes for MMC resins,and the magical balance of the two binding modes leads to the special salt-tolerant adsorption properties with the specially-designed ligands.The results indicate that TA-B is more suitable for HSA separation from crude feedstock with medium-to-high conductivity.
3.6.Adsorption selectivity of rHSA from P.pastoris broth
The design of MMC resins with tryptophan analogues as functional ligands in the presentwork aimed to capture HSA from crude feedstock,such as rHSAfrom yeastfermentation.HereP.pastorisbroth was used to test the selectivity of HSA with the five MMC resins prepared.pH 5.0 was chosen as the optimized adsorption condition for rHSA.The collected fractions were analyzed by SEC-HPLC after adsorption equilibrium.Fig.5(a)shows that there were observable low-molecular-weight(LMW)impurities in the feedstock(corresponding to the retention time of 18.5-30 min).As shown in Fig.5(b)-(f),the five MMC resins developed showed good adsorption selectivity for rHSA.Most rHSA inP.pastorisbroth was adsorbed,while near all LMW impurities still retained in the feedstock.In general,rHSA adsorption with L-TRP-B,L-TEE-B and TA-B was higher than that of L-NAT-B and 4-HIPA-B.The impurity adsorption ratios varied from 3.2%to 11.6%,which was much higher for the ligands possessed more partial charges,such as L-TRP-B,L-TEE-B and L-NAT-B.The highest impurities uptake was found with L-TEE-B(11.6%).Not more than 5%of impurities were adsorbed by TA-Band 4-HIPA-B.Therefore,the five MMC resins developed in the present work could be used to capture rHSA from crude yeast fermentation with high efficiency.Due to the highest adsorption capacity,best salttolerant property and good selectivity,TA-B is a promising potential for rHSA purification.
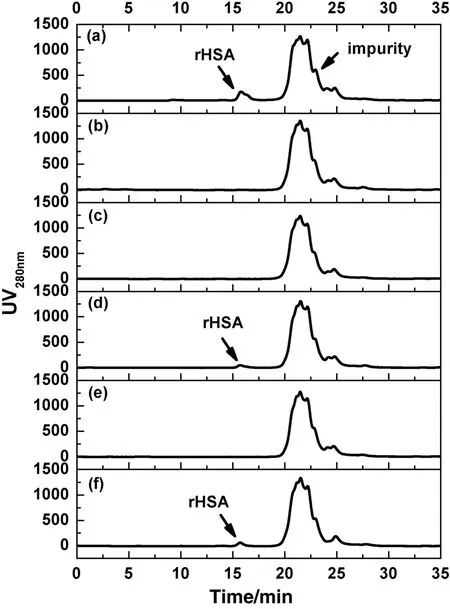
Fig.5.Comparison of feedstock(a)and the supernatants after adsorption with L-TRP-B(b),L-TEE-B(c),L-NAT-B(d),TA-B(e)and 4-HIPA-B(f)by SEC-HPLC analysis.

Fig.6.Molecular docking of the tryptamine ligand onto the surface of HSA.(a)Potential binding domains on HSA for tryptamine;(b)tryptamine in Site IIof HSA;(c)key residues around tryptamine in Site II.Tryptamine is shown by balland stick.Hydrophobic residue and negatively charged residue are shown in brown and red,respectively.Black dash stands for hydrogen bond.
3.7.Molecular docking of tryptamine and HSA
The molecular interactions between tryptamine and HSA were further studied with molecular docking.After docking scanning on the whole surface ofHSAwith LibDock,544 binding posesofthe tryptamine ligand were found.The LibDockScore was then used to evaluate the tryptamine-HSA binding at different sites,which considered the geometry,energy and chemical environment.Finally,95 binding poses with high score(more than 90)were identified,which mainly located on four regions named Domain 1,Domain 2,Domain 3 and Domain 4 as shown in Fig.6(a).These four domains composed the potential binding sites of tryptamine ligand on the surface of HSA molecule,where hydrophobic pockets or space cavities could be found to accommodate the ligand.The numbers of binding modes on the four domains were 40,19,18 and 18,respectively.Domain 1 seemed to be the most favorable binding sites for tryptamine,which occupied about 40%high-score binding modes and was two times higher than other three domains.As shown in Fig.6(b),Domain 1 was located on Site II(indole-binding site)ofHSA,which is a hydrophobic pocketwith ionic group located nearthe entrance,and has a preference for accommodating neutral lipophilic and acidic drug-like ligands[39].The cationic antimicrobial peptides were also found to bind onto Site II of HSA[40].Fig.6(c)shows the hot residues around the tryptamine ligand including hydrophobic and negative residues in Site II.Four leucine residues surround the hydrophobic indole ring of tryptamine dominated the binding,while hydrogen bonds between ASN391 and hydroxyl of tryptamine ligand could provide an assistant effect.The molecular simulation results indicate that tryptamine is favorable to bind on the indole-binding site of HSA,which might be the main reasons on the adsorption selectivity of five tryptophan analogues with typical indole ring for HSA.
4.Conclusions
Mixed-mode resins with tryptophan analogues including L-TRP,L-TEE,L-NAT-B,TA and 4-HIPA as the functional ligands were prepared.A typical pH-dependent adsorption was found and the highest adsorption capacity and affinity were found at pH 5.0 for all of the five resins tested.The highestQmwas 138.02 mg·g-1for the TA-Bresin.Molecular interaction analysis indicates that hydrophobic interactions dominated HSA adsorption atpH 5.0,while electrostatic attraction also plays an assistant role due to the asymmetrical charge distribution on HSA surface and partial charges on the mixed-mode ligands.When pH was lower than 4.0,the adsorption capacity decreased due to electrostatic repulsion between positively-charged ligands and HSA.The addition of NaCl and(NH4)2SO4reduced HSA adsorption due to electrostatic interaction shield between HSA and ligands.Suitable combination of hydrophobic and electrostatic interactions could control protein binding and lead to special adsorption properties.In addition,binding selectivity of HSA was evaluated with rHSA inP.pastorisfermentation broth.The five MMC resins prepared showed good adsorption selectivity for rHSA.Moleculardocking results oftryptamine ligand and HSA indicated that the tryptamine ligand was favorable to bind on Site II(indolebinding site)of HSA.In general,the TA-B resin showed the best salttolerant property and high adsorption capacity,which is a potential MMC resin for HSA separation from crude feedstock.
[1]S.C.Burton,N.W.Haggarty,D.R.K.Harding,One step purification of chymosin by mixed mode chromatography,Biotechnol.Bioeng.56(1)(1997)45-55.
[2]S.J.Yao,D.Gao,D.Q.Lin,Novel technology in bioseparation process:Mixed-mode chromatography,J.Chem.Ind.Eng.(China)(09)(2007)2169-2177(in Chinese).
[3]B.L.Johansson,M.Belew,S.Eriksson,G.Glad,O.Lind,J.L.Maloisel,N.Norrman,Preparation and characterization of prototypes for multi-modal separation aimed for capture of positively charged biomolecules at high-salt conditions,J.Chromatogr.A1016(1)(2003)35-49.
[4]G.F.Zhao,X.Y.Dong,Y.Sun,Ligands for mixed-mode protein chromatography:Principles,characteristics and design,J.Biotechnol.144(1)(2009)3-11.
[5]Q.L.Zhang,T.T.Zhuang,H.F.Tong,H.Y.Wang,D.Q.Lin,S.J.Yao,Experimental and in silico studies on three hydrophobic charge-induction adsorbents for porcine immunoglobulin purification,Chin.J.Chem.Eng.24(1)(2016)151-157.
[6]G.E.Hamilton,F.Luechau,S.C.Burton,A.Lyddiatt,Development of a mixed mode adsorption process for the direct product sequestration of an extracellular protease from microbial batch cultures,J.Biotechnol.79(2)(2000)103-115.
[7]R.Bischoff,L.W.Mclaughlin,Nucleic-acid resolution by mixed-mode chromatography,J.Chromatogr.296(Jul)(1984)329-337.
[8]R.C.Ruaan,A.Yang,D.Hsu,Bifunctional adsorbents for hydrophobic displacement chromatography of proteins,Biotechnol.Prog.16(6)(2000)1132-1134.
[9]J.Zhao,S.J.Yao,D.Q.Lin,Adsorbents for expanded bed adsorption:Preparation and functionalization,Chin.J.Chem.Eng.17(4)(2009)678-687.
[10]M.A.Rothschild,M.Oratz,S.S.Schreiber,Serum albumin,Am.J.Digest.Dis.14(10)(1969)711-744.
[11]G.E.Hastings,P.G.Wolf,The therapeutic use of albumin,Arch.Fam.Med.1(2)(1992)281-287.
[12]E.Marth,B.Kleinhappl,Albumin is a necessary stabilizer of TBE-vaccine to avoid fever in children after vaccination,Vaccine20(3-4)(2001)532-537.
[13]K.Kobayashi,N.Nakamura,A.Sumi,T.Ohmura,K.Yokoyama,The development of recombinant human serum albumin,Ther.Apher.2(4)(1998)257-262.
[14]X.M.He,D.C.Carter,Atomic structure and chemistry of human serum albumin,Nature358(6383)(1992)209-215.
[15]D.C.Carter,X.M.He,S.H.Munson,P.D.Twigg,K.M.Gernert,M.B.Broom,T.Y.Miller,Three-dimensional structure of human serum albumin,Science244(1989)4.
[16]D.C.Carter,X.M.He,Structure of human serum albumin,Science249(1990)2.
[17]U.Kragh-Hansen,V.T.G.Chuang,M.Otagiri,Practical aspects of the ligand-binding and enzymatic properties of human serum albumin,Biol.Pharm.Bull.25(6)(2002)695-704.
[18]R.H.Mcmenamy,J.L.Oncley,The specific binding of L-tryptophan to serum Albumin,J.Biol.Chem.233(6)(1958)1436-1447.
[19]W.E.Müller,U.Wollert,Benzodiazepines:Specific competitors for the binding of L-tryptophan to human serum albumin,Naunyn Schmiedeberg's Arch.Pharmacol.288(1)(1975)17-27.
[20]U.Kraghhansen,Octanoate binding to the indole-and benzodiazepine-binding region of human serum albumin,Biochem.J.273(1991)641-644.
[21]R.H.Mcmenamy,C.C.Lund,J.L.Oncley,J.Vanmarck,Binding of L-tryptophan in human plasma at 37 degrees C,Arch.Biochem.Biophys.93(1)(1961)135-159.
[22]R.H.Mcmenamy,Association ofindole analogues to defatted human serum albumin,Arch.Biochem.Biophys.103(3)(1963)409-417.
[23]J.Vajda,E.Mueller,E.Bahret,Dual salt mixtures in mixed mode chromatography with an immobilized tryptophan ligand in fluence the removal of aggregated monoclonal antibodies,Biotechnol.J.9(4)(2014)555-565.
[24]G.F.Zhao,G.Y.Peng,F.Q.Li,Q.H.Shi,Y.Sun,5-Aminoindole,a new ligand for hydrophobic charge induction chromatography,J.Chromatogr.A1211(1-2)(2008)90-98.
[25]Q.C.Wu,D.Q.Lin,W.Shi,Q.L.Zhang,S.J.Yao,A mixed-mode resin with tryptamine ligand for human serum albumin separation,J.Chromatogr.A1431(2016)145-153.
[26]S.C.Burton,D.R.K.Harding,Preparation of chromatographic matrices by free radical addition ligand attachment to allyl groups,J.Chromatogr.A796(2)(1998)273-282.
[27]S.C.Burton,D.R.K.Harding,High-density ligand attachment to brominated allyl matrices and application to mixed mode chromatography of chymosin,J.Chromatogr.A775(1-2)(1997)39-50.
[28]Z.M.Chen,D.Q.Lin,S.J.Yao,New mixed-mode adsorbent and its application for separation of bovine colostrum immunoglobulin,CIESC J.08(2012)2453-2459(in Chinese).
[29]H.F.Xia,D.Q.Lin,L.P.Wang,Z.J.Chen,S.J.Yao,Preparation and evaluation of cellulose adsorbents for hydrophobic charge induction chromatography,Ind.Eng.Chem.Res.47(23)(2008)9566-9572.
[30]R.Z.Wang,D.Q.Lin,H.F.Tong,S.J.Yao,Molecular insights into the binding selectivity of a synthetic ligand DAAG to Fc fragment of IgG,J.Mol.Recognit.27(5)(2014)250-259.
[31]H.L.Lu,D.Q.Lin,D.Gao,S.J.Yao,Evaluation of immunoglobulin adsorption on the hydrophobic charge-induction resins with different ligand densities and pore sizes,J.Chromatogr.A1278(2013)61-68.
[32]T.Liu,D.Q.Lin,H.L.Lu,S.J.Yao,Preparation and evaluation of dextran-grafted agarose resin for hydrophobic charge-induction chromatography,J.Chromatogr.A1369(2014)116-124.
[33]H.F.Xia,D.Q.Lin,Z.M.Chen,S.J.Yao,In fluences of ligand structure and pH on the adsorption with hydrophobic charge induction adsorbents:A case study of antibody IgY,Sep.Sci.Technol.46(12)(2011)1957-1965.
[34]D.Gao,D.Q.Lin,S.J.Yao,Mechanistic analysis on the effects of saltconcentration and pH on protein adsorption onto a mixed-mode adsorbent with cation ligand,J.Chromatogr.B859(1)(2007)16-23.
[35]P.Li,G.Xiu,V.G.Mata,C.A.Grande,A.E.Rodrigues,Expanded bed adsorption/desorption of proteins with Streamline Direct CST I adsorbent,Biotechnol.Bioeng.94(6)(2006)1155-1163.
[36]Q.H.Shi,Z.Cheng,Y.Sun,4-(1H-imidazol-1-yl)aniline:A new ligand of mixedmode chromatography for antibody purification,J.Chromatogr.A1216(33)(2009)6081-6087.
[37]D.Gao,D.Q.Lin,S.J.Yao,Protein adsorption kinetics of mixed-mode adsorbent with benzylamine as functional ligand,Chem.Eng.Sci.61(22)(2006)7260-7268.
[38]Q.L.Zhang,F.Schimpf,H.L.Lu,D.Q.Lin,S.J.Yao,Binary adsorption processes of albumin and immunoglobulin on hydrophobic charge-induction resins,J.Chem.Eng.Data61(3)(2016)1353-1360.
[39]J.Ghuman,P.A.Zunszain,I.Petitpas,A.A.Bhattacharya,M.Otagiri,S.Curry,Structural basis of the drug-binding specificity of human serum albumin,J.Mol.Biol.353(1)(2005)38-52.
[40]A.Sivertsen,J.Isaksson,H.K.S.Leiros,J.Svenson,J.S.Svendsen,B.O.Brandsdal,Synthetic cationic antimicrobial peptides bind with their hydrophobic parts to drug site II of human serum albumin,BMC Struct.Biol.14(2014).
杂志排行
Chinese Journal of Chemical Engineering的其它文章
- Suppression of gold nanoparticle agglomeration and its separation via nylon membranes☆
- Effects of solubility parameter differences among PEG,PVP and CA on the preparation of ultra filtration membranes:Impacts of solvents and additives on morphology,permeability and fouling performances
- Experimental detection of bubble-wall interactions in a vertical gas-liquid flow☆
- Horizontal gas mixing in rectangular fluidized bed:A novel method for gas dispersion coefficients in various conditions and distributor designs
- Synthesis of clay-supported nanoscale zero-valent iron using green tea extract for the removal of phosphorus from aqueous solutions
- Hydrodynamic dispersion ofreactive solute in a Hagen-Poiseuille flow of a layered liquid
