Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer
2016-03-31YiwenFangMelissaFullwood
Yiwen Fang, Melissa J. Fullwood
Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer
Yiwen Fang1,a, Melissa J. Fullwood1,2,3,4,*,b
1Cancer Science Institute of Singapore, National University of Singapore, Singapore 117599, Singapore
2School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore
3Institute of Molecular and Cell Biology, Agency for Science, Technology and Research (A*STAR), Singapore 138673, Singapore
4Yale-NUS Liberal Arts College, Singapore 138527, Singapore
Received 10 July 2015; revised 31 August 2015; accepted 17 September 2015
Available online 12 February 2016
Handled by Xiangdong Fang
E-mail: melissa.fullwood@nus.edu.sg (Fullwood MJ).aORCID: 0000-0002-2611-8424.bORCID: 0000-0003-0321-7865.
Peer review under responsibility of Beijing Institute of Genomics,
Chinese Academy of Sciences and Genetics Society of China.
http://dx.doi.org/10.1016/j.gpb.2015.09.006
1672-0229ⓒ2016 The Authors. Production and hosting by Elsevier B.V. on behalf of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
KEYWORDS
lncRNAs;
Chromatin;
Transcription regulation;
Antisense oligonucleotides;
Cancer
Abstract Long non-coding RNAs (lncRNAs) play important roles in cancer. They are involved in chromatin remodeling, as well as transcriptional and post-transcriptional regulation, through a variety of chromatin-based mechanisms and via cross-talk with other RNA species. lncRNAs can function as decoys, scaffolds, and enhancer RNAs. This review summarizes the characteristics of lncRNAs, including their roles, functions, and working mechanisms, describes methods for identifying and annotating lncRNAs, and discusses future opportunities for lncRNA-based therapies using antisense oligonucleotides.
Introduction
Recent advances in sequencing technologies enabling more indepth genomic and transcriptomic analyses have revealed that as much as 85% of the human genome is transcribed [1–3]. This was surprising, as studies of mammalian genomes have shown that a drastically low population of RNA transcripts code for protein products. Notably, from the recent Encyclopedia of DNA Elements (ENCODE) work, out of 41,204 called genes, only 56 genes (0.1%) showed mass spectrometric evidence consistent with protein expression, suggesting that the majority of RNA transcripts are non-coding [4]. The generation of such a large population of non-coding RNA (ncRNA) transcripts indicates that RNAs have a larger and more diverse role in biological processes than initially anticipated. ncRNAs can be roughly classified into two groups based on their size. One group includes short RNAs less than 200 nucleotides (nt) in length, such as microRNAs (miRNAs) that are small RNA (sRNA) molecules around 21–24 nt in length, as well as other classes such as piwi-interacting RNAs (piRNAs) [5]. The other group includes long ncNAs (lncRNAs) of around 200 nt or more [6]. While miRNAs have been heavily studied and are well understood for their function in gene regulation [7], lncRNAs in contrast are less understood.
Many lncRNAs have been functionally associated with human diseases, in particular, cancers [8] (Table 1). Dysregulation of lncRNAs has been implicated in glioblastoma [9–11],breast cancer [12], colorectal cancer [9,13], liver cancer [14,15], and leukemia [9,16]. Commonly, dysregulation of lncRNAs exerts impacts on cellular functions such as cell proliferation, resistance to apoptosis, induction of angiogenesis, promotion of metastasis, and evasion of tumor suppressors [8,17].

Table 1 Examples of lncRNAs in cancer
An emerging view of lncRNAs is that they are fundamental regulators of transcription. This view has led to an intense focus on elucidating the molecular mechanisms that underlie their function [18]. Many lncRNAs have been characterized and several models of action have been proposed, such as functioning as signal, decoy, scaffold, guide, enhancer RNAs, and short peptides [19,20], as mentioned below. The main function of a signal lncRNA is to serve as a molecular signal to regulate transcription in response to various stimuli. Thus its production and presence can serve as an indicator of transcriptional activity [21]. Decoy lncRNAs limit the availability of regulatory factors by presenting‘decoy”binding sites. These lncRNAs modulate transcription by sequestering regulatory factors including transcription factors, catalytic proteins, subunits of larger chromatin modifying complexes, as well as miRNAs, thereby reducing their availability [22]. Transcripts from the scaffold class of lncRNAs play a structural role by providing platforms for assembly of multiple-component complexes, such as ribonucleoprotein (RNP) complexes [23]. In the case of RNP complexes, once the complexes have been fully assembled, either transcriptional activation or repression could be conferred depending on the nature of proteins and RNAs present [24,25]. Guide lncRNAs interact with RNPs and direct them to specific target genes. These guide lncRNAs are essential for the proper localization of RNPs [26]. Next, enhancer RNAs (eRNAs) are produced from enhancer regions and may work by influencing the 3-dimensional (3D) organization of DNA, also known as‘chromatin interactions”. One hypothesized model of action is that these lncRNAs may possibly work as‘tethers”as they may not be released from the enhancer regions when functioning, thus tethering the interacting proteins to enhancer regions [19]. In addition, lncRNAs can encode short peptides, which may also have functions [27]. It is likely that additional mechanisms will be discovered in the future.
This review introduces lncRNAs, focusing on lncRNAs with cancer-associated roles, and discusses proposed mechanisms by which these lncRNAs function in chromatin remodeling, chromatin interactions, and as competing endogenous RNAs (ceRNAs). In addition, we highlight approaches for the identification and annotation of lncRNAs, cross-talk between these mechanisms, and methods for perturbing specific lncRNAs, which could eventually provide lncRNA-based therapies for diseases. We then conclude by highlighting challenges and future research topics.
Characteristics of lncRNAs
lncRNAs are defined as RNA molecules with more than 200 nucleotides. This distinction, while somewhat arbitrary and based on technical aspects of RNA isolation methods, serves to distinguish lncRNAs from miRNAs and other sRNAs. lncRNAs are present in large numbers in genome [28,29]. They typically do not possess functional open reading frames (ORFs). However, this distinction is blurred by the discovery of bifunctional RNAs that can have both protein-coding and coding-independent functions [30,31], raising the possibility that many protein-coding genes may also have non-coding functions. Many lncRNAs are lowly expressed [32], posing a challenge in terms of exploration of lncRNAs and explaining why lncRNAs had been thought to be only‘transcriptional noise”until recently. RNA-seq studies in different tetrapods show that most (81%) lncRNAs are poorly conserved in DNA sequence and are primate-specific. However, it should be noted that several lncRNAs are ultra-conserved in DNA sequence—3% of lncRNAs appear to have originated more than 300 million years ago and can be found from organisms ranging from Xenopus and chicken to man [33]. It is possible that lncRNAs might be fast-evolving RNA species that can play key roles in specifying lineages. In support of this idea, a comparison of matched tissues in Mus musculus domesticus, Mus musculus castaneus, and Rattus norvegicus shows that the emergence or extinction of intergenic lncRNAs is associated with changes in transcription levels of proximal protein-coding genes [34]. In addition, there are examples of lncRNAs exhibiting conserved biological function but low sequence conservation, such as megamind/TUNA, which is associated with brain development in zebrafish, mouse, and human [35,36], as well as X-inactive specific transcript (Xist), which is involved in X-inactivation [37]. It is possible that RNA molecules need less sequence conservation to retain their function compared to proteins. Conversely, there is high sequence conservation of lncRNA promoters, which is even higher than that of protein-coding gene promoters [29], suggesting that regulation of lncRNA expression is important.
Many lncRNAs possess features reminiscent of proteincoding genes, such as 5'cap and alternative splicing [32].In fact, many lncRNA genes have two or more exons [32] and about 60% of lncRNAs have polyA+ tails [38] (Figure 1). Additionally, while there are long intergenic RNAs (lincRNAs) [39] including eRNAs from gene-distal enhancers [40], the majority of lncRNA genes are located within 10 kb of protein-coding genes [41] and many lncRNAs are antisense to coding genes or intronic [42].
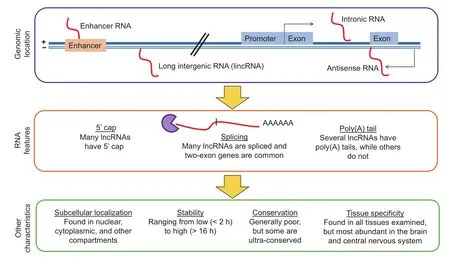
Figure 1 Characteristics of lncRNAs
In line with a wide range of functions that lncRNAs are involved in, lncRNAs can be found in many tissues, although the brain and central nervous system appear to have the highest diversity of expressed lncRNAs [43]. Also, lncRNAs can be found in different cellular compartments including both the cytoplasm and nucleus to which they seem to predominantly localize [32]. lncRNAs have been generally thought to be unstable due to their low expression levels and the existence of known classes of unstable transcripts such as the promoter upstream transcripts (PROMPTs) [44]. Interestingly, a recent study indicates that only a minority (240 out of 823 lncRNAs; 29%) of lncRNAs are unstable with half-lives less than 2 h, while 51 (6%) were extremely stable with half-lives of over 12 h [45].
Acting mechanisms of lncRNAs
In cancer, lncRNAs work through multiple mechanisms such as chromatin remodeling, chromatin interactions, ceRNAs, and natural antisense transcripts (NATs) (Figure 2). lncRNAs can interface with chromatin remodeling machinery in several ways, including acting as signal lncRNAs or scaffold lncRNAs. Recently, another mechanism of action by which lncRNAs function to regulate transcription has been posited. This involves the production of lncRNAs from enhancer regions in the genome (eRNAs), which function to stabilize as well as maintain chromatin loops [40,46–48]. Chromatin looping enables distally-located enhancers (with some located several hundred kilobases away) to interact with their target gene promoters [49–51]. In addition, several studies aimed at elucidating the biological functions of miRNAs have previously used artificial miRNA sponges that compete with native targets for interaction with the miRNAs of interest [52–54]. Similarly, studies have also demonstrated that some lncRNAs modulate transcription by sequestering regulatory factors, including transcription factors and catalytic proteins or subunits of larger chromatin-modification complexes, as well as miRNAs [22]. They are known as ceRNAs [55–57]. Transcribed from the opposite DNA strand in relative to the sense transcripts, NATs overlap with their sense transcripts [58] and are thought to regulate the expression of the sense transcripts as well. We will discuss examples of these mechanisms in the next sections. Nonetheless, it should be noted that the mechanisms described here are not exhaustive, and future work is likely to provide new insights in this fast-moving field.
lncRNAs in chromatin remodeling
lncRNAs can act through chromatin remodeling to achieve transcriptional regulation. One example is the potassium voltage-gated channel subfamily KT member 1 opposite strand/antisense transcript 1 (KCNQ1OT1), which is upregulated in colon cancer [59]. KCNQ1OT1 acts as a signal lncRNA by recruiting G9a histone methyltransferases and polycomb repressive complex 2 (PRC2) [21], which mediate the gene-silencing-associated marks, i.e., dimethylation of lysine 9 (H3K9me2) and lysine 27 on histone 3 (H3K27me3) [60]. Through chromatin remodeling, KCNQ1OT1 induces transcriptional silencing of genes, which may occur in cis (for targets close by) or in trans (for distal targets).
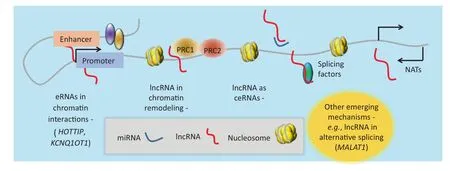
Figure 2 Acting mechanisms of lncRNAseRNA, enhancer RNA; ceRNA, competing endogenous RNA; NAT, natural antisense transcript; PRC1, polycomb repressive complex 1.
As another example, ANRIL/CDKN2B, the antisense ncRNA in the INK4 locus, works as a scaffold in mediating transcriptional silencing of the INK4b-ARF-INK4a locus by recruiting and interacting with PRC1 and PRC2 [24,25]. PRC2 recruited to this gene locus then mediates the spread of the methylation marks that are distinct for transcriptionally-silent genes. Several lncRNAs have been demonstrated to interact with chromosome-modification complexes and direct them to specific target genes. These guide lncRNAs are essential for the proper localization of the chromosome-modification complexes including PRC2 and subsequent repression of gene expression as demonstrated by the lncRNA fetal-lethal non-coding developmental regulatory RNA (FENDRR), which serves to bring PRC2 in close proximity to the promoters of genes associated with the formation and differentiation of the lateral mesoderm lineage, such as forkhead box F1 (FOXF1) and paired-like homeodomain2 (PITX2) genes [26]. We hypothesize that other guide lncRNAs may also function to regulate transcription by targeting other chromatin-modification complexes, in addition to PRC2, to their target genes.
Finally, the HOX transcript antisense RNA (HOTAIR) lncRNA functions cooperatively with PRC2 in mediating the repression of the homeobox D cluster (HOXD) locus through spreading H3K27me3 marks, which are associated with gene silencing [61]. HOTAIR forms multiple double stem-loop structures that bind to lysine-specific demethylase 1 (LSD1) and PRC2 histone-modification complexes [12]. Other lncRNAs also operate in a similar manner, with as much as 20% of lncRNAs known to associate with PRC2 [62]. For example, the lncRNA TUG1 is associated with PRC2, and depletion of TUG1 expression in the developing mouse eye leads to the blockage of retinal development [62]. While PRC2 has been found to interact with many lncRNAs, other chromatin remodelers have been implicated and it is likely that more interactions with other chromatin remodelers remain to be discovered. Moreover, given that cancers are associated with aberrant levels of PRC2, H3K27me3, and mutated enhancer of zeste homolog 2 (EZH2), which is a component of the PRC2 complex [63], we hypothesize that lncRNA-based mechanisms provide an explanation as to how these alterations in PRC2 levels can give rise to cancer (Table 1). Taken together, it is becoming increasingly clear that certain lncRNAs can associate with chromatin-modification complexes to carry out cellular functions.
lncRNAs in chromatin looping in cancers
Another mechanism of action by which lncRNAs function to regulate transcription is via enhancer lncRNAs (eRNAs) associated with chromatin loops. To study chromatin looping, techniques including chromosome conformation capture (3C) [64] and fluorescence in situ hybridization (FISH) [65], have been employed. More recent technical developments have focused on higher-throughput analyses and these include circular chromosome conformation capture (4C) [66,67], chromosome conformation capture carbon copy (5C) [68], combined 3C-ChIP-cloning (6C) [69], chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) [70], and Hi-C [71]. Using ChIA-PET, a correlation is revealed between expression level of elncRNAs and estrogen receptor α(ERα)-associated chromatin interactions [72]. The experimental downregulation of these eRNAs leads to a loss of chromatin loops and a corresponding change in expression of the genes targeted by ERα[46].
In addition, a relationship is found between the levels of eRNAs produced by upstream enhancers of the prostatespecific antigen (PSA) gene and the actual levels of PSA gene expression, suggesting a possible link between eRNAs and chromatin interactions [73]. Melo et al. show the existence of enhancer regions that bind the transcription factor p53. These enhancer regions produce RNAs and display chromatin interactions with multiple neighboring genes. Ablation of these eRNAs leads to reduced transcription at neighboring genes and reduced p53-dependent cell cycle arrest [74]. More recently, a role for Integrator in the biogenesis of eRNAs is demonstrated [75]. Integrator is a complex associated with RNA polymerase II (RNAPII), and possesses RNA endonuclease activity, which is required for 3'end processing of non-polyadenylated nuclear RNA genes [76]. These studies show that depletion of Integrator leads to a decrease in the induction of eRNAs, which is accompanied by the loss of enhancer-promoter chromatin looping [75].
One of the best examples of lncRNAs that regulate chromatin interactions is the HOXA transcript at the distal tip (HOTTIP). HOTTIP is a lncRNA shown to regulate chromatin interactions in the HOX cluster, from which it is produced. HOTTIP is necessary to coordinate activation of HOX genes, through binding WD repeat-containing protein 5 (WDR5), an adaptor protein [77]. WDR5 interacts withthe mixed lineage leukemia (MLL) complexes for substrate recognition and genomic targeting. The MLL complexes catalyze H3K4 methylation, which is a mark of actively transcribed genes [77]. HOTTIP is necessary for the maintenance of a specific pattern of WDR5/MLL complexes across the HOXA locus to facilitate gene transcription [77].
Additionally, there is cross-talk between the different mechanisms: a novel RNA chromosome conformation capture (R3C) strategy demonstrated that KCNQ1OT1, which regulates genes in the KCNQ1 imprinting region, is involved in imprinting-associated chromatin interactions [78]. EZH2, which catalyzes the deposition of gene silencing-associated H3K27me3 marks, is recruited by the KCNQ1OT1 [78]. Depletion of KCNQ1OT1 leads to depletion of the loop and loss of imprinting [78]. These examples all demonstrate a role for eRNA-associated chromatin interactions in transcriptional suppression, enhancement, and coordination of gene expression, as well as other functions such as imprinting. A hypothesized model of action is that eRNAs may help to tether different genomic regions together, recruiting factors such as PRC2 and other proteins at the 5'end, while remaining tethered to the location of production [79] (Figure 3). Other factors might help to bring together other DNA regions, or the length of the eRNA might enable it to tether and guide other DNA regions together, thus forming a chromatin interaction. Other factors may bind to the chromatin directly or indirectly through the bound proteins, stabilizing and maintaining the chromatin even after the RNA may have been degraded. For example, cohesin has been shown to play such a role in the case of ERα-associated RNAs [46]. Adding support to this idea is the finding that transcriptional repressor CCCTC-binding factor (CTCF), which is involved in chromatin interactions, can bind to ncRNAs. For example, the steroid receptor RNA activator (SRA) binds to CTCF and enhances its functioning [80].
lncRNAs as ceRNAs in cancers
The lncRNA HULC is highly upregulated in hepatocellular carcinoma (HCC) [81]. There are multiple miR-372-binding sites present in HULC, and the overexpression of HULC can reduce miR-372 expression. This leads to reduced translational repression of its target transcript PRKACB, thereby inducing the phosphorylation of the cAMP-responsive element (CRE)-binding protein (CREB) [81]. On the other hand, papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) is highly downregulated in thyroid cancers and its overexpression leads to reduced expression of oncogenic miR-574-5p, resulting in growth inhibition, cell-cycle arrest, and increased apoptosis [82]. Studies identifying additional molecular targets of decoy lncRNAs will add to our understanding of diseases while presenting possible therapeutic interventions for which drugs may be designed.
lncRNAs as NATs
NATs are surprisingly common in the mammalian genome—over 20% of human transcripts form sense–antisense pairs [83]. A later estimate in mice based on large RNA-seq datasets shows that up to 72% of genes may have transcription-related activity on the opposite strand [58]. NATs can be produced against both protein-coding and non-coding genes [58] and some NATs are protein-coding genes while others are lncRNAs [58]. In addition, the genomic organization of NAT-sin relative to their sense transcripts varies with different configurations. One common configuration, called‘divergent”, is that the promoter generates bidirectional transcription [58,84]. Another common configuration, called‘convergent”, arises whereby the NAT starts from a different promoter, and transcribes a region on the opposite strand from the protein-coding transcript [58]. For example, the NAT could start from the 3'end of the sense transcript and transcribe toward its 5'end [58].
NATs exert varied influence on their sense transcripts, either suppression or activation. The expression levels of sense/antisense pairs are generally concordant, but reciprocal expression is also observed [58]. NATs can work through a variety of mechanisms. Other than employing mechanisms similar to other lncRNAs such as by scaffolding proteins, NATs that overlap with sense transcripts can work through a particular mechanism, that is, transcriptional collision. RNAPII complexes transcribing on opposite DNA strands cannot bypass each other [85]. Head-to-head collision results in stalling of RNAPII and subsequent removal of collided RNAPII by ubiquitin-directed proteolysis [85]. Therefore, convergent sense and antisense transcripts could lead to sense strand suppression.

Figure 3 Hypothesized acting mechanism for eRNAs to initiate chromatin interactionseRNA, enhancer RNA; CTCF, CCCTC-binding factor.
One example of a NAT in cancer is WD40-encoding RNA antisense to p53 (Wrap53) [86]. As a NAT of the important oncogene TP53, Wrap53 can induce TP53 expression by targeting the 5'untranslated region of the TP53 mRNA [86]. Blocking this interaction between the Wrap53 lncRNA and TP53 mRNA reduces basal levels of TP53 and prevents induction of TP53 after DNA damage [86]. Wrap53 is overexpressed
in cancer cell lines; interestingly, overexpression of Wrap53 results in cellular transformation while ablation leads to apoptosis [87]. Additionally, Wrap53 overexpression is correlated with poor prognosis in head and neck squamous cell carcinoma [87]. Taken together, Wrap53 is an oncogenic lncRNA that regulates TP53 expression.
lncRNA networks and cross-talk in cancer
An interesting aspect of lncRNAs is that there can be crosstalk between different mechanisms, leading to the formation of complex networks, particularly in cancer. For example, the transcription factor p53 regulates many genes [88], including lncRNAs such as lncRNA-p21, PANDA, H19, and loc285184, which serve as effectors of p53 by leading to p53-associated cellular functions such as cell cycle arrest and apoptosis [89]. p53 is self-regulated not only by well-known regulators such as MDM2, an E3 ubiquitin ligase, but also by lncRNAs MALAT1, and MEG3, as well as the TP53NATWrap53 as discussed in the previous section [89]. MALAT1 is upregulated in several cancers [90] and may be used to predict survival and metastasis in non-small cell lung cancer [91]. And MALAT1 is expressed in a cell cycle-dependent manner and is required for G1/S and mitotic progression [92]. Paradoxically, depletion of MALAT1 leads to TP53 activation [92]. However, the cells show reduced oncogenic transcription factor B-MYB, leading to increased cellular proliferation via B-MYB [92]. For more details on these lncRNAs, readers are referred to the excellent review by Zhang and colleagues [89]. Interestingly, c-Myc, another important cancer-associated transcription factor, has a similar network [93]. It remains an open question whether other cancer-associated transcription factors possess a similar network consisting of lncRNAs that lead to altered cellular functions and ultimately cancer. Given that transcription factors are generally considered difficult to target by small molecule inhibitors due to the intrinsically disordered nature of their binding sites (meaning they lack stable secondary and/or tertiary structure under physiological conditions in vitro) and their binding promiscuity [94], lncRNAs that regulate the transcription factors may open up new avenues for targeting cancers with aberrant transcription factor signaling.
Approaches for the identification and annotation of lncRNAs
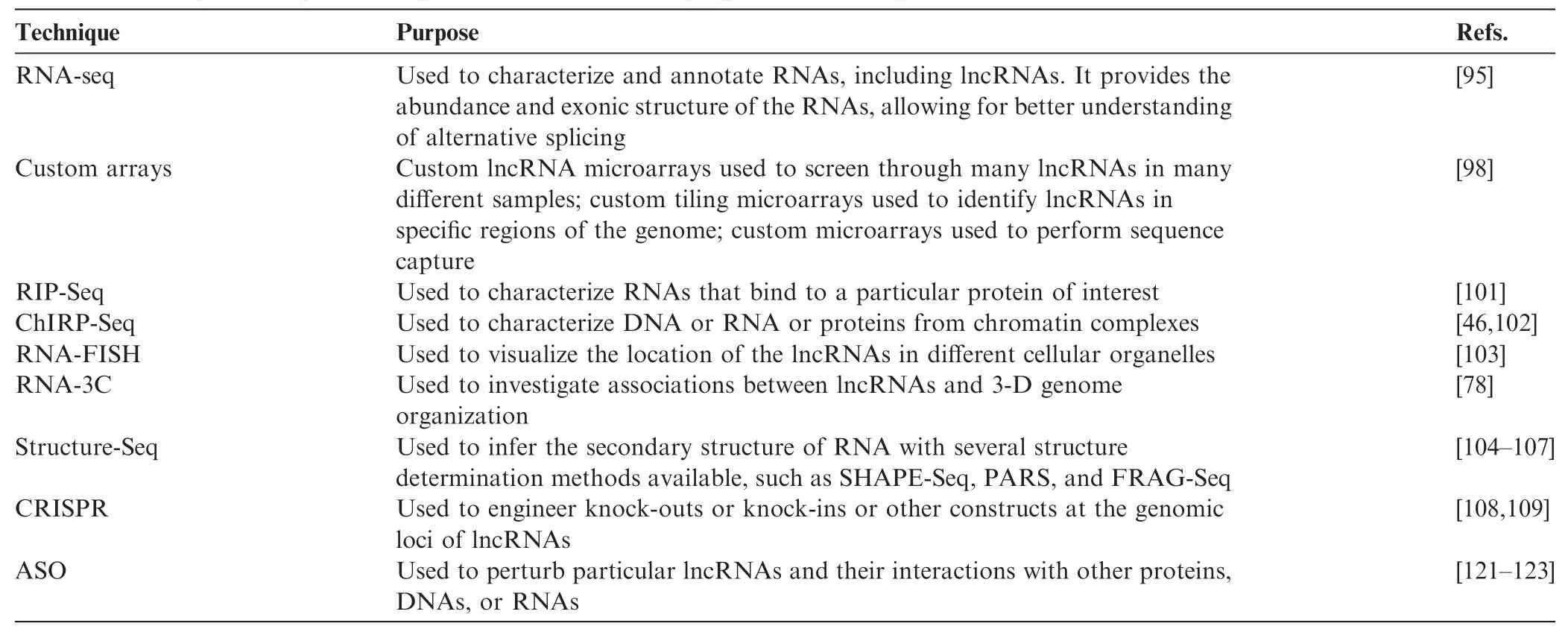
Table 2 Examples of key technologies and tools for identifying and annotating lncRNAs
In general, the expression levels of lncRNAs are low in abundance and lncRNAs are often not polyA-tailed, posing several challenges for their identification and analysis because previous methods of microarray and mRNA-seq rely on polyA+ isolation and tend to favor the detection of transcripts that are abundant in expression. In spite of these challenges, a number of effective methods have been developed for lncRNA identification and annotation (Table 2). Today, RNA-seq is one of the most common methods for identifying novel lncRNAs, because of the dropping sequencing costs and the single-nucleotide resolution nature of RNA-seq [95]. RNA-seq involves converting RNA into cDNA, followed by fragmentation, and ultra-high-throughput sequencing by methods such as Illumina HiSeq. As lncRNAs have low expression levels, high sequencing depths of 100–150 million reads or more are needed to discover these rare lncRNAs [96]. In addition, strand-specific sequencing methods are also needed to distinguish antisense lncRNAs from sense transcripts and cDNA conversion must be performed using random hexamers as opposed to oligo-dT methods [96]. RNA-seq experiments to detect lncRNAs have to be performed through rRNA-depletion to enrich for mRNAs and lncRNAs or by sequencing both polyA+ and polyA-fractions [96]. Owing to the high sequencing depth, RNA-seq becomes expensive when investigating some of the rarest lncRNAs. Hence researchers have also turned to customlncRNA microarrays to screen through many lncRNAs in many different samples [97], as well as custom tiling microarrays to identify lncRNAs in specific regions of the genome [61]. Other than that, microarray capture followed by sequencing is also used for quantitative gene profiling and annotation of specific lncRNAs [98].
Several new sequencing methods are becoming available, such as Pacific Biosciences (PacBio) sequencing, which features extremely long (on the order of kilobases) reads [99,100]. Pac-Bio sequencing allows for read-through of complete gene sequences, facilitating the understanding of alternative splicing. While this method is currently only cost-effective for small genomes such as bacteria, in the near-future, it will probably be applicable to analysis of human samples.
There is a battery of experimental methods for annotating lncRNAs. RNA immunoprecipitation (RIP)-Seq involves sequencing RNAs that are associated with a particular RNA-binding protein of interest with immunoprecipitation [101]. A related method, chromatin isolation by RNA purification (ChIRP)-Seq involves the design of multiple biotin-tagged oligonucleotide probes that recognize a particular RNA [46,102]. Cross-linked chromatin complexes with RNA of interest are then isolated using streptavidin magnetic beads recognizing the biotin tag [46,102]. DNA, RNA, and protein can be isolated from these complexes and subjected to sequencing or mass spectrometry for identification [46,102]. Another method, RNA-FISH, involves designing fluorescent probes to RNA and performing the hybridization in cells followed by microscopic imaging. It is used to visualize the location of the lncRNAs in different cellular organelles and investigate how lncRNA localization is altered in response to different stimuli [103]. In addition, RNA-3C involves double-stranded cDNA synthesis using biotinylated oligonucleotides to obtain chromatin complexes with biotinylated cDNA, followed by digestion and proximity ligation, generating DNA-cDNA constructs. These constructs can then be pulled down using the biotin group and analyzed by PCR. RNA-3C has been used to investigate associations between lncRNAs and 3-D genome organization [78].
In addition, there are a variety of sequencing methods for analyzing the secondary structure of RNA. These include selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq), whereby RNA is isolated, barcoded, and folded in vitro [104]. A SHAPE reagent, 1M7, is added to the isolated, barcoded, and folded RNA, which blocks the reverse transcriptase reaction whereby the 1M7 is included in the RNA, and hence leads to a series of truncated products to be sequenced, allowing for reconstruction of the original structure [104]. A similar method, parallel analysis of RNA structure (PARS) involves digestion of RNAs using RNAses specific for double-stranded and single-stranded RNAs. The fragments are then reversely transcribed and compared with each other, allowing for deduction of RNA structures in vivo [105,106]. A third method, fragmentation sequencing (FRAG-Seq), is similar to PARS except that it uses P1 nuclease instead of RNAses [107].
Perturbing lncRNA expression levels can be achieved in different ways, including clustered regularly interspaced short palindromic repeat (CRISPR) genomic editing of lncRNA genes [108,109]. In addition, new antisense oligonucleotide (ASO) approaches can be applied to perturb both cytoplasmic and nuclear lncRNAs, which will be discussed in more details in the next section. CRISPR genomic editing comes from the immune defenses of bacteria and archaea, which use short RNA to degrade invading nucleic acids [110,111]. Being one component of the CRISPR immune defense system, Cas9 effector nuclease is the first known nuclease that is capable of binding to specific short RNA and thereby directing cleavage at complementary genomic loci [110]. CRISPR/Cas9 represents a giant leap forward in terms of ease of use and efficiency in comparison with traditional methods for excising DNA such as homologous recombination and zinc finger nucleases (ZFNs) [110]. This method has since been widely adopted by many labs around the world, in a wide range of different cell types and organisms [110]. CRISPR editing can also be used to excise lncRNAs for functional studies. In addition, CRISPR can be modified for other purposes such as upregulating gene expression by combining a Cas9 unable to cleave nucleic acids with transcription activators such as VP64 activator domains [110,112,113]. This system could be used to upregulate lncRNA expression levels to understand their functions.
There are several databases available for lncRNA research (Table 3). ENCODE [28], FANTOM [114], and TCGA [115] have all embarked on massive RNA-seq efforts in different tissues including patient samples, which have yielded an unprecedented collection of lncRNAs. In addition, several groups have developed curated lists of lncRNAs by mining the existing literature and by predictions. For example, lncRNome [116] can serve as a general resource, while LncRNA Disease [117] presents lncRNA–disease associations. Other groups have also developed bioinformatics tools to infer functions of lncRNAs. These include looking for co-expressed genes and lncRNAs as featured in the lncRNAtor database [118], looking at functional similarity patterns (motifs) and predicting RNA structures as featured in LNCipedia and lncRNome [116,119], and integrating epigenomic information as featured in lncRNome [116]. For a detailed discussion and comparison of lncRNA databases, we refer readers to the article by Fritah and colleagues [120].
ASOs for modulation of lncRNA
The importance of lncRNAs in health and diseases (Table 1) indicates a need to find methods for modulating lncRNAs. To do this, ASOs are used (Figure 4), which are short DNA sequences and complementary to an RNA of interest. The oligonucleotide works by hybridizing to the RNA, which then blocks the action of the RNA. While unmodified oligonucleotides are available, certain chemical modifications such as 2'-O-(2-methoxy) ethyl oligonucleotides increase the lifespan of the oligonucleotide in the complex milieu of nucleases within the cell, and reduce degradation products, which may also have effects on the cells [121]. Two ASOs have been approved as drugs by the Food and Drug Administration (FDA) of the United States: fomivirsen, which is used to treat cytomegalovirus retinitis [122], and mipomersen, which is used to treat homozygous familial hypercholesterolemia [123]. ASOs can enter the nucleus and knock down nuclear lncRNAs [124], enabling all classes of lncRNAs to be explored. In contrast, siRNA and shRNA are not effective in targeting nuclear lncRNAs because the RNAi machinery is located in the cytoplasm.
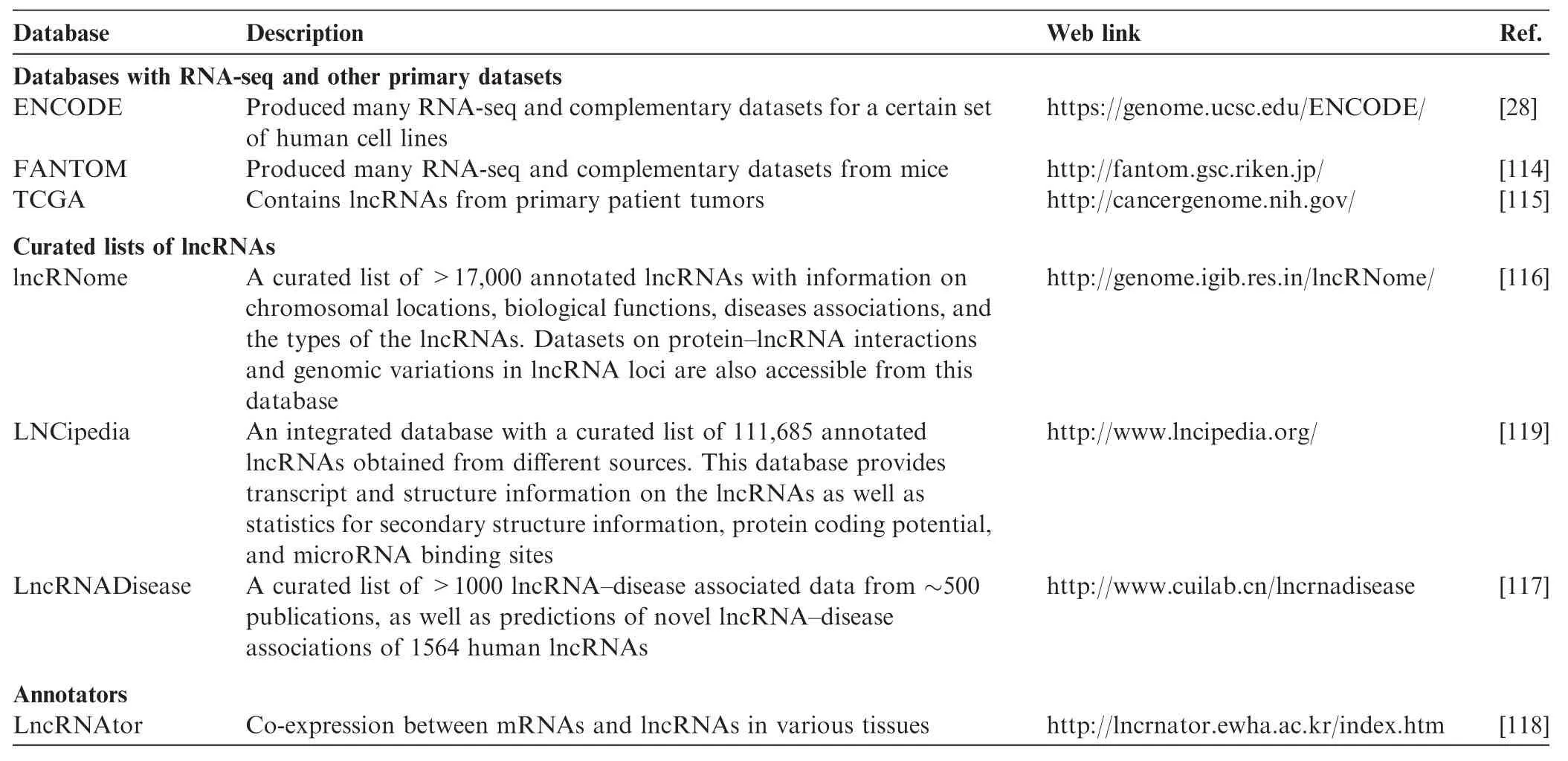
Table 3 Key databases of lncRNAs
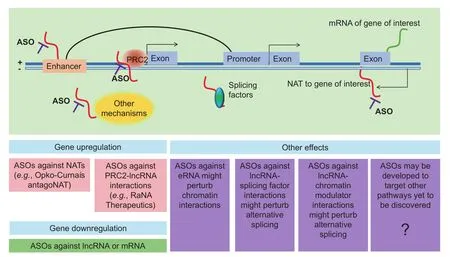
Figure 4 Targeting lncRNAs for therapeutic applicationsNAT, natural antisense transcript; PRC2, polycomb repressive complex 2; ASO, antisense oligonucleotide; eRNA, enhancer RNA.
A major interest for antisense therapy is to upregulate gene expression. Several lncRNAs work as NATs to genes of therapeutic interest. Classic small molecule drugs work by inhibiting gene expression; therefore genes that need to be upregulated have been almost undruggable, although enzyme replacement therapy has been used as a strategy, such as lysosomal enzymes in the case of lysosomal storage diseases [125]. Antisense targeting of natural antisense lncRNAs, or ‘antagoNATs”, provides a very specific way for gene upregulation [126,127]. Taken together, ASOs constitute a very promising tool that can be used to translate basic discoveries about lncRNAs to the clinic. For instance, Opko-Curna is a company taking forward this technology into the clinic [126].
Similarly, company RaNa Therapeutics aims to upregulate gene expression, but using a different approach [128]. Using RIP-Seq, thousands of lncRNAs were found to interact with PRC2, which represses transcription of the genes targeted bylncRNAs [101]. Notably, specific PRC2-lncRNA interactions can be disrupted using locked nucleic acids, resulting in upregulation of the target genes [129]. Locked nucleic acids are RNA derivatives similar in principle to ASOs [130]. Therefore, the company is keen to de-repress the expression of genes of interest by using locked nucleic acids against specific lncRNAs that interact with PRC2.
Given that lncRNAs have been found to act in other pathways, such as eRNAs, they have additional potential in terms of therapeutic mechanisms that can be targeted by ASOs. For example, transcription factors, chromatin modulators, and chromatin interactions are difficult pathways to target, as these factors are in the nucleus, which is difficult for small molecule inhibitors to enter. ASOs against lncRNAs, which interact with factors that can enter the nucleus, might offer a feasible mechanism for future targeting of these important pathways.
Future directions
A major challenge to work on lncRNAs is that despite their importance, the molecular mechanisms underlying their functions are not yet fully understood. Further insight into the biological significance and functioning of lncRNAs will require additional studies to be conducted, which may lead to the discovery of yet more mechanisms of action. Several functions are just starting to be appreciated, such as roles in alternative splicing. For example, MALAT1 controls alternative splicing by regulating the phosphorylation and distribution of serine/ arginine splicing factors in nuclear speckle domains [131]. It is likely that other lncRNAs will be found that regulate alternative splicing through other mechanisms. Moreover, this review did not cover novel mechanisms and forms of lncRNAs such as circular RNA [132]. These novel forms will need to be explored in more details, and their possible relevance to cancer pathways will also need to be examined.
Further confounding factors in our understanding of lncRNAs is that lncRNAs can have more than one mechanisms of action to confer transcriptional activation or repression of their target genes. For example, KCNQ1OT1 can function as both a signal lncRNA [133] and a guide lncRNA [21]. In addition, HOTAIR acts via at least three mechanisms namely signal, decoy, and guide [134]. These studies suggest that for many lncRNAs, even lncRNAs that have been characterized, new mechanisms may yet be uncovered.
Moreover, it is unclear whether there exists correlation between particular functioning mechanisms of lncRNAs and their roles in cancer. For example, XIST is a lncRNA with a well-established role in dosage compensation in the fruitfly [135] and mammals [136]. XIST lncRNA is exclusively produced by the XIST gene located on the inactive X-chromosome [137]. Notably, such inactivation is mediated by the ability of XIST to recruit chromatin-modification complexes [37]. At the same time, aberrant expression of XIST has been linked to a variety of cancers [138]. One mechanism that XIST may function through is as a miRNA sponge for miR-152 [11]. This raises interesting questions such as whether this is the only mechanism of action for XIST in cancer? Does the role of XIST in dosage compensation also have anything to do with its role in cancer?
Another direction is to understand how different lncRNAs cross talk with each other, and how aberrant cross-talk may be regulated in cancer. Laying the ground for further work are new technologies, for example new sequencing methods that can directly sequence RNA and RNA modifications without the need for reverse transcription. In addition, miniaturization of sequencing devices could enable the development of handheld RNA-seq devices that rapidly analyze miRNAs and lncRNAs of interest in patient blood samples or biopsies, quickly providing insights into RNA biology that may be dysregulated in individual patients, thereby helping pave the way to an era of precise, personalized evaluation of patient health and disease.
In conclusion, enhanced understanding of lncRNAs in cancer will shed light into disease etiology and will help guide future diagnosis as well as therapeutic options. In future, with the potential therapeutic options for modulating lncRNAs in the form of ASOs as well as other technologies that may arise, lncRNA-based therapies could become an important healthcare strategy for consideration.
Competing interests
MJF is a co-inventor on 2 patents related to chromatin interactions. There are no other conflicts of interest to be declared.
Acknowledgments
We would like to thank members of the Fullwoodlab for helpful suggestions and comments. This research is supported by the National Research Foundation (NRF) of Singapore through an NRF fellowship awarded to MJF (Grant No. NRFF2012-054), NTU startup funds, and Yale-NUS startup funds awarded to MJF. In addition, this research is supported by funds given to the Cancer Science Institute (CSI), National University of Singapore (NUS), by the NRF and the Ministry of Education - Singapore under the Research Center of Excellence funding. This research is also supported by the RNA Biology Center at the CSI, NUS, as part of the funding under the Tier 3 grants of the Ministry of Education, Singapore.
References
[1] Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep 2009;10:973–82.
[2] Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature 2012;489:101–8.
[3] Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 2013;9:e1003569.
[4] Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74.
[5] Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol 2007;23:175–205.
[6] Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–74.
[7] He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5:522–31.
[8] Gutschner T, Diederichs S. The hallmarks of cancer: a long noncoding RNA point of view. RNA Biol 2012;9:703–19.
[9] Ellis BC, Molloy PL, Graham LD. CRNDE: a long non-coding RNA involved in cancer, neurobiology, and development. Front Genet 2012;3:270.
[10] Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem 2012;113:1868–74.
[11] Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett 2015;359:75–86.
[12] Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6.
[13] Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb–dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320–6.
[14] Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 2011;18:1243–50.
[15] Quagliata L, Matter M, Piscuoglio S, Makowska Z, Heim M, Tornillo L, et al. Hoxa13 and Hottip expression levels predict patients’survival and metastasis formation in hepatocellular carcinoma. J Hepatol 2013;58:S39–40.
[16] Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 2013;152:727–42.
[17] Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol 2012;13:R75.
[18] Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155–9.
[19] Li XL, Wu ZQ, Fu XB, Han WD. LncRNAs: insights into their function and mechanics in underlying disorders. Mutat Res Rev Mut Res 2014;762:1–21.
[20] Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 2012;40:6391–400.
[21] Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008;32:232–46.
[22] Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013;52:101–12.
[23] Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 2014;39:35–43.
[24] Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res 2011;71:5365–9.
[25] Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011;30:1956–62.
[26] Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206–14.
[27] Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147:789–802.
[28] Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816.
[29] Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559–63.
[30] Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 2004;566:43–7.
[31] Warden CD, Kim SH, Yi SV. Predicted functional RNAs within coding regions constrain evolutionary rates of yeast proteins. PLoS One 2008;3:e1559.
[32] Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775–89.
[33] Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014;505:635–40.
[34] Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet 2012;8:e1002841.
[35] Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147:1537–50.
[36] Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014;53:1005–19.
[37] Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992;71:515–26.
[38] Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005;308:1149–54.
[39] Ulitsky I, Bartel DP. LincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46.
[40] Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 2014;39:170–82.
[41] Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 2010;16:1478–87.
[42] Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–66.
[43] Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 2006;16:11–9.
[44] Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 2008;322:1851–4.
[45] Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res 2012;22:885–98.
[46] Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013;498:516–20.
[47] Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 2013;154:1190–3.
[48] Ounzain S, Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc Med 2015;25:592–602.
[49] Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature 2012;489:109–13.
[50] Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012;148:84–98.
[51] Visel A, Rubin EM, Pennacchio LA. Genomic views of distantacting enhancers. Nature 2009;461:199–205.
[52] Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 2007;25:1457–67.
[53] Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–8.
[54] Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007;4:721–6.
[55] Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011;147:382–95.
[56] Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011;147:344–57.
[57] Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353–8.
[58] Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science 2005;309:1564–6.
[59] Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, et al. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci 2006;97:1147–54.
[60] Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 2002;111:185–96.
[61] Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311–23.
[62] Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667–72.
[63] Volkel P, Dupret B, Le Bourhis X, Angrand PO. Diverse involvement of EZH2 in cancer epigenetics. Am J Transl Res 2015;7:175–93.
[64] Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science 2002;295:1306–11.
[65] Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2001;2:292–301.
[66] Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 2006;38:1348–54.
[67] Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intraand interchromosomal interactions. Nat Genet 2006;38:1341–7.
[68] Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 2006;16: 1299–309.
[69] Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res 2008;18:1171–9.
[70] Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 2009;462:58–64.
[71] Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013;502:59–64.
[72] Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 2013;23:1210–23.
[73] Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A 2014;111:7319–24.
[74] Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, et al. ERNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 2013;49:524–35.
[75] Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature 2015;525:399–403.
[76] Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005;123:265–76.
[77] Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin domain to coordinate homeotic gene expression. Nature 2011;472:120–4.
[78] Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol 2014;204: 61–75.
[79] Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 2009;23:1831–42.
[80] Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 2010;24:2543–55.
[81] Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB upregulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366–83.
[82] Fan M, Li XY, Jiang W, Huang Y, Li JD, Wang ZM. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med 2013;5:1143–6.
[83] Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, et al. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 2004;32:4812–20.
[84] Lepoivre C, Belhocine M, Bergon A, Griffon A, Yammine M, Vanhille L, et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics 2013;14:914.
[85] Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell 2012;48:365–74.
[86] Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell 2009;33:462–71.
[87] Mahmoudi S, Henriksson S, Farnebo L, Roberg K, Farnebo M. WRAP53 promotes cancer cell survival and is a potential target for cancer therapy. Cell Death Dis 2011;2:e114.
[88] Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 2008;9:402–12.
[89] Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol 2014;6:181–91.
[90] Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One 2011;6:e25915.
[91] Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031–41.
[92] Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368.
[93] Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol 2014;35:9497–503.
[94] Dunker AK, Uversky VN. Drugs for‘protein clouds’: targeting intrinsically disordered transcription factors. Curr Opin Pharmacol 2010;10:782–8.
[95] Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63.
[96] Ilott NE, Ponting CP. Predicting long non-coding RNAs using RNA sequencing. Methods 2013;63:50–9.
[97] Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 2011;10:141.
[98] Clark MB, Mercer TR, Bussotti G, Leonardi T, Haynes KR, Crawford J, et al. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Methods 2015;12:339–42.
[99] Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014;32:834–41.
[100] Rhoads A, Au KF. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 2015;13:278–89.
[101] Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 2010;40:939–53.
[102] Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011;44:667–78.
[103] Dunagin M, Cabili MN, Rinn J, Raj A. Visualization of lncRNA by single-molecule fluorescence in situ hybridization. Methods Mol Biol 2015;1262:3–19.
[104] Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, et al. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc Natl Acad Sci U S A 2011;108:11063–8.
[105] Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010;467:103–7.
[106] Wan Y, Qu K, Ouyang Z, Chang HY. Genome-wide mapping of RNA structure using nuclease digestion and high-throughput sequencing. Nat Protoc 2013;8:849–69.
[107] Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 2010;7:995–1001.
[108] Han J, Zhang J, Chen L, Shen B, Zhou J, Hu B, et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 2014;11:829–35.
[109] Ho TT, Zhou N, Huang J, Koirala P, Xu M, Fung R, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res 2015;43:e17.
[110] Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods 2013;10:957–63.
[111] Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–6.
[112] Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442–51.
[113] Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 2013;10:977–9.
[114] Consortium F, the RP, Clst, Forrest AR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature 2014;507:462–70.
[115] Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20.
[116] Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B, et al. LncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database 2013;2013:bat034.
[117] Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013;41:D983–6.
[118] Park C, Yu N, Choi I, Kim W, Lee S. LncRNAtor: a comprehensive resource for functional investigation of long non-coding RNAs. Bioinformatics 2014;30:2480–5.
[119] Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 2013;41: D246–51.
[120] Fritah S, Niclou SP, Azuaje F. Databases for lncRNAs: a comparative evaluation of emerging tools. RNA 2014;20:1655–65.
[121] Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 2002;1:347–55.
[122] Geary RS, Henry SP, Grillone LR. Fomivirsen: clinical pharmacology and potential drug interactions. Clin Pharmacokinet 2002;41:255–60.
[123] Lee RG, Crosby J, Baker BF, Graham MJ, Crooke RM. Antisense technology: an emerging platform for cardiovascular disease therapeutics. J Cardiovasc Transl Res 2013;6:969–80.
[124] Zong X, Huang L, Tripathi V, Peralta R, Freier SM, Guo S, et al. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol Biol 2015;1262:321–31.
[125] Baldo BA. Enzymes approved for human therapy: indications, mechanisms and adverse effects. BioDrugs 2015;29:31–55.
[126] Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013;12:433–46.
[127] Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 2012;30:453–9.
[128] Sheridan C. RaNA therapeutics. Nat Biotechnol 2012;30:909.
[129] Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A 2010;107:22196–201.
[130] Braasch DA, Corey DR. Locked nucleic acid (LNA):fine-tuning the recognition of DNA and RNA. Chem Biol 2001;8:1–7.
[131] Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925–38.
[132] Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 2015;6:563–79.
[133] Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, et al. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development 2012;139:2792–803.
[134] Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353a–68a.
[135] Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell 1999;4:117–22.
[136] Gendrel AV, Heard E. Fifty years of X-inactivation research. Development 2011;138:5049–55.
[137] Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991;349:38–44.
[138] Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg 2011;35:1751–6.
[139] Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroen Hepat 2009;21:688–92.
[140] Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 2012;287:26302–11.
[141] Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007;132:330–42.
[142] Sirchia SM, Tabano S, Monti L, Recalcati MP, Gariboldi M, Grati FR, et al. Misbehaviour of XIST RNA in breast cancer cells. PLoS One 2009;4:e5559.
[143] Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res 2014;74:6890–902.
[144] Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 2012;29:1810–6.
[145] Schmidt LH, Spieker T, Koschmieder S, Humberg J, Jungen D, Bulk E, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011;6:1984–92.
[146] Xu CA, Yang MH, Tian J, Wang XY, Li ZG. MALAT-1: A long non-coding RNA and its important 3’end functional motif in colorectal cancer metastasis. Int J Oncol 2011;39:169–75.
[147] Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysis. Int J Clin Exp Med 2015;8:5499–505.
[148] Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening noncoding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res 2011;39:2119–28.
[149] Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013;32:1616–25.
[150] Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012;72:1126–36.
[151] Yap KL, Li SD, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 Lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662–74.
[152] Chung SY, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci 2011;102:245–52.
[153] Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-Iail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007;2:e845.
ORIGINAL RESEARCH
*Corresponding author.
杂志排行
Genomics,Proteomics & Bioinformatics的其它文章
- A Bipartite Network-based Method for Prediction of Long Non-coding RNA–protein Interactions
- EDISON-WMW: Exact Dynamic Programing Solution of the Wilcoxon–Mann–Whitney Test
- Translational Bioinformatics: Past, Present, and Future
- Single-cell Transcriptome Study as Big Data
- Understanding Spatial Genome Organization: Methods and Insights
- Call for Papers Special Issue on‘‘Biomarkers for Human Diseases and Translational Medicine’’
