马铃薯3个StSnRK2 基因启动子的克隆与序列分析
2016-02-05范阿棋毛娟杨宏羽张俊莲白江平
范阿棋,毛娟,杨宏羽,张俊莲,,白江平
马铃薯3个StSnRK2 基因启动子的克隆与序列分析

【目的】 克隆马铃薯StSnRK2.1、StSnRK2.2和StSnRK2.4基因的启动子片段,为马铃薯StSnRK2基因表达与功能研究奠定基础.【方法】 以马铃薯(SolanumtuberosumL.)品种‘陇薯3号’为试材,采用PCR技术从马铃薯基因组DNA中分离得到了3个SnRK2基因启动子.通过PCR扩增,酶切鉴定,测序,利用PlantCARE和PLACE在线分析软件预测分析所得序列.【结果】 获得与预期大小基本一致的目的片段,分别为StSnRK2.1基因启动子、StSnRK2.2基因启动子和StSnRK2.4基因启动子.分析软件预测分析表明,启动子调控序列中除含有典型的真核生物核心启动子外,还含有多个CAAT-box、TATA-box等启动子元件,以及相应的ABRE、DRE/CRT、LTRE等与激素应答、逆境胁迫相关的顺式作用元件.【结论】 这些结果预示StSnRK2.1、StSnRK2.2和StSnRK2.4基因调控机制相对复杂,可能参与了ABA,干旱,盐等多种逆境胁迫信号的转导过程,在马铃薯逆境胁迫应答过程中具有重要的功能作用.
马铃薯;StSnRK2基因;启动子;克隆;序列分析
干旱、极温、氧化以及盐碱等非生物胁迫严重制约着植物的生长发育.植物在不断的进化过程中产生一种复杂的生理和遗传机制来感受外部刺激,并对逆境做出应答[1].因而,阐明植物在逆境胁迫条件下的抗性机制和提高植物在各种逆境环境下的抗性水平是各国科学家研究的主要目标.近年来已有研究表明,蔗糖非酵解型蛋白激酶(sucrose non-fermenting1-related protein kinase,SnRK)是广泛存在于植物中的一类Ser/Thr类蛋白激酶,参与植物体内多种信号途径的转导,在植物的抗逆境生理过程中具有重要作用[2-4].根据核酸和蛋白质序列相似性,植物的SNF/SnRK相关蛋白激酶基因分为3个家族:SnRK1、SnRK2和SnRK3[5].其中SnRK2为植物特有,主要参与对环境因素如干旱和盐胁迫等的应答过程.第一个SnRK2成员是从ABA处理的小麦胚胎cDNA文库中分离得到的PKABA1[6].PKABA1的表达除受ABA诱导外,还可被脱水胁迫诱导.目前,已在拟南芥,小麦,水稻等植物中发现和克隆出SnRK2基因[7-8],随后的研究表明,这类蛋白激酶还可以被高渗透胁迫诱导,并且参与ABA信号途径[9,10].SnRK2作为一类蛋白激酶,探索其基因调控机制成为研究热点.
启动子是基因表达调控的重要元件,它能够活化RNA聚合酶并使之与模板DNA相互结合,继而启动下游基因的表达[11].因而启动子是控制基因表达开启的核心,克隆基因启动子序列并对其中的特征信号进行分析是了解基因转录调控表达模式及其调控机制的关键.目前,许多植物的SnRK2基因已被克隆,但有关SnRK2基因启动子克隆和功能分析的报道还很少,有关马铃薯SnRK2基因的研究仅限于基因的克隆和遗传转化[12],马铃薯SnRK2基因家族的3个成员,StSnRK2.1、StSnRK2.2和StSnRK2.4已经被克隆和鉴定,并对其序列进行了分析[13-15].本研究采用PCR克隆策略从马铃薯基因组中克隆获得3个SnRK2基因家族启动子,并采用生物信息学分析方法分析其序列,对其功能调控元件进行分析,以揭示马铃薯SnRK2基因家族的转录机制,从而为该基因家族在马铃薯抗逆转基因分子育种中的应用奠定基础.
1 材料与方法
1.1 材料及试剂

1.2 试验方法
1.2.1 马铃薯基因组DNA的提取 将马铃薯陇薯3号新鲜幼叶置于液氮预冷的研钵内,充分研磨.采用CTAB法[16]提取试验材料的基因组DNA.0.8%琼脂凝胶电泳鉴定所提取的DNA质量.置于-20 ℃冰箱保存待用.
1.2.2StSnRK2基因启动子克隆 从马铃薯基因组数据库(http://solanaceae.plantbiology.msu.edu/) 2 kb的StSnRK2.1、StSnRK2.2 和StSnRK2.4基因启动子序列.依据该区域的序列,应用Primer Premier 5.0设计用于扩增启动子片段的引物,并且根据载体构建的需要,在启动子序列5’端和3’端分别加入HindⅢ和BamHⅠ酶切位点设计引物(表1).以马铃薯基因组DNA为模板,进行PCR扩增.扩增反应体系为:Dream Taq Green PCR Master Mix 25 μL,上游引物 1 μL,下游引物 1 μL,DNA 1 μL,ddH2O 22 μL,共50 μL体系.扩增条件为:95 ℃ 预变性5 min,95 ℃变性30 s,55 ℃退火1 min,72 ℃延伸1 min,40个循环,72 ℃延伸10 min.取6 μL PCR扩增产物进行1.0%的琼脂糖凝胶电泳分析结果.蓝白斑筛选阳性克隆菌落[17],菌液PCR,提取阳性菌液质粒酶切鉴定后送北京华大基因公司测序.
1.2.3 序列分析 利用PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html)[18]和PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)[19]分析克隆获得的3个基因启动子StSnRK2.1、StSnRK2.2和StSnRK2.4的序列内部顺式调控元件.
表1 启动子引物序列
Tab.1 Primer sequences of promoters

名称上游引物序列(5'→3')下游引物序列(5'→3')产物长度/bpStSnRK2.1GCAAGCTTGGCAAGTTTAAGGGGCTGTATGCGGATCCCGTAGTACCTGCCAAGAAGATC977StSnRK2.2GCAAGCTTGGCATTTTCGTATCTGACCAAGCGGATCCGAATCACACACCCACAAA961StSnRK2.4GCAAGCTTTCACTCATTTTGACCCGAATCGCGGATCCCCCTTCAATTCAAAACCACAA932
2 结果与分析
2.1 启动子片段克隆
提取马铃薯试管苗基因组DNA,琼脂糖凝胶电泳显示DNA条带清晰,完整,DNA没有降解,能够用于PCR扩增.以DNA为模板,采用常规PCR扩增启动子,结果如图1所示,能够扩增出与预期大小相符的特异性条带.回收目的片段经连接转化后菌液PCR扩增筛选阳性克隆,提取质粒酶切鉴定,结果能够切下预期大小片段,送去测序后获得完整的上游启动子序列,分别得到977、961、932 bp大小的片段.
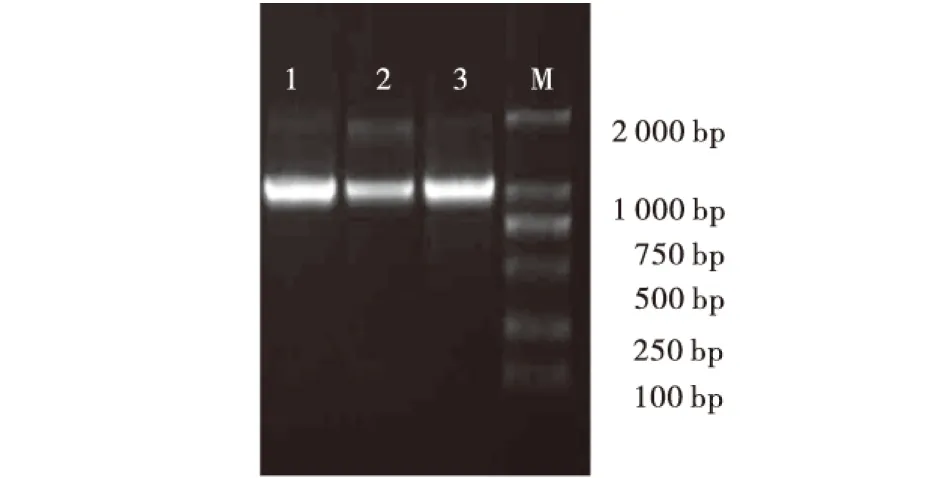
1-3:分别为StSnRK2.1、StSnRK2.2、StSnRK2.4基因启动子;M:DNA Marker.图1 PCR扩增结果Fig.1 The electrophoresis profile of PCR amplification products
2.2 启动子的序列分析
将测得序列与其他植物序列经BLAST比对,在GenBank中均没有显著同源的序列.将所得3个基因启动子序列进行同源性比对,如图2所示,其同源性为56.69%.利用BDGP (http://www.fruitfly.org/seq_tools/promoter.html)在线分析软件分别对StSnRK2.1、StSnRK2.2和StSnRK2.4的5’调控区序列进行分析,结果表明,这3个启动子都含有多个50 bp的典型真核生物核心启动子区域,初步判断这些克隆序列具有启动子结构.根据与翻译起始位点的距离以及转录起始位点上游的一般特征判断,初步确定这3个启动子的核心区域,如表2所示.StSnRK2.1的核心启动子区域位于-76~-26 bp,其转录起始位点的碱基为腺嘌呤;StSnRK2.2的核心启动子区域位于-402~-352 bp,其转录起始位点的碱基也是腺嘌呤;StSnRK2.4的核心启动子区域位于-73~-23 bp,其转录起始位点的碱基是胸腺嘧啶(图3),该启动子与一般所认为的转录起始位点碱基为嘌呤有所不同.
利用PLACE和PlantCARE软件分别对StSnRK2.1、StSnRK2.2和StSnRK2.4基因启动子序列进行顺式作用元件分析,这些序列正、负链在翻译起始位点上游存在多种顺式作用元件,其中包括真核生物典型的启动子基本元件CAAT-box、TATA-box外,还具有众多与激素、胁迫应答等相关的顺式作用元件(表3,图3).这些顺式作用元件可能在StSnRK2.1、StSnRK2.2和StSnRK2.4基因在响应ABA、干旱胁迫、逆境胁迫等各种环境刺激时发挥着重要调控作用.

表2 启动子核心启动子区Tab.2 Core sequence of promoters

A、B、C:分别为StSnRK2.1、StSnRK2.2、StSnRK2.4基因启动子.图2 StSnRK2.1、StSnRK2.2和StSnRK2.4基因启动子序列同源性比对Fig.2 Homologous analysis of StSnRK2.1,StSnRK2.2 and StSnRK2.4 gene promoter sequences
3 讨论
在非生物胁迫下,植物不断进化形成各种适应性机制,其中一种机制就是通过蛋白质的磷酸化与去磷酸化来调控植物抗逆相关基因的表达,从而提高植物的抗逆性.SnRK2是植物体内特有的一类抗逆相关蛋白激酶,主要参与ABA、高盐、低温等非生物胁迫的调节机制[20-21].启动子作为精确调控外源基因在植物体内表达的重要元件[22],决定基因转录的起始和转录起始特异性.因而本试验根据已获得的StSnRK2.1、StSnRK2.2和StSnRK2.4基因序列设计特异性引物,通过PCR扩增技术,从马铃薯基因组DNA文库中扩增获得了这3个基因的启动子序列.启动子区含有一系列特异的蛋白质结合序列,统称为顺式作用元件,不同启动子的顺式作用元件决定了基因具有不同的表达特性[23],通过生物软件进行生物信息学分析,这3个基因启动子除了含有真核生物典型的基本启动子元件外,还含有与ABA调控有关的元件和大量与逆境相关的顺式作用元件[24-25]。ABA在植物响应多种非生物胁迫中发挥重要作用,而ABRE元件是对ABA起响应的主要顺式作用元件,存在于许多胁迫诱导基因的启动子区,同样响应ABA的顺式作用元件还有DPBFCOREDCDC3[26]、E-box[27]、MYB1AT[28]等.另外还存在众多响应逆境胁迫的顺式作用元件,如干旱元件ACGTATERD1[29]、CBEHV[30]、DRE2COREZMRAB17[31]、DRECRTCOREAT[32]等,低温响应元件CRTDREHVCBF2[33]、LTRE1HVBLT49[34]、LTREATLTI78[35]、LTRECOREATCOR15[36]等,盐胁迫响应元件GT1GMSCAM4[37].已有研究证明这些作用位点与其相应转录因子在ABA 信号转导和非生物胁迫应答中具有关键作用.


A、B、C:分别为StSnRK2.1、StSnRK2.2、StSnRK2.4基因启动子;下划线表示上下游引物,起始密码子ATG的A记为+1位点,TATA-box,CAAT-box用粗体表示,部分响应逆境相关元件用 表示,预测的转录起始位点用→表示。图3 启动子序列分析Fig.3 Sequence analysis of promoter sequences
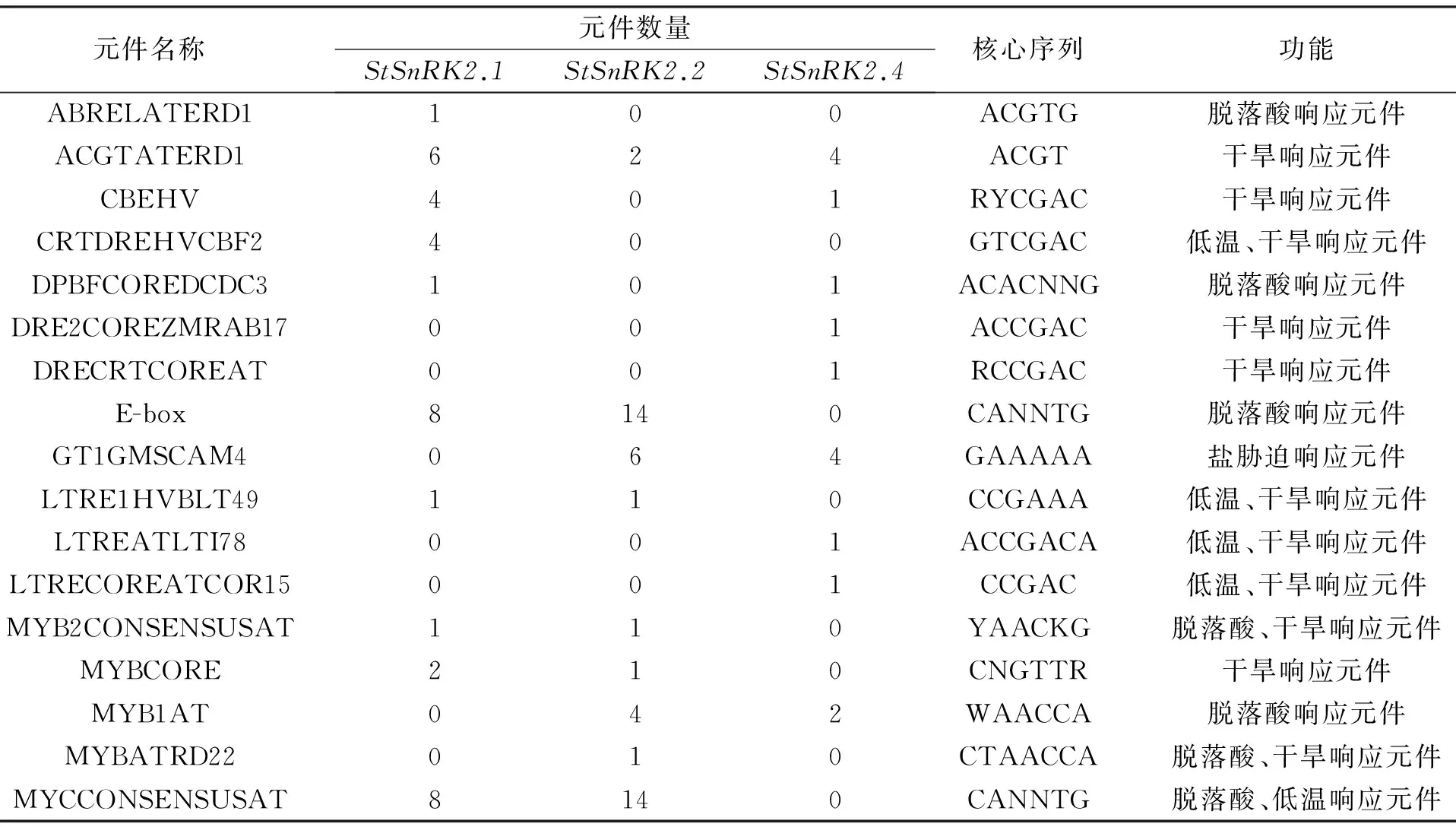
表3 启动子顺式作用元件的功能预测Tab.3 Cis-element of the promoters function prediction
目前,毛娟等[12]对马铃薯试管苗用ABA、NaCl和PEG胁迫处理,以未受胁迫处理的马铃薯试管苗作为对照,通过qRT-PCR进行了SnRK2基因家族成员的诱导表达特性,结果显示在PEG和NaCl诱导胁迫后,马铃薯内部StSnRK2.1、StSnRK2.2和StSnRK2.4基因的相对表达量明显增加.ABA诱导胁迫后StSnRK2.2和StSnRK2.4基因的相对表达量虽有所增加,但不明显,且StSnRK2.1基因的相对表达量反而有些下降.Huai等[38]在玉米中验证,SnRK2基因家族分为3个亚群,每个亚群的基因表达特性有相似之处,StSnRK2.1、StSnRK2.2与ZmSnRK2.3在同一亚群,StSnRK2.4与ZmSnRK2.6在同一亚群.ZmSnRK2.3和ZmSnRK2.6在受到NaCl胁迫后其基因表达量明显增加,说明其响应NaCl诱导胁迫.这与上述马铃薯3个基因表达结果基本一致,因此,可以推断StSnRK2.1、StSnRK2.2和StSnRK2.4同样响应PEG和NaCl诱导胁迫.ABA诱导后,ZmSnRK2.3和ZmSnRK2.6基因表达量均不高,这与StSnRK2.1、StSnRK2.2和StSnRK2.4基因表达结果相似.推断StSnRK2.1、StSnRK2.2和StSnRK2.4可能响应ABA诱导,但是并不敏感.本试验软件分析结果StSnRK2.1和StSnRK2.4基因启动子分别含有4个CRT元件和2个DRE元件,StSnRK2.2虽不含有这2个主要的响应干旱元件,但是含有其它6个响应干旱元件,这些元件的存在可能参与了SnRK2基因在PEG胁迫下的表达.StSnRK2.1含有3个ABRE元件,但是该基因对ABA的响应并不敏感,可能是由于SnRK2基因家族中大部分成员所参与的植物抗逆途径属于ABA非依赖型.StSnRK2.2和StSnRK2.4基因虽然不含有ABRE元件,但是含有其它ABA响应元件,可能在ABA信号转导中起到一定作用,从而使该基因在ABA诱导下相对表达量有所增加.此外,StSnRK2.2和StSnRK2.4基因启动子分别含有6个和4个盐胁迫响应元件GT1GMSCAM4,而StSnRK2.1不含有该元件.毛娟试验中提到StSnRK2.2和StSnRK2.4基因表达量明显增加,是对照表达量的3倍,而StSnRK2.1基因的相对表达量则增加的不明显,说明该元件的存在可能与盐胁迫诱导基因表达过程中起到重要作用.综上所述,可以确定马铃薯StSnRK2.1、StSnRK2.2和StSnRK2.4基因调控机制相对复杂,可能参与了ABA,干旱,盐等多种逆境胁迫信号的转导过程,在马铃薯逆境胁迫应答过程中具有重要的功能作用.这些启动子的获得,为利用其介导植物转基因研究奠定了基础.
[1] Yamaguchi-Shinozaki K,Shinozaki K.Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses[J].Annu Rev Plant Biol,2006,57:781-803
[2] 王永波,高世庆,唐益苗,等.植物蔗糖非发酵-1相关蛋白激酶家族研究进展[J].生物技术通报,2010(11):7-18
[3] Boudsocq M,Droillard M J,Barbier-Brygoo H,et al.Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid[J].Plant Molecular Biology,2007,63(4):491-503
[4] Di dhiou C J,Popova O V,Dietz K J,et al.The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice[J].BMC Plant Biology,2008,8(1):49-61
[5] Hrabak E M,Chan C W M,Gribskov M,et al.The Arabidopsis CDPK-SnRK superfamily of protein kinases[J].Plant Physiology,2003,132(2):666-680
[6] Anderberg R J,Walker-Simmons M K.Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases[J].Proceedings of the National Academy of Sciences,1992,89(21):10183-10187
[7] Gonzalez-Ballester D,Pollock S V,Pootakham W,et al.The central role of a SnRK2 kinase in sulfur deprivation responses[J].Plant physiology,2008,147(1):216-227
[8] Fujii H,Verslues P E,Zhu J K.Identification of two protein kinases required for abscisic acid regulation of seed germination,root growth,and gene expression inArabidopsis[J].The Plant Cell,2007,19(2):485-494
[9] Kobayashi Y,Yamamoto S,Minami H,et al.Differential activation of the rice sucrose nonfermenting1 related protein kinase2 family by hyperosmotic stress and abscisic acid[J].The Plant Cell,2004,16(5):1163-1177
[10] Yoshida R,Hobo T,Ichimura K,et al.ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis[J].Plant and Cell Physiology,2002,43(12):1473-1483
[11] 毛娟,陆璐,陈佰鸿,等.甜瓜CmACO启动子组织特异性表达研究[J].园艺学报,2014,40(6):1101-1109
[12] Mao J,Bai J P,Fan A Q,et al.Identification and characterization of eight sucrose non-ferment 1-related protein kinases 2 activated by obiotic stress in potato[J].Joukull Journal,2013,63(12):449-576
[13] 刘思妍,毛娟,范阿棋,等.马铃薯StSnRK2.1基因克隆与生物信息学分析[J].生物技术通报,2013(3):83-89
[14] 范阿棋,毛娟,刘思妍,等.马铃薯StSnRK2.2基因克隆与生物信息学分析[J].植物研究,2013,33(3):294-301
[15] 李洋,白江平,毛娟,等.马铃薯StSnRK2.4基因克隆与序列特征分析[J].甘肃农业大学学报,2014,182(02):77-83
[16] Bustamante C A,Civello P M,Martínez G A.Cloning of the promoter region of β-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression ofFaXyl1 in strawberry fruit [J].Plant Science,2009,177(1):49-56
[17] Sambrook J,Fritsh E F,Maniatis T.分子克隆实验指南[M].2版.北京:科学出版社,1992
[18] Kenichi H,Yoshihiro U,Masao I,et al.Plant cis-acting regulatory DNA elements (PLACE) database [J].Nucleic Acids Res,1999,27(1):297-300
[19] Lescot M,D hais P,Thijs G,et al.PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J].Nucleic Acids Research,2002,30(1):325-327
[20] Wang P,Xue L,Batelli G,et al.Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action [J].Proceedings of the National Academy of Sciences,2013,110(27):11202-11210
[21] Xie T,Ren R B,Zhang Y Y,et al.Molecular mechanism for inhibition of a critical component in theArabidopsisthalianaabscisic acid signal transduction pathways,SnRK2.6,by protein phosphatase ABI1[J].The Journal of Biological Chemistry,2012,287(1):794-802
[22] 魏桂民,张金文,王蒂,等.马铃薯sgt2基因启动子的克隆与活性分析[J].甘肃农业大学学报,2014,53(1):41-47
[23] 朱玉贤,李毅.现代分子生物学第二版[M].北京:高等教育出版社,2007
[24] Shinozaki K,Yamaguchi-Shinozaki K.Molecular response to dehydration and low temperature:differences and cross-talk between two stress signaling pathways[J].Current Opinion in P1ant Biology,2000,3(3):217-223
[25] 王舟,刘建秀.DREB/CBF类转录因子研究进展及其在草坪草和牧草抗逆基因工程中的应用[J].草业学报,2011,20(1):222-236
[26] Lopez-Molina L,Chua NH.A null mutation in a bZIP factor confers ABA-insensitivity inArabidopsisthaliana[J].Plant Cell Physiol,2000,41(5):541-547
[27] Hartmann U,Sagasser M,Mehrtens F,et al.Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB,BZIP,and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes[J].Plant Mol Biol,2005,57(2):155-171
[28] Abe H,Urao T,Ito T,et al.ArabidopsisAtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling [J].Plant Cell,2003,15(1):63-78
[29] Simpson SD,Nakashima K,Narusaka Y,et al.Two different novel cis-acting elements of erd1,a clpA homologousArabidopsisgene function in induction by dehydration stress and dark-induced senescence [J].Plant J,2003,33(2):259-270[30] Svensson J T,Crosatti C,Campoli C,et al.Transcriptome analysis of cold acclimation in barleyalbinaandxanthamutants [J].Plant Physiol,2006,141(1):257-270
[31] Dubouzet J G,Sakuma Y,Ito Y,et al.OsDREBgenes in rice,OryzasativaL,encode transcription activators that function in drought-,high-salt- and cold-responsive gene expression [J].Plant J,2003,33(4):751-763
[32] Skinner J S,Von Zitzewitz J,Szucs P,et al.Structural,functional,and phylogenetic characterization of a largeCBFgene family in barley[J].Plant Mol Biol,2005,59(4):533-551
[33] Xue G P.The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature [J].Plant J,2003,33(2):373-383
[34] Dunn M A,White A J,Vural S,et al.Identification of promoter elements in a low-temperature-responsive gene from barley[J].Plant Mol Biol,1998,38(4):551-564
[35] White A J,Dunn M A,Brown K,et al.Comparative analysis of genomic sequence and expression of a lipid transfer protein gene family in winter barley[J].J Exp Bot,1994,45(12):1885-1892
[36] Kim H J,Kim Y K,Park J Y,et al.Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) inArabidopsisthaliana[J].Plant J,2002,29(6):693-704
[37] Park H C,Kim M L,Kang Y H,et al.Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor [J].Plant Physiol,2004,135(4):2150-2161
[38] Huai J L,Wang M,He J G,et al.Cloning and characterization of theSnRK2 gene family fromZeamays[J].Plant Cell Reports,2008,28:1861-1868
(责任编辑 李辛)
Cloning and bioinformatic analysis of three StSnRK2 gene promoters in potatio(Solanum tuberosum)
(1.Gansu Key Laboratory of Crop Genetic and Germplasm Enhancement,Gansu Provincial Key Laboratory of Aridland Crop Science,Lanzhou 730070,China;2. College of Agronomy,Gansu Agricultural University,Lanzhou 730070,China;3.College of Horticulture,Gansu Agricultural Unicersity,Lanzhou 730070,China)
【Objective】StSnRK2.1,StSnRK2.2 andStSnRK2.4 gene promoter fragments of potato were cloned by molecular cloning,and it laid the foundation of further studies in potatoStSnRK2 gene expression and function.【Method】 Potato ‘cv. Longshu 3’ (SolanumtuberosumL.) was the test material,threeSnRK2 gene promoters were cloned from potato genomic DNA by PCR,restriction enzyme,sequencing. PlantCARE and PLACE were used for analyzing the obtained sequences.【Result】We obtained the target fragments were consistent with expected size using PCR,restriction enzyme digestion and sequencing,and namedStSnRK2.1 gene promoter,StSnRK2.2 gene promoter andStSnRK2.4 gene promoter,respectively. PlantCARE and PLACE online analysis software prediction analysis showed that the promoter regulatory region includes typical eukaryotic core promoter region,in addition to containing multiple CAAT-box,TATA-box. promoter elements and other cis-acting elements were involved in the response to phytohormone and osmotic stresses,such as ABRE,DRE/CRT,LTRE.【Conclusion】StSnRK2.1,StSnRK2.2 andStSnRK2.4 gene regulation mechanism is relatively complex,may be involved in the transduction of ABA,drought ,salinity and other abiotic stress signal,expression may have relation to potato resistance.
potato;StSnRK2 gene;promoter;clone;sequence analysis
范阿棋(1989-),女,硕士,从事马铃薯逆境分子生物学研究.E-mail:15095371268@163.com
白江平,男,博士,副教授,从事马铃薯抗逆基因工程研究.E-mail:baijp@gsau.edu.cn
国家自然科学基金项目(31460369),甘肃省自然科学基金项目(1308RJIA131),甘肃省干旱生境作物学重点实验室开放基金项目(GSCS-2012-04).
2015-10-29;
2015-12-22
S 531
A
1003-4315(2016)06-0036-08
