基于线粒体控制区Dloop序列的长臀种群遗传结构分析
2016-02-23谢少林吕子君李正光石忍耐邹记兴
谢少林, 王 超,, 吕子君, 李正光, 石忍耐, 邹记兴
(1 华南农业大学 动物科学学院,广东 广州 510642; 2 清远市兴渔水产科技有限公司,广东 清远 511510)
谢少林1, 王超1,2, 吕子君1, 李正光1, 石忍耐2, 邹记兴1
(1 华南农业大学 动物科学学院,广东 广州 510642; 2 清远市兴渔水产科技有限公司,广东 清远 511510)
摘要:【目的】了解长臀Cranoglanis3个种群的野生资源状况,并对3个种群的物种有效性进行分析。【方法】采用线粒体控制区Dloop基因序列测定技术, 分析了珠江水系、海南水系和越南红河水系长臀种群的群体遗传结构及其变异。【结果】在检测的84个个体中共得到43个单倍型,呈现出较高的单倍型多样性与较为贫乏的核苷酸多样性,其中海南长臀C.multiradiatus群体的单倍型多样性(Hd=0.871)和核苷酸多样性(Pi=0.006 4)最低;Tajima’sD中性检验以及核苷酸不配对分析均表明,3个长臀群体趋于稳定,未经历过大规模的种群扩张。Fst分析发现,海南长臀同珠江长臀C.bouderius、红河长臀C.henrici产生了一定的遗传分化,而珠江和红河群体未发现明显遗传分化,从遗传距离来看,珠江和红河长臀净遗传距离为0.000。【结论】长臀野生资源较为贫乏,且海南群体最为严重;认为应将珠江长臀和红河长臀归为同一亚种长臀C.bouderius,而海南长臀作为长臀的另一个亚种。
关键词:长臀; 线粒体控制区; Dloop基因序列; 物种有效性; 遗传结构; 资源状况
鱼类线粒体基因是遗传信息重要的载体,具有母系遗传、进化速度快等特点,是研究种群遗传结构的理想材料[1-2];线粒体控制区又称D-环区(Dloop),其进化速度最快,是mtDNA其他区段的5~10倍[3],遗传变异大,是种群遗传多样性研究的常用标记之一,很适合做种内、种群或个体间的遗传分化研究[4]。


1材料与方法
1.1材料


1.2方法

2结果与分析


表1 长臀3个种群Dloop序列特征




Tab.2The distribution of 43 haplotypes in Cranoglanis populations

个

表3 长臀种群的遗传变异参数统计1)
1)*表示达统计显著水平 (P<0.05)。
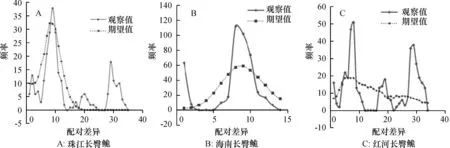
图1 长臀群体Dloop基因序列的核苷酸不配对分布
Tab.4Pairwise genetic distances within population (diagonal), net genetic distance (above diagonal) and fixation index (Fst) (below diagonal) between Cranoglanis population

种群珠江长臀海南长臀红河长臀珠江长臀0.0150.0010.000海南长臀0.114*0.0060.003红河长臀0.0150.170*0.018
1)*表示达统计显著水平 (P<0.05)。

Z:珠江长臀C.bouderius;H:海南长臀C.multiradiatus;Y:红河长臀C.henrici。
3讨论






参考文献:
[1]唐文乔,胡雪莲,杨金权. 从线粒体控制区全序列变异看短颌鲚和湖鲚的物种有效性[J].生物多样性,2007, 15(3): 224-231.
[2]牛素芳,苏永全,钟声平,等.福建近海2 个鲐鱼群体遗传结构与遗传多样性分析[J].台湾海峡,2012,31(4):509-515.
[3]ROSEL P E, DIZON A E, HAYGOOD M G. Variability of the mitoehondrial control region in populations of the harbour porpoise,Phoeoena,on interoeeanic and regional scales[J]. Can J Fish Aquat Sci, 1995,52(6):1421-1429.
[4]乐小亮.中国野生香鱼(Plecogzossusaltivelis)遗传多样性分析[D].广州:暨南大学, 2010.
[7]乐佩琦, 陈宜瑜. 中国濒危动物红皮书:鱼类[M]. 北京:科学出版社, 1998:240-243.

[11]KOLLER O.Fishe von der Insel Hainan[J]. Anna des Naturh Mus Wien, 1927,41:25-49.
[12]MYERS G S.On the fishes described by Koller from Hainan in 1926 and 1927[J].Lingnan Sci J,1931, 10:255-262.
[13]JAYARAM K C.Taxonomic status of the Chinese catfish family Cranoglanideae Myers[J]. Proc Natn Inst Sci India, 1995,15:85-87.
[14]褚新洛.郑葆珊,成定远,等.中国动物志:硬骨鱼纲·鲇形目[M].北京:科学出版社,1999:74-77.
[15]NG H H,KOTTELAT M.Cranoglanishenrici(Vaillant, 1893), a valid species of cranoglanidid catfish from Indochina (Teleostei, Cranglanididae) [J]. Zoosystema,2000,22(4): 847-852.
[17]THOMPSON J D,GIBSON T J,PLEWNIAK F,et al.The CLUSTAL_X window s interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Res, 1997,25 (24):876-4882.
[18]TAMURA K,PETERSON D,PETERSON N, et al.MEGA5:Molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods[J]. Mol Biol Evol, 2011,28(10): 2731-2739.
[19]LIBRADO P, ROZAS J. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data [J]. Bioinformatics, 2009,25(11): 1451-1452.
[20]EXCOFFIER L, LISCHER H E L.Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows[J]. Mol Ecol Resour,2010,10(3):564-567.
[21]陈大庆,张春霖,鲁成,等.青海湖裸鲤繁殖群体线粒体基因组Dloop区序列多态性[J].中国水产科学, 2006,13(5):800-806.
[22]袁希平,严莉,徐树英,等.长江流域铜鱼和圆口铜鱼的遗传多样性[J].中国水产科学, 2008,15(3):380-383.
[23]杨金权,胡雪莲,唐文乔,等.长江口邻近水域刀鲚的线粒体控制区序列变异与遗传多样性[J].动物学杂志, 2008,43(1):8-15.
[24]袁娟,张其中,李飞,等.铜鱼线粒体控制区的序列变异和遗传多样性[J].水生生物学报,2010,34(1):13-16.
[25]ROGERS A R, HARPENDING A.Population growth makes waves in distribution of pairwise genetic differences [J].Mol Biol Evol, 2006, 9: 552-569.
【责任编辑柴焰】
Analysis of population genetic structure ofCranoglanisbased on
mitochondrial Dloop sequences
XIE Shaolin1, WANG Chao1,2, LÜ Zijun1, LI Zhengguang1, SHI Rennai2, ZOU Jixing1
(1 College of Animal Science, South China Agricultural University, Guangzhou 510642, China;
2 Qingyuan Xingyu Fishery Science Co., Ltd., Qingyuan 511510, China)
Abstract:【Objective】 In order to understand the current situation of wild resources for threeCranoglanispopulations and to evaluate the species validities of three species.【Method】The genetic structure and genetic variation of threeCranoglanispopulations (C.multiradiatus,C.bouderiusandC.henrici) were investigated by sequencing the Dloop genes in the mitochondrial control region. 【Result】Among 84 individuals from the three populations, fourty-three hapotypes were detected, indicating a relatively high haplotype diversity and low nucleotide diversity. The lowest hapotype diversity (Hd) and nucleotide diversity (Pi) were both found inC.multiradiatusgroup(Hd=0.871, Pi=0.006 4). Tajima’sDneutral test and nucleotide mismatch distribution analysis showed that the three populations were getting stable without experincing any large-scale expansion. Fst analysis suggested thatC.multiradiatushad certain genetic differentiation compared toC.bouderiusandC.henrici, while the latter two did not have any obvious differentiation from each other. The genetic distance betweenC.bouderiusandC.henriciwas 0.000. 【Conclusion】The wild resources forCranoglanisare relatively poor and the case inC.multiradiatusis the most serious.C.bouderiusandC.henriciare considered to be one subspecies, as well asC.multiradiatusbeing another subspecies ofCranoglanis.
Key words:Cranoglanisbouderius; mitochondrial control region; Dloop gene sequence;species validity; genetic structure; resource status
中图分类号:Q953
文献标志码:A
文章编号:1001-411X(2016)01-0008-06
基金项目:广东省海洋渔业科技推广专项科技攻关与研发项目(A201301F03);广东省海洋渔业科技与产业发展专项(A201501A02);科技型中小企业技术创新基金(14C26214402655)
作者简介:谢少林(1988—),男,博士研究生,E-mail:xieshaolinscau@163.com;通信作者:邹记兴(1966—),男,教授,博士, E-mail:zoujixing@scau.edu.cn
收稿日期:2015-01-28优先出版时间:2015-12-07
优先出版网址:http://www.cnki.net/kcms/detail/44.1110.s.20151207.1115.004.html

