过表达三角褐指藻苹果酸酶基因提高E.coli脂肪酸合成能力研究❋
2016-06-16吕娜娜朱葆华杨官品潘克厚
吕娜娜, 朱葆华❋❋, 鹿 琳, 杨官品, 潘克厚,3
(1. 中国海洋大学海水养殖教育部重点实验室应用微藻生物学研究室,山东 青岛 266003;2. 中国海洋大学海洋生命学院,山东 青岛 266003;3. 国家实验室海洋渔业与食物产出功能实验室,山东 青岛 266100)
过表达三角褐指藻苹果酸酶基因提高E.coli脂肪酸合成能力研究❋
吕娜娜1, 朱葆华1❋❋, 鹿琳1, 杨官品2, 潘克厚1,3
(1. 中国海洋大学海水养殖教育部重点实验室应用微藻生物学研究室,山东 青岛 266003;2. 中国海洋大学海洋生命学院,山东 青岛 266003;3. 国家实验室海洋渔业与食物产出功能实验室,山东 青岛 266100)
摘要:为验证三角褐指藻(Phaeodactylum tricornutum)苹果酸酶基因的功能,本研究将PtME1插入pET-30a中得到重组质粒pET30a-PtME1。IPTG诱导后,携带pET30a-PtME1的大肠杆菌BL21(DE3)高效表达一分子量约为72 kDa的可溶性重组蛋白。重组蛋白经Ni SephroseTM6 Fast Flow系统纯化,酶活力达75.18 U/mg。GC-MS分析显示表达PtME1提高了大肠杆菌脂肪酸合成能力,其C14∶0、C16∶0、C18∶1及总脂肪酸含量较对照分别提高了34.8%、69.9%、54.2%和50.2%,C16∶1产量是对照的5.6倍。研究结果表明,NADP依赖型苹果酸酶能为大肠杆菌脂肪酸合成及脂肪酸去饱和提供充足的NADPH,为进一步研究该酶在藻体内的功能奠定了基础。
关键词:三角褐指藻; 苹果酸酶; 脂肪酸; 原核表达
引用格式:吕娜娜, 朱葆华, 鹿琳, 等. 过表达三角褐指藻苹果酸酶基因提高E.coli脂肪酸合成能力研究[J]. 中国海洋大学学报(自然科学版), 2016, 46(5): 65-69.
LV Na-Na, ZHU Bao-Hua, LU Lin, et al. Overexpression of malic enzyme gene fromPhaeodactylumtricornutumpromotes fatty acids production inEscherichiacoli[J]. Periodical of Ocean University of China, 2016, 46(5): 65-69.
NADP 依赖型苹果酸酶(NADP-malic enzyme, EC1.1.1.40)普遍存在于各种生物中,参与多种代谢途径。该酶催化苹果酸氧化脱羧,生成丙酮酸和CO2,还原NADP+[1]。植物中,NADP-ME可分为光合型和非光合型[2-3]。光合型NADP-ME主要为C4植物Rubisco 酶提供CO2, 非光合型 NADP-ME则参与植物防御反应,稳定细胞内pH,为细胞合成代谢提供 NADPH 和丙酮酸[4]。富油微生物、植物NADP-ME参与油脂积累的报道较多。例如: 蓖麻子非光合型NADP-ME为长链脂肪酸合成提供碳和NADPH[5-6]。拟南芥基因组存在4个NADP-ME基因,其中,NADP-ME4参与脂肪酸合成[7]。卷枝毛霉(Mucorcircinelloides)有6种NADP-ME亚型,其中,亚型IV与油脂积累有关[8]。多种产油微生物及植物苹果酸酶基因已被克隆并验证了功能[9-12]。然而,关于藻类苹果酸酶基因的克隆和功能验证却鲜有报道, Shang等[13]克隆并分析了巴夫杜氏藻(Dunaliellaparva)的苹果酸酶基因,但未进行深入研究。
三角褐指藻(Phaeodactylumtricornutum)全基因组测序已完成[14],细胞壁硅含量很少,被广泛用于硅藻生态学、生理学、生物化学和分子生物学研究。该藻油脂含量较高,能大量合成并积累PUFAs,特别是EPA[15],是规模化生产生物柴油的优良候选藻种[14-15]。
为验证三角褐指藻苹果酸酶基因功能,本研究用RT-PCR技术分离了三角褐指藻苹果酸酶基因cDNA (PtME1),并在大肠杆菌中进行了表达;对融合蛋白进行了纯化,据纯化蛋白底物特异性和大肠杆菌脂肪酸组成变化验证了三角褐指藻苹果酸酶基因功能。研究结果为深入探索苹果酸酶在三角褐指藻脂肪酸合成过程中的作用,进而为用基因工程手段调控三角褐指藻油脂积累研究奠定了基础。
1材料与方法
1.1 三角褐指藻培养
三角褐指藻(P.tricornutumPT-01)由中国科学院水生生物研究所馈赠,用无菌f/2海水培养基在(20±1)℃、27.5~37.5 (mol photons·m-2·s-1光强(12 h光照∶12 h黑暗)下静置培养。
1.2 引物设计与合成
据P.tricornutum苹果酸酶基因设计PCR引物,并引入酶切位点NcoI和SaII (见表1),下游引物加入6xHis标签,便于重组蛋白纯化。引物由上海生工生物工程技术服务有限公司合成。
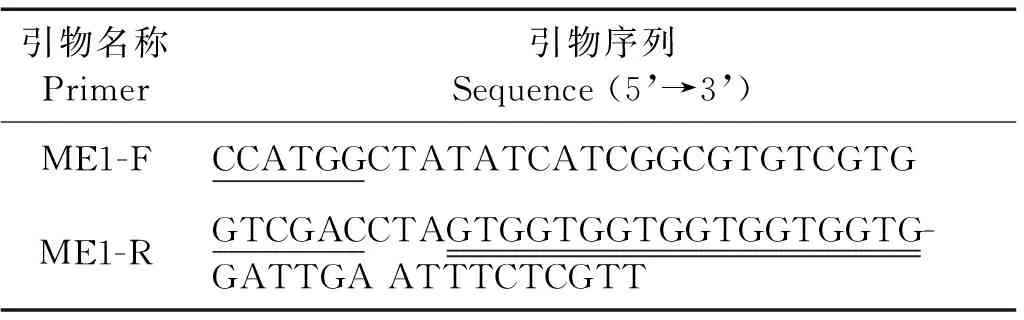
表1 PtME1扩增引物
注:下划线示引入的限制性酶切位点,双下划线示加入的6个His的密码子。Underlined bases indicate the digested nucleotide sequences of the restriction enzyme, double underlined ones show the joined six His codons.
1.3 PtME1克隆及表达载体构建
三角褐指藻总RNA提取采用Total RNA Kit I试剂盒(Omega公司)。用TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (全式金公司)反转录合成cDNA。用TaqDNA polymerase(Takara公司)扩增PtME1。用pET30a(由中国科学院青岛生物能源与过程研究所咸漠老师馈赠)构建重组质粒。宿主菌E.coliBL21(DE3)购自全式金公司。限制性内切酶及胶回收试剂盒购自Fermentas公司。
PCR反应条件为94 ℃预变性1 min,94 ℃ 30 s,63 ℃ 30 s,72 ℃ 2 min, 30个循环,最后72 ℃延伸10 min。PCR产物用胶回收试剂盒(Omega公司)切胶纯化,构建重组质粒pET30a-PtME1,连同pET30a质粒分别转化至E.coliBL21(DE3),在含100μg/mL卡那霉素LB平板上37℃过夜培养。挑取抗性克隆,提取重组质粒酶切鉴定,测序验证。
1.4 重组蛋白诱导表达和纯化
将重组菌株(含pET30a-PtME1质粒)、含pET30a质粒菌株(作为对照)分别按1∶50接种量接种到含50μg/mL卡那霉素LB液体培养基中,37 ℃,200 r/min震荡培养至OD600nm约0.6~0.8,加入IPTG至终浓度0.5 mmol/L,30 ℃诱导4 h。在4℃,8 000 r/min离心8 min收集菌体,PBS重悬洗涤一次,沉淀加10 mL磷酸缓冲液(20 mmol/L 磷酸钠,500 mmol/L NaCl,pH=7.4),冰浴超声,处理3 s,停8 s,共10 min,功率80 W,上清即为粗酶液,取部分上清进行SDS-PAGE检测,剩余部分用Ni SephroseTM6 Fast Flow系统(GE公司)纯化蛋白。
1.5 重组ME酶活性的测定
按文献[18]配置酶活性测定最适体系(50 mmol/L, pH=7.5 Tris-HCl, 1 mmol/L MgCl2, 0.5 mmol/L NADP+, 10 mmol/L L-malate)。取870 μL反应溶液和30 μL纯化酶液于1 mL石英比色皿中,混匀,立即置于紫外分光光度计中,在室温下连续监测1 min内340 nm 下的吸收值的变化。每分钟催化产生1 μmol NADPH 定义为一个酶活力单位。蛋白浓度采用 Bradford 蛋白浓度定量试剂盒(索莱宝公司)。
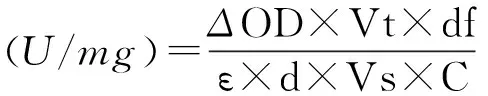
其中:Vt为反应体系总体积(9×10-4L) ;Vs为加入样品体积(0.03 mL);ε:为在测定条件下NADPH的摩尔消光系数,6.22×103L·mol-1·cm-1;d为光程(1 cm);df为稀释因子,106;C为在溶液中酶的浓度,mg/mL。
1.6 重组子脂肪酸组成分析和含量测定
将2种重组菌(分别含pET30a质粒和 pET30a-PtME1质粒)接种至M9基本培养基中,37 ℃,200 r/min震荡培养至OD600nm约0.6~0.8,加入IPTG至终浓度0.5 mmol/L,同时加入苹果酸至终浓度15 mmol/L,30℃诱导24 h。离心收集菌液,冷冻干燥用于脂肪酸的测定。取40 mg菌粉于15 mL试管中,参照文献[19]加入3 mL萃取剂(甲醇∶氯仿 = 1∶2),充分涡旋震荡萃取细胞总脂,再加入5 mL皂化试剂(水∶甲醇=1∶4,含6%NaOH),同时加入100 μL浓度为2 mg/mL的十五烷酸(Sigma公司)作为内标进行定量,60℃水浴1 h,最后加入2 mL甲酯化试剂(12%三氟化硼-甲醇溶液),60 ℃水浴30 min,冷却后用1 mL色谱纯正己烷震荡萃取脂肪酸甲酯,然后在气相-质谱仪上检测(GCMS-QP2010,岛津,日本)。气相条件,色谱柱:Rxi-1MS(30 m×0.25 mm,0.25 μm)毛细管柱;升温程序:初始温度 150 ℃ ,以15 ℃/min 升至200 ℃ ,再以2 ℃/min升至250 ℃ ;进样口温度:250 ℃ ;载气(He) 流量:1 mL/min;自动进样,进样体积1 μL,分流比20∶1;溶剂切除时间:2.5 min。质谱条件:电子轰击离子源,离子源温度230 ℃ ,接口温度280 ℃ ,电子能量70 eV,质量扫描范围45~500 m/z。
2结果与分析
2.1 三角褐指藻苹果酸酶基因的克隆
PtME1(XM_002177854.1,见图1(a))的DNA扩增片段与预期大小一致。重组质粒双酶切获得的片段与菌落PCR产物长度长度一致(见图1(c)),说明重组质粒所连接的为目的条带,且酶切完全。
2.2 重组蛋白的诱导表达及纯化
将重组质粒pET30a-PtME1转化至E.coliBL21(DE3) 感受态细胞,IPTG诱导4 h后,破菌上清经SDS-PAGE检测发现在72 kD处有一条特异条带(见图2),与预期蛋白大小相符,对照无特异条带,表明PtME1已在大肠杆菌BL21(DE3)中正确表达。
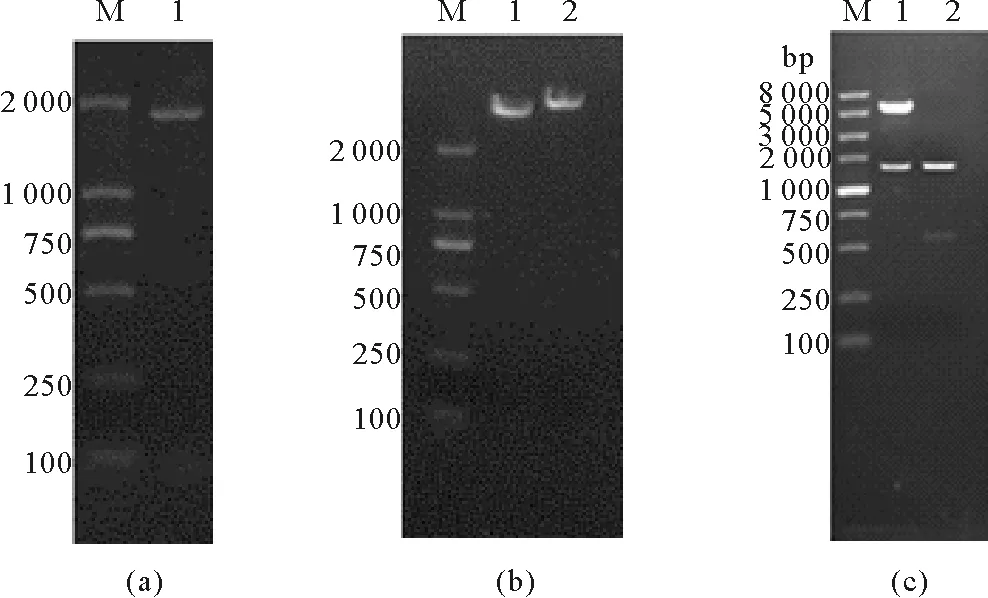
((a)基因组扩增产物;M,DNA marker DL 2000;1,PCR产物。(b)质粒;M,DNA marker DL 2000;1,质粒pET30a;2,重组质粒pET30a-PtME1。(c) 重组质粒酶切和PCR扩增;M,DNA marker DL 2000;1,重组质粒pET30a-PtME1双酶切;2,PCR产物(目的片断)。(a)The genome amplification products: M, DNA marker DL 2000; lane 1, PCR products. (b) Plasmid: M, DNA marker DL 2000; lane 1, plasmid pET30a; lane 2, recombinant plasmid pET30a-PME1. (c) Enzyme digestion of recombinant plasmid and PCR amplification: M, DNA marker DL 2000; lane 1,double digestion of recombinant plasmid pET30a-PtME1; lane2, PCR products.)
图1基因组ME1扩增(a),质粒图谱(b),重组
质粒pET30a-ME1酶切(c)
Fig.1PtME1 amplified fromP.tricornutum
genomic DNA (a), plasmid (b), and double digestion
of recombinant plasmid pET30a-PtME1 (c)
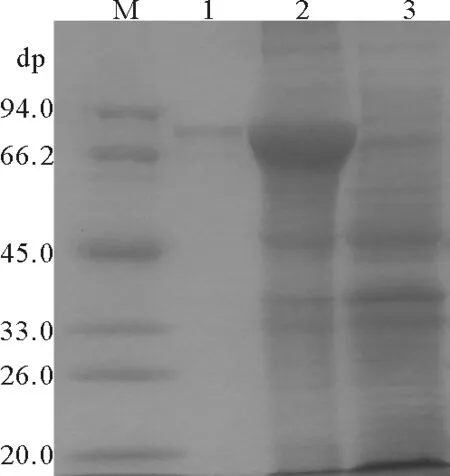
(M.蛋白质marker; 1. 纯化His-PtME1; 2. 含pET30a-PtME1重组子经0.5mmol/L IPTG诱导后的细胞破碎上清液; 3. 含 pET-30a对照经0.5mmol/L IPTG诱导后的细胞破碎上清液。M, Protein marker; lane 1, the purified His-PME1; lane 2, the supernatant of cell disruption of the recombinant inducing by 0.5mmol/LIPTG; lane 3, the supernatant of cell disruption of the control inducing by 0.5mmol/LIPTG.)
图2三角褐指藻ME1融合蛋白的诱导表达和纯化
Fig.2 Induced expression and purification of recombinant
protein fromP.tricornutumME1 gene
2.3 重组ME酶活力测定
大肠杆菌BL21(DE3)也能合成NADP-ME,重组NADP-ME1活性需要纯化后测定。粗酶液经Ni SephroseTM6 Fast Flow纯化系统纯化得到目的蛋白,用建立的苹果酸酶酶活性测定体系,测定340 nm下吸光度酶活力扫描图(见图3),表明重组酶具有很高活性,通过计算酶活力可达75.18 U/mg。
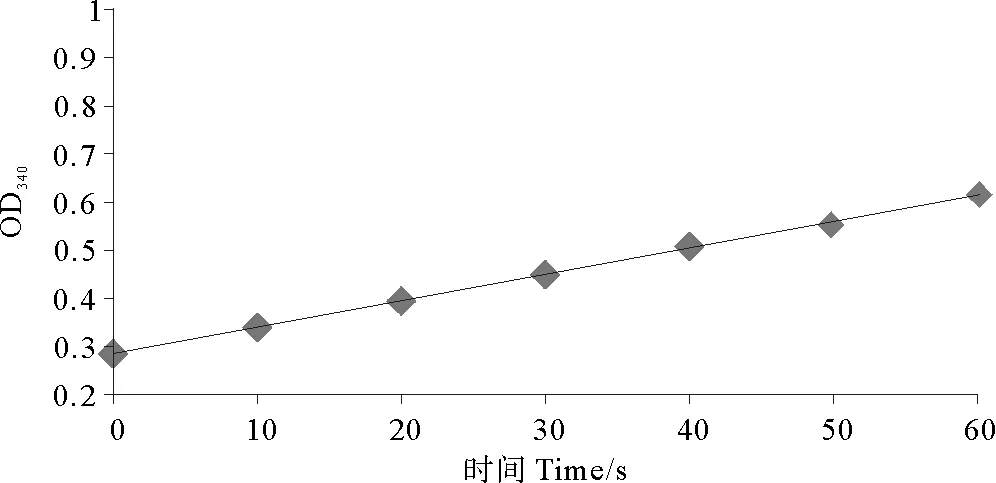
图3 苹果酸酶活力曲线
2.4 重组子脂肪酸组成和含量
以C15脂肪酸作为内标,对2种重组菌株脂肪酸产量进行定量分析。结果表明,重组菌株和对照菌株主要合成4种脂肪酸,即C14∶0、C16∶0、C16∶1和C18∶1(见图4)。重组菌株脂肪酸C14∶0、C16∶0、C18∶1及总脂肪酸含量较对照分别提高了34.8%、69.9%、54.2%和50.2%,胞内总脂产量达到122.68 mg/g, 重组菌株C16∶1产量是对照的5.6倍。结果表明PtME1表达不改变大肠杆菌脂肪酸种类,但能够显著提高脂肪酸含量。
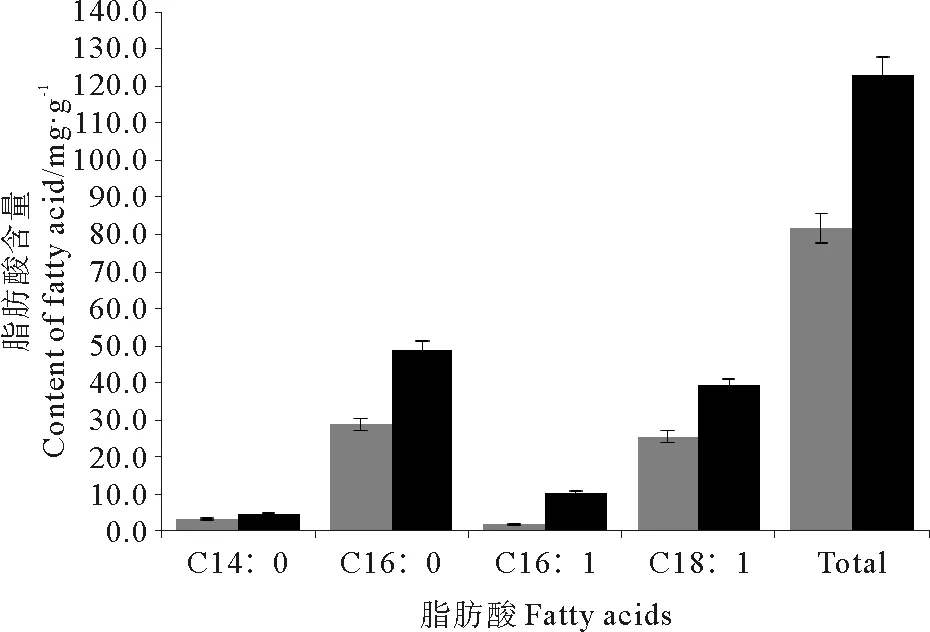
(黑色柱和灰色柱分别代表重组菌株和对照。 Black and grey bars represent recombinant and the control, respectively.)
图4PtME1表达对大肠杆菌脂肪酸组成和含量的影响
Fig.4Effects of expressingPtME1 on the
composition and content of fatty acids inE.coliBL21 (DE3)
3讨论
脂肪酸合成过程中碳链的延伸不仅需要连续供给乙酰CoA,还需要提供足够的NADPH[19]。在新生脂肪酸碳链延伸时每一个乙酰CoA需消耗2个NADPH用于还原反应。但脂肪酸合成及去饱合所需NADPH主要由NADP依赖型苹果酸酶提供。这一结论在酵母和真菌中得到广泛证实。如在构巢曲霉(Aspergillusnidulans)中敲除ME后,在低氮环境下不含ME菌株的油脂积累量只有野生菌株的一半[20]。在卷枝毛霉(Mucorcircinelloides)中过表达ME基因不仅使油脂的含量增加了2.5倍,同时不饱和脂肪酸的比例也有增加[21],而在卷支毛霉的培养基中加入苹果酸酶的抑制剂-芝麻酚后,该菌几乎失去油脂积累能力,从不加抑制剂的24%降低到1%~2%[22]。这些研究结果表明当苹果酸酶活性被抑制时,油脂积累量也大幅降低。而在富油微生物(霉菌、细菌、微藻)中对油脂代谢途径和转基因研究中也发现ME是提高油脂含量最有潜力的目标基因之一[23-24]。但针对三角褐指藻苹果酸酶的研究还没有报道。因此,探索三角褐指藻苹果酸酶功能对全面认识NADPH对脂肪酸积累作用至关重要。
本研究分离了一条三角褐指藻苹果酸酶基因的cDNA,并在大肠杆菌中进行了表达,通过镍柱纯化获得目的蛋白并验证了其底物特异性,证明该蛋白是NADP依赖型ME且具有较高的酶活力。GC-MS分析显示,大肠杆菌重组子胞内总脂含量提高了50.2%,产量达到122.68 mg/g, 这与文献中报道的结果一致[18-19],即过量表达苹果酸酶基因可提高宿主菌的总脂肪酸含量。本研究中,重组细菌饱和脂肪酸占总脂肪酸的百分含量由对照的36.3%提高到39.7%,不饱和脂肪酸的百分含量由对照的9.3%提高到15.5%(数据未显示),其中重组细菌C16∶1的含量显著提高,这表明过量表达的苹果酸酶确实为脂肪酸的合成以及碳链的去饱和提供了足够的NADPH。这些结果虽已证明ME与大肠杆菌油脂积累有关,但ME与油脂积累的直接关系尚不明确。一方面,脂肪酸积累所需的NADPH并非全部由苹果酸酶提供[21],另一方面,苹果酸酶催化苹果酸氧化脱羧反应的产物既有NADPH还有丙酮酸,过表达苹果酸酶基因提高大肠杆菌油脂含量,不仅与胞内NADPH含量的增加有关,可能也与丙酮酸的含量提高有关[25]。一般认为,脂肪酸合成的碳源主要来自糖酵解产生的丙酮酸氧化脱羧生成的乙酰辅酶A,因此,苹果酸酶催化生成的丙酮酸能否为脂肪酸的合成提供碳源,有多大的比例能用来合成脂肪酸都有待进一步实验研究。
对三角褐指藻苹果酸酶功能初步探索使我们清楚地认知了异源表达该酶可提高细菌总脂肪酸含量。但该蛋白的细胞定位以及在藻体内对脂肪酸积累的影响等需要更进一步研究。我们拟在三角褐指藻细胞中调控该基因表达,遗传修饰三角褐指藻,提高其脂肪酸合成效率。
参考文献:
[1]Chang G G, Tong L. Structure and function of malic enzymes, a new class of oxidative decarboxylases [J]. Biochemistry, 2003, 42(44): 12721-12733.
[2]Edwards G E, Andreo C S. NADP-malic enzyme from plants [J]. Phytochemistry, 1992, 31(6): 1845-1857.
[3]Drincovich M F, Casati P, Andreo C S. NADP-malic enzyme from plants: A ubiquitous enzyme involved in different metabolic pathways [J]. FEBS Letters, 2001, 490(1): 1-6.
[4]Casati P, Drincovich M F, Edwards G E, et al. Malate metabolism by NADP-malic enzyme in plant defense [J]. Photosynthesis Research, 1999, 61(2): 99-105.
[5]Smith R G, Gauthier D A, Dennis D T, et al. Malate-and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm [J]. Plant Physiology, 1992, 98(4): 1233-1238.
[6]Shearer H L, Turpin D H, Dennis D T. Characterization of NADP-dependent malic enzyme from developing castor oil seed endosperm [J]. Archives of Biochemistry and Biophysics, 2004, 429(2): 134-144.
[7]Wheeler M C G, Tronconi M A, Drincovich M F, et al. A comprehensive analysis of the NADP-malic enzyme gene family ofArabidopsis[J]. Plant Physiology, 2005, 139(1): 39-51.
[8]Song Y, Wynn J P, Li Y, et al. A pre-genetic study of the isoforms of malic enzyme associated with lipid accumulation inMucorcircinelloides[J]. Microbiology, 2001, 147(6): 1507-1515.
[9]Zhang Y, Adams I P, Ratledge C. Malic enzyme: The controlling activity for lipid production? Overexpression of malic enzyme inMucorcircinelloidesleads to a 2. 5-fold increase in lipid accumulation [J]. Microbiology, 2007, 153(7): 2013-2025.
[10]Wynn J P, bin Abdul Hamid A, Ratledge C. The role of malic enzyme in the regulation of lipid accumulation in filamentous fungi [J]. Microbiology, 1999, 145(8): 1911-1917.
[11]Santos M M, Raghevendran V, Kötter P, et al. Manipulation of malic enzyme inSaccharomycescerevisiaefor increasing NADPH production capacity aerobically in different cellular compartments [J]. Metabolic Engineering, 2004, 6(4): 352-363.
[12]Wheeler M C G, Tronconi M A, Drincovich M F, et al. A comprehensive analysis of the NADP-malic enzyme gene family ofArabidopsis[J]. Plant Physiology, 2005, 139(1): 39-51.
[13]Shang C H, Zhu S N, Yuan Z H, et al. Molecular cloning and characterization analysis of malic enzyme gene fromDunaliellaparva[C].//Advanced Materials Research, 2012, 347: 2536-2540.
[14]Bowler C, Allen A E, Badger J H, et al. ThePhaeodactylumgenome reveals the evolutionary history of diatom genomes [J]. Nature, 2008, 456(7219): 239-244.
[15]Tonon T, Harvey D, Larson T R, et al. Identification of a very long chain polyunsaturated fatty acid Δ4-desaturase from the microalgaPavlovalutheri[J]. FEBS Letters, 2003, 553(3): 440-444.
[16]Horst I, Parker B M, Dennis J S, et al. Treatment ofPhaeodactylumtricornutumcells with papain facilitates lipid extraction [J]. Journal of Biotechnology, 2012, 162(1): 40-49.
[17]Dillschneider R, Steinweg C, Rosello-Sastre R, et al. Biofuels from microalgae: Photoconversion efficiency during lipid accumulation [J]. Bioresource Technology, 2013, 142: 647-654.
[18]孟鑫, 尚宏丽, 郑益. 过量表达苹果酸酶对E.coli脂肪酸合成能力的影响 [J]. 食品工业科技, 2013, 34(12): 207-209.
Meng X, Shang H L, Zheng Y. Effect of overexpression of malic enzyme on fatty acid production ofEscherichiacoli[J]. Science and Technology of Food Industry, 2013, 34(12): 207-209.
[19]Meng X, Yang J, Cao Y, et al. Increasing fatty acid production inE.coliby simulating the lipid accumulation of oleaginous microorganisms [J]. Journal of Industrial Microbiology & Biotechnology, 2011, 38(8): 919-925.
[20]Wynn J P, Ratledge C. Malic enzyme is a major source of NADPH for lipid accumulation byAspergillusnidulans[J]. Microbiology, 1997, 143(1): 253-257.
[21]Ratledge C. The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: A reappraisal and unsolved problems [J]. Biotechnology Letters, 2014, 36(8): 1557-1568.
[22]Wynn J P, Kendrick A, Ratledge C. Sesamol as an inhibitor of growth and lipid metabolism inMucorcircinelloidesvia its action on malic enzyme [J]. Lipids, 1997, 32(6): 605-610.
[23]程万, 林辉, 赵宇华, 等. 苹果酸酶调控微生物油脂积累的研究进展 [J]. 科技通报, 2010, 26(6): 853-857.
Cheng W, Lin H, Zhao Y H, et al. Research Progress on malic enzyme regulating the accumulation of microbial oils [J]. Bulletin of Science and Technology, 2010, 26(6): 853-857.
[24]Radakovits R, Jinkerson R E, Darzins A, et al. Genetic engineering of algae for enhanced biofuel production [J]. Eukaryotic Cell, 2010, 9(4): 486-501.
[25]Sauer U, Eikmanns B J. The PEP—pyruvate—oxaloacetate node as the switch point for carbon flux distribution in bacteria: We dedicate this paper to Rudolf K. Thauer, Director of the Max-Planck-Institute for Terrestrial Microbiology in Marburg, Germany, on the occasion of his 65th birthday [J]. FEMS Microbiology Reviews, 2005, 29(4): 765-794.
责任编辑高蓓
Overexpression of Malic Enzyme Gene fromPhaeodactylumtricornutumPromotes Fatty Acids Production inEscherichiacoli
LV Na-Na1, ZHU Bao-Hua1, LU Lin1, YANG Guan-Pin2, PAN Ke-Hou1, 3
(1.Lab of Applied Microalgae Biology, The Key Laboratory of Aquaculture, Ministry of Education, Ocean University of China, Qingdao 266003, China; 2. College of Marine Life Sciences, Ocean Univesity of China, Qingdao 266003, China; 3.Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266100, China)
Abstract:The marine diatom Phaeodactylum tricornutum has been shown to be a potential producer of biodiesel due to its fast growth, lipid accumulation capability and established genetic tools. Thus, it is possible to genetically manipulate the key genes involved in fatty acids synthesis in microalgae to improve traits to achieve both high lipid and high biomass for industrial production. Malic enzyme (ME) catalyzes the oxidative decarboxylation of L-malate to yield pyruvate, CO2 and NADPH in the presence of a divalent metal ion. It is a widely distributed enzyme involved in different metabolic pathways in prokaryotic and eukaryotic microorganisms. To date, there have been a few studies that have focused on the role of MEs in lipid accumulation, mainly in plants and mammals; however, little is known about the role of these enzymes in microalgae. The full-length cDNA of malic enzyme gene was isolated from P. tricornutum and named as PtME1. It is 1 917 bp in length, encoding 437 amino acids with a molecular mass of 72 kD. In order to verify its function, the recombinant plasmid pET30a-PtME1 was built by inserting PtME1 into pET-30a. Upon IPTG induction, soluble recombinant protein was obtained with high efficiency in E.coli BL21 (DE3) harboring pET30a-PtME1. Recombinant protein was purified by Ni SephroseTM6 Fast purification Flow system and showed a single band about 72 kDa on SDS-PAGE gel. The specific activity of purified enzyme protein was measured, which reached 75.18 U per milligram protein. GC-MS analysis revealed that increased expression of the ME gene leads to increased biosynthesis of fatty acids in the recombinant strain, the contents of C14∶0, C16∶0, C18∶1 and total fatty acids were increased by 34.8%, 69.9%, 54.2% and 50.2%, respectively. The content of C16∶1 was increased by 5.6 fold compared with that of the control. Research results indicated that over-expression of PtME1 in E.coli has improved the capacity of fatty acid synthesis in Escherichia coli. The NADP-ME in lipid biosynthesis is to supply enough NADPH for both biosynthesis and desaturation of fatty acids in E.coli. These results also laid foundation for further research of the malic enzyme in P. tricornutum.
Key words:P. tricornutum; malic enzyme gene; fatty acid; prokaryotic expression
基金项目:❋ 国家重点基础研究发展计划项目(2011CB200901);国家科技支撑计划项目(2011BAD14B01)资助
收稿日期:2015-09-16;
修订日期:2015-11-10
作者简介:吕娜娜(1988-),女,硕士生。E-mail: lvnanabest@163.com ❋❋通讯作者: E-mail: zhubaohua@ouc.edu.cn
中图法分类号:Q341
文献标志码:A
文章编号:1672-5174(2016)05-065-05
DOI:10.16441/j.cnki.hdxb.20150322
Supported by the Major State Basic Research Development Program of China (2011CB200901); the National Technical Supporting Project Foundation (2011BAD14B01)
