两个3-甲基-2-乙酰吡嗪缩4-苯基氨基脲双核铜配合物的晶体结构及与DNA的相互作用
2017-02-16林龙李先宏张波张战营吴伟娜王元
林龙李先宏张波张战营吴伟娜王元
(1河南理工大学材料科学与工程学院,焦作454000)
(2河南理工大学数学与信息科学学院,焦作454000)
(3河南理工大学化学化工学院,焦作454000)
林龙1,2李先宏1张波1张战营1吴伟娜*,3王元*,3
(1河南理工大学材料科学与工程学院,焦作454000)
(2河南理工大学数学与信息科学学院,焦作454000)
(3河南理工大学化学化工学院,焦作454000)
合成并通过单晶X射线衍射、元素分析及红外光谱表征了配合物[Cu2(L)2Cl2](1)和[Cu2(L)2(OAc)2](2)的结构(HL为3-甲基2-乙酰吡嗪缩4-苯基氨基脲)。单晶衍射结果表明,2个配合物中,每个拥有四方锥配位构型的Cu离子与来自1个阴离子配体L-的N2O电子供体和2个阴离子配位(1中为氯离子,2中为醋酸根离子),其中1个阴离子为μ2桥联配位模式。荧光光谱结果表明,配合物与DNA的相互作用强于配体。
缩氨基脲;铜配合物;吡嗪;晶体结构;DNA相互作用
In the past few decades,Schiff bases and their metal complexes have been a focus of chemists and biologists because of their noteworthy antibacterial, antifungal,anticancer,urease inhibition,antioxidant and antiglycation activities[1-8].It has been demonstrated that the presence of heterocyclic ring in the synthesized Schiff bases plays a major role in extending their pharmacological properties[8].As a result,a sizable numberoftransitionmetalcomplexeswith acylhydrazones and thiosemicarbazones derived fromactyl-pyridine/pyrazinehavebeenextensively investigatedaspotentialanticanceragents[3,8-10]. However,astheirstructurallyanalogous, semicarbazones have been paid much less attention[6].
On the other hand,the previous studies revealed that Cucontaining anticancer agents are promising leadsfornextgenerationmetal-basedanticancer agents because Cuplays a significant role in biological systems[1,5].Therefore,in this paper,two Cucomplexes with a semicarbazone ligand derived from2-acetyl-3-methylpyrazineand4-phenylsemicarbazidehavebeensynthesizedand structuraldeterminedbysingle-crystalX-ray diffraction.In addition,the interactions between three compoundsandct-DNAhavebeenstudiedby ethidium bromide(EB)fluorescence probe.
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for synthesis were purchasedcommerciallyandusedasreceived. Elemental analysis was carried out on an Elemental Vario EL analyzer.The IR spectra(ν=4000~400cm-1) were determined by the KBr pressed disc method on a Bruker V70FTIR spectrophotometer.1H NMR spectra ofLwasacquiredwithBrukerAV400NMR instrument in DMSO-d6solution with TMS as internal standard.The interactions between three compounds and ct-DNA are measured using literature method[11]via emission spectra on a Varian CARY Eclipse spectrophotometer.
1.2 Preparations of the ligand HL,complexes 1and 2
As shown in Scheme 1,the ligand HL was produced by condension of 2-acetyl-3-methylpyrazine (1.36g,0.01mol)and 4-phenylsemicarbazide(1.51g, 0.01mol)in ethanol solution(30mL)with continuous stirring at room temperature for 5h.The white solid was filtered and washed three times by cold ethanol. Yield:2.21g(85%).m.p.178~180℃.Elemental analysis Calcd.for C14H15N5O(%):C 62.44,H 5.61,N 26.01.Found(%):C 62.56,H 5.39,N 25.89.FTIR (cm-1):ν(C=O)semicarbazone1702,ν(C=N)1604,ν(C=N)pyrazine1593.1H NMR(400MHz,DMSO-d6):δ 10.03(1H,s,NH),8.77(1H,s,NH),8.45~8.48(2H,m, pyrazine-H),7.54~7.56(2H,m,phenyl-H),7.23~7.27(2H,m,phenyl-H),6.95~6.99(1H,m,phenyl-H),2.75(3H,s,CH3),2.28(3H,s,CH3). The complexes 1and 2were generated by reaction of the ligand HL(5mmol)with equimolar of CuCl2·2H2O and Cu(OAc)2·H2O in methanol solution (10mL),respectively.Crystals suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
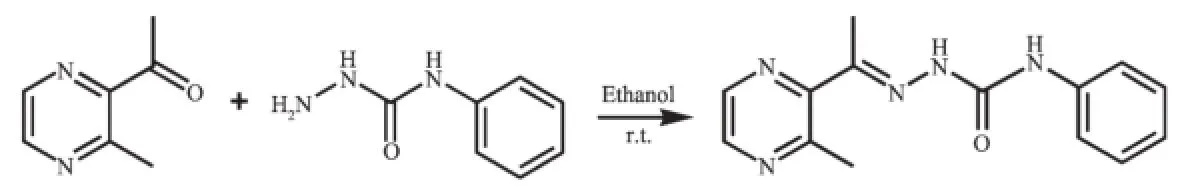
Scheme 1Synthesis route of HL
1:Greenblocks.Anal.Calcd.for C28H28N10O2Cl2Cu2(%):C 45.78,H 3.84,N 19.07. Found(%):C 45.65,H 4.00,N 18.94.FTIR(cm-1): ν(N=C-O)1619,ν(C=N)1594,ν(C=N)pyrazine1548.
2:Black blocks.Anal.Calcd.for C32H34N10O6Cu2(%):C 49.16,H 4.38,N 17.92.Found(%):C 49.36, H 4.52,N 17.74.FTIR(cm-1):ν(N=C-O)1595,ν(C= N)1579,ν(C=N)pyrazine1544,νas1(COO-)1509, νas4(COO-)1435and 1352.
1.3.1 X-ray crystallography
The X-ray diffraction measurement for complexes 1and 2were performed on a Bruker SMART APEXⅡCCD diffractometer equipped with a graphite monochromatized Mo Kα radiation(λ=0.071073nm) by using φ-ω scan mode.Semi-empirical absorption correction was applied to the intensity data using the SADABS program[12].
The structures were solved by direct methods and refined by full matrix least-square on F2using the SHELXTL-97program[13].All non-hydrogen atoms were refined anisotropically.All the H atoms were positioned geometrically and refined using a ridingmodel.Detailsofthecrystalparameters,data collection and refinements for complexes 1and 2are summarized in Table 1.
CCDC:1455421,1;1455422,2.

Table 1Crystal data and structure refinement for complexes 1and 2
2 Results and discussion
2.1 Crystal structure description
A diamond drawing for complexes 1and 2is shown in Fig.1.Selected bond distances and angles are listed in Table 2.The lengths of C-O bond of the semicarbazone moiety are 0.1279(5)and 0.1269(3)nm in complexes 1and 2,respectively,clearly showing that the ligand HL has enolizated and deprotonated in both complexes[8].
As shown in Fig.1a,complex 1contains one discrete dimeric Cumolecule in the unit cell.Two Cu atoms of the dimer were separated by 0.3524nm and doubly bridged by two chloride anions to form an ideal planar four-membered Cu2Cl2core.Each of the Cuions is penta-coordinated by one independent anionic ligand with N2O donor set and two chloride anions,one of which acts as a μ2-bridge,thus giving a distorted square pyramid coordination geometry(τ= 0.122)[14].In the solid state,the discrete Cudimers of 1were further linked into a one-dimensional chain along a axis(Fig.1c)by intermolecular N-H…Cl(N5-H5A…Cl1iii,with D…A distance being 0.3431(4)nm, D-H…A angle being 170.1°,Symmetry codes:iiix+1, y,z)hydrogen bonds between the amine nitrogen atoms from one dimer and chloride anions from the adjacent one.
The structure of 2is similar as that of 1,while the chloride anion is replaced by monodentate acetate, in which one oxygen atom of the carboxyl group bridges two Cuions to form μ-O.One-dimensional chain(Fig.1d)along a axis formed by intermolecular N-H…O(N5-H5A…O3iv,with D…A distance being 0.2886(3)nm,D-H…A angle being 157.8°,Symmetry codes:iv2-x,2-y,2-z)hydrogen bonds are also present in the crystal of 2.

Fig.1Diamond drawing of 1(a)and 2(b)with 30%thermal ellipsoids and extend 2D supramolecular structure along a axis in complexes 1(c)and 2(d)
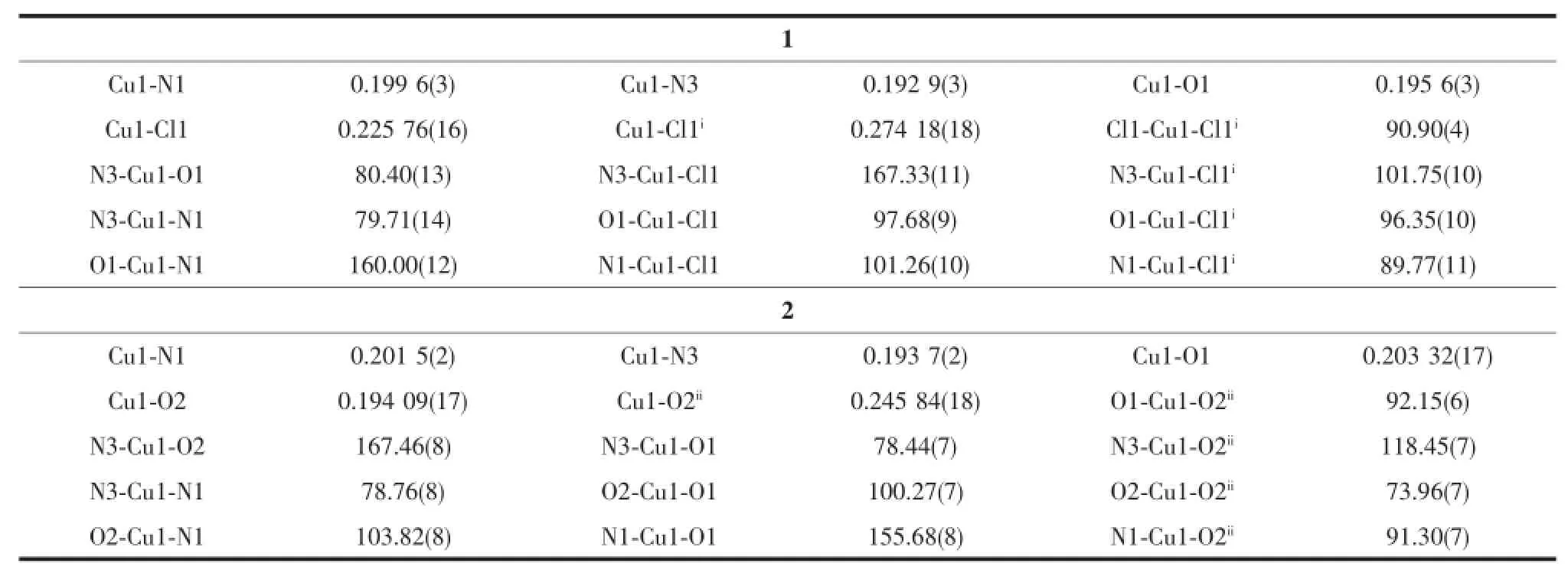
Table 2Selected bond lengths(nm)and angles(°)in complexes 1and 2
2.2 IR spectra
The νsemicarbazone(C=O)of the free ligand is 1748cm-1,while it is disappeared in both complexes, meanwhile,new N=C-O stretching vibration absorption is observed at 1619and 1595cm-1in complexes 1and 2,respectively,revealing that the C=O in O=C-N moiety has enolized and the oxygen atom coordinates to the metal ions in both complexes[6-7].The ν(C=N) bands of the imine group and pyrazine ring in the ligand HL shift to lower frequency values in the complexes,indicating that the N atoms of both units take part in the coordination[8].Furthermore,the bands at 1509,1435and 1352cm-1in complex 2could be assigned to the split bands νas1(COO-)and νas4(COO-)of the acetate group,respectively,showing that there are bridged and monodentate carboxyl groups in the complex 2[15].It is in accordance with the crystal structure study.
2.3 EB-DNA binding study by fluorescence spectrum
ItiswellknownthatEBcanintercalate nonspecifically into DNA,which causes it to fluoresce strongly.Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[16].The effects of the ligand and complexes on the fluorescence spectra ofEB-DNAsystemarepresentedinFig.2,the fluorescence intensities of EB bound to ct-DNA at about 600nm show remarkable decreasing trends with the increasing concentration of the tested compounds, indicating that some EB molecules are released into solution after the exchange with the compounds.The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation: I0/I=1+Ksqr[11],whereI0andIrepresentthe fluorescence intensities in the absence and presence of quencher,respectively,Ksqis the linear Stern-Volmer quenching constant,r is the ratio of the concentration of quencher and DNA.In the quenching plots of I0/I versus r,Ksqvalues are given by the slopes.The Ksqvalues are 0.268,0.723and 0.780for the ligand HL,complexes 1and 2,respectively.The results indicate that interactions of the complexes with DNA are stronger than that of the ligand HL,because the complexes have higher rigidity to bind the base pairsalongDNA,whichincreasestheirbinding abilities.In addition,the complexes 1and 2have similarKsqvalues,showingthatthecoordination anionsarealmostirresponsiblefortheDNA interaction.
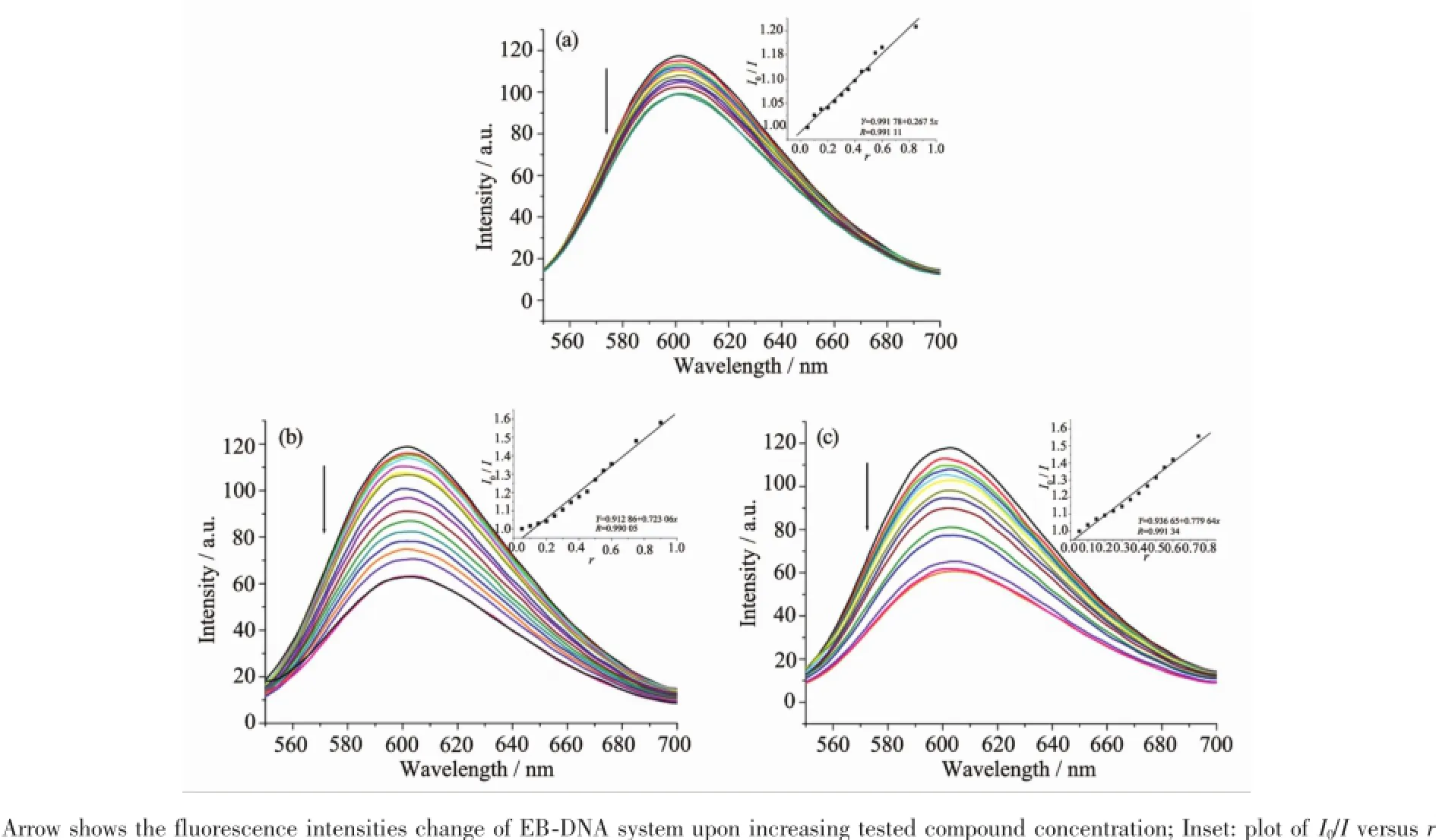
Fig.2Emission spectra of EB-DNA system in the absence and presence of ligand HL(a),complexes 1(b)and 2(c)
[1]Ye X P,Zhu T F,Wu W N,et al.Inorg.Chem.Commun., 2014,47:60-62
[2]Alagesan L,Bhuvanesh N S P,Dharmaraj N.Dalton Trans., 2013,42:7210-7223
[3]CHEN Yan-Min(陈延民),XIE Qing-Fan(解庆范),LIU Jin-Hua(刘金花),et al.Chinese J.Inorg.Chem.(无机化学学报),2015,31:74-80
[4]Singh P,Singh D P,Singh V P.Polyhedron,2014,81:56-65
[5]Nath M,Vats M,Roy P.Eur.J.Med.Chem.,2013,59:310-321
[6]Milenkovi M,Cantoni G,Bacchi A,et al.Polyhedron,2014, 80:47-52
[7]Milenkovi M,Bacchi A,Cantoni G,et al.Inorg.Chim.Acta,2013,395:33-43
[8]Chang H Q,Jia L,Xu Z Q,et al.Inorg.Chem.Commun., 2015,57:8-10
[9]Li M X,Zhang L Z,Yang M,et al.Bioorg.Med.Chem. Lett.,2012,22:2418-2433
[10]Li M X,Zhang L Z,Zhang D,et al.Eur.J.Med.Chem., 2011,46:4383-4390
[11]SHEN Wei(沈伟),HU Wei-Ji(胡未极),WU Xiao-Yong (吴小勇),et al.Chinese J.Inorg.Chem.(无机化学学报), 2016,32:1101-1110
[12]Sheldrick G M.SADABS,University of Göttingen,Germany, 1996.
[13]Sheldrick G M.SHELX-97,Program for the Solution and the Refinement of Crystal Structures,University of Göttingen, Germany,1997.
[14]WU Wei-Na(吴伟娜),WANG Yuan(王元),TANG Ning (唐宁).Chinese J.Inorg.Chem.(无机化学学报),2012,28: 425-428
[15]Nakamoto K.Infrared and Raman Spectra of Inorganic and CoordinationCompounds.4thEd.NewYork:Wiley,1986:257[16]Yang Z Y,Wang Y,Wang Y.Bioorg.Med.Chem.Lett., 2007,17:2096-2101
LIN Long1LI Xian-Hong1ZHANG Bo1ZHANG Zhan-Ying1WU Wei-Na*,2WANG Yuan*,2
(1School of Materials Science and Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
(2School of Mathematics and Informatics,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
(3College of Chemistry and Chemical Engineering,Henan Polytechnic University,Jiaozuo,Henan 454000,China)
Two binuclear complexes,namely[Cu2(L)2Cl2](1)and[Cu2(L)2(OAc)2](2)(where HL is 1-(3-methylpyrazin-2-yl)ethylidene-4-phenylsemicarbazide),have been synthesized and characterized by single crystal X-ray diffraction, elemental analysis and IR spectroscopy.X-ray diffraction analysis results show that in both binuclear complexes, each central Cuion with a distorted square pyramid coordination geometry is surrounded by one independent anionic ligand with N2O donor set and two coordinated anions(chloride for 1,whilst acetate for 2),one of which acts as a μ2-bridge.In addition,the fluorescence spectra indicate that the interactions of the complexes with DNA are stronger than that of the ligand HL.CCDC:1455421,1;1455422,2.
semicarbazone;Cucomplex;pyrazine;crystal structure;DNA interaction
O614.121
A
1001-4861(2017)01-0143-06
10.11862/CJIC.2016.283
2016-07-15。收修改稿日期:2016-10-27。
国家自然科学基金(No.21001040,21404033,21401046)、河南省科技厅基础与前沿项目(No.162300410011)和教育厅自然科学基金(No.12B150011,14B150029)资助。
*通信联系人。E-mail:wuwn08@hpu.edu.cn,wangyuan08@hpu.edu.cn;会员登记号:S06N6704M1112。
