胃癌差异表达蛋白的蛋白质组学分析
2015-12-28,,,,*
,,,,*
(1.南华大学肿瘤研究所,湖南 衡阳421001;2.南华大学医学院解剖教研室)
·基础医学·
胃癌差异表达蛋白的蛋白质组学分析
李素云1,2,陈苏琼1,谢远杰1,张志伟1,贺修胜1*
(1.南华大学肿瘤研究所,湖南 衡阳421001;2.南华大学医学院解剖教研室)
目的筛选与鉴定胃癌差异表达蛋白质分子。方法采用定量蛋白组学方法,通过同位素iTRAQ标记胃癌与正常胃黏膜组织总蛋白,然后用强阳离子交换/反相液相色谱(SCX/nanoHPLC)进行电喷雾串联质谱分析(ESI-MS/MS),获得两组样本的多肽及相对丰度信息,通过数据库搜索与比对,鉴定差异表达蛋白质。
结果共鉴定出319个蛋白质,其中表达差异在2倍以上的88个,在胃癌中呈高表达的12个、呈低表达的76个。按蛋白质功能分为七大类:细胞骨架蛋白、分子伴侣、信号传导、生物代谢、凋亡,蛋白质合成与代谢类及其他蛋白质。结论成功筛选了胃癌差异表达蛋白质分子,这些蛋白质的表达改变,可能参与了胃癌的发生和发展。
胃癌; iTRAQTM同位素标记相对和绝对定量; 多维液相色谱; 串联质谱
我国是胃癌的高发区,每年死于胃癌的人数约为16万人,因此,提高胃癌的诊疗水平,是我国肿瘤基础及临床研究工作者所面临的挑战[1]。现今临床所使用的肿瘤诊断标记物有CA19-9、CEA、CA72-4[2-3],但这些标记物对于早期胃癌检测缺乏特异性,仅能用作为术后疗效监测[4]。
本研究将LCM技术纯化后的组织,胰酶裂解,iTRAQ试剂分别标记多肽,强阳离子反相液相色谱进行肽段分离,结合电喷雾串联质谱对多肽进行定性定量,通过数据库搜索鉴定胃腺癌的差异表达蛋白质,寻找胃癌早期诊断的特异分子标志物,对胃癌的早期诊断、防治及预后评估、发病机制提供理论和实验基础。
1 材料与方法
1.1 材料
1.1.1 标本 20例手术切除的新鲜胃腺癌组织及配对的正常胃黏膜上皮组织,取自湖南省衡阳市南华大学附属第一医院肿瘤外科,所有取材严格按照实验要求操作,所取组织经激光捕获显微切割纯化。
1.1.2 试剂 trypsin为 promega Madison WI,乙腈(ACN )为Fisher scientific pittsburg,PA,0.1%甲酸购自sigma公司,三氟乙酸(TFA,色谱纯) 购自Merck公司,iTRAQ Reagentkit购自Applied Biosystems公司。
1.2 胃癌细胞和正常胃黏膜上皮细胞蛋白质的消化和标记
1.2.1 蛋白沉淀还原与半胱氨酸封闭 每组样品100 μg蛋白质,分别置于洁净的微量离心管中,分别加入4倍体积的丙醇(-20 ℃)和12%三氯乙酸(TCA)沉淀蛋白,将两组样品离心管置于-20 ℃孵育过夜; 两组样品离心管离心 12 000 g,4 ℃,30 min,弃上清,同样的试剂再次沉淀,-20 ℃孵育2 h,弃上清让沉淀自然干燥,再溶解在缓冲溶液中;分别加入2 μL还原剂[三(2-羰基乙基)磷盐酸盐]; 60 ℃中孵育2.5 h,离心使样品沉于管底,弃上清液;分别加入2 μL封闭剂半胱氨酸 (硫甲磺酸S-甲醋),涡旋,室温下孵育20 min。
1.2.2 胰蛋白酶消化蛋白 每个样品管中按1∶20(胰酶∶蛋白质)的比例加入胰蛋白酶,旋涡混合,在37 ℃下过夜(12~16 h)。
1.2.3 iTRAQ试剂标记消化蛋白 标记使用4标试剂盒进行2标标记,使用116,117标记。向116、117报告基团的iTRAQ试剂瓶内加入70 μL无水乙醇,混匀,使iTRAQ试剂充分溶解备用;将含116报告基团的iTRAQ试剂与胃正常黏膜上皮细胞组蛋白样品管混合,117报告基团的iTRAQ试剂与胃癌细胞蛋白样品管混合,室温下孵育l h;将标记好的消化蛋白样品混合入统一管中,旋转真空离心浓缩干燥(Speed Vac)混合后的蛋白。
1.3 全自动2D-Nano-LC-ESI-MS/MS分析肽段
肽段使用强阳离子交换/反相纳升级液相色谱(SCX/nanoHPLC)进行电喷雾串联质谱分析(ESI-MS/MS)。
1.4 数据分析
所有的串级谱图通过SEQUEST搜索引擎 [v.28 (revision 12),Thermo Electron Corp.]进行数据库检索。数据库为Swiss-Prot人种属数据库(Release 2010_04,20331条序列)。为了减少假阳性结果,在该数据库中增加一个含有所有蛋白反转序列的诱饵数据库。搜索参数设置如下:胰蛋白酶(Trypsin),半酶切方式,最大漏切为2。固定修饰为半胱氨酸(Cysteine)的甲基硫代磺酸甲酯化(methyl methanethiosulfonate,MMTS),可变修饰为甲硫氨酸(Methionine)的氧化,赖氨酸(lysine)和肽段N端的iTRAQ试剂标记。单一同位素模式,肽段质量误差50 ppm,碎片离子质量误差0.05 Da。
使用Trans Proteomic Pipeline 软件(revision 4.0)(Institute of Systems Biology,Seattle,WA),通过Peptide Prophet(P≥0.90)和Protein Prophet(P≥0.95)来过滤肽段和蛋白结果,获得置信度在0.95以上的蛋白结果。根据肽段结果通过定量计算方法,计算得到各个蛋白的定量结果。
2 结 果
2.1 蛋白质的质谱分析
所有的串级谱图通过SEQUEST搜索引擎进行Swiss-Prot人种属数据库检索。使用Trans Proteomic Pipeline 软件来过滤肽段和蛋白,获得置信度在0.95以上的蛋白结果。共发现鉴定了319个蛋白,其中差异蛋白为88个,12个蛋白在胃腺癌中高表达,76个蛋白在胃腺癌中低表达(表1)。
2.2 已鉴定的差异蛋白的亚细胞定位和功能分类
根据UniprotKB/Swiss-prot数据库中蛋白质的相对定位和功能信息,对人胃低分化腺癌所鉴定得到的蛋白质进行亚细胞定位和功能分类。从图1中可以看出已鉴定出的蛋白有17.4%位于细胞浆中;27.7%定位于线粒体,大部分都是氧化还原和呼吸链相关的酶类;5.1%为核糖体蛋白;12.2%定位于细胞外基质,基本上都是分泌蛋白;14.2%为细胞骨架蛋白;4.0%的蛋白仅定位于细胞核;位于细胞浆和细胞核的蛋白质为9.1%;10.2%定位于细胞膜上,与细胞连接和信号传导相关。
对于蛋白质的功能分类见图2,已鉴定的蛋白质中27.4%与生物代谢相关如Creatine kinase B-type等;另一大类(17.9%)为信号传导通路蛋白质如Ras-related protein Rab-44等;蛋白合成与代谢类也占16.8%如60S ribosomal protein L8等;还有14.7%是细胞骨架类蛋白质如Myosin-13;4.2%的分子伴侣类;8.4%的细胞凋亡类。
表1Nano-RPLC-MS/MS鉴定的GAC和NGEC差异在2倍以上的蛋白质
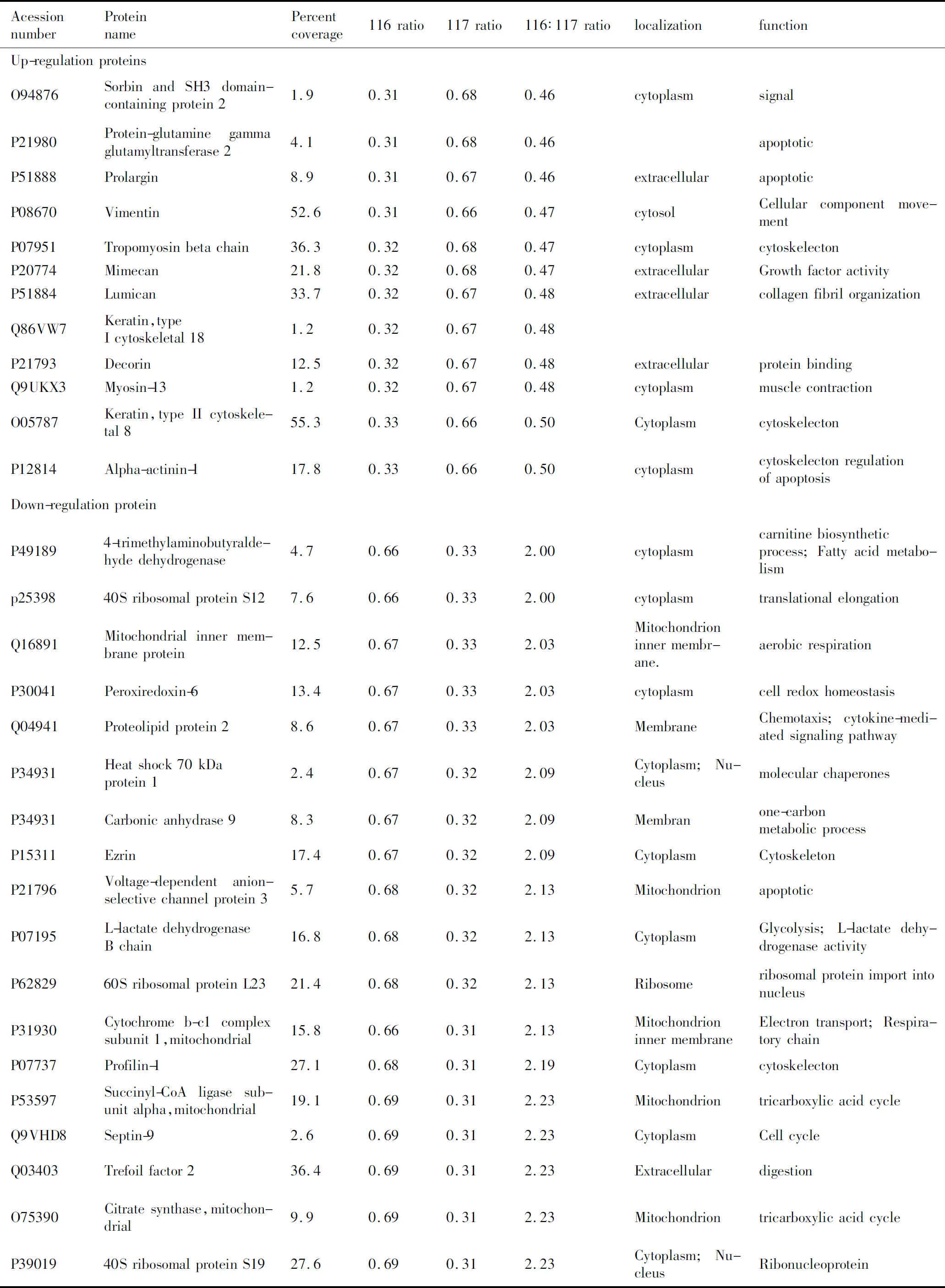
AcessionnumberProteinnamePercentcoverage116rati-o117rati-o116∶117rati-olocalizationfunctionUp-regulationproteinsO94876SorbinandSH3domain-containingprotein21.90.310.680.46cytoplasmsignalP21980Protein-glutaminegam-maglutamyltransferase24.10.310.680.46apoptoticP51888Prolargin8.90.310.670.46extracellularapoptoticP08670Vimentin52.60.310.660.47cytosolCellularcomponentmove-mentP07951Tropomyosinbetachain36.30.320.680.47cytoplasmcytoskelectonP20774Mimecan21.80.320.680.47extracellularGrowthfactoractivityP51884Lumican33.70.320.670.48extracellularcollagenfibrilorganizationQ86VW7Keratin,typeIcytoskeletal181.20.320.670.48P21793Decorin12.50.320.670.48extracellularproteinbindingQ9UKX3Myosin-131.20.320.670.48cytoplasmmusclecontractionO05787Keratin,typeIIcytoskel-etal855.30.330.660.50CytoplasmcytoskelectonP12814Alpha-actinin-117.80.330.660.50cytoplasmcytoskelectonregulationofapoptosisDown-regulationproteinP491894-trimethylaminobutyr-aldehydedehydrogenase4.70.660.332.00cytoplasmcarnitinebiosyntheticprocess;Fattyacidmetab-olismp2539840SribosomalproteinS127.60.660.332.00cytoplasmtranslationalelongationQ16891Mitochondrialinnermembraneprotein12.50.670.332.03Mitochondrioninnermembr-ane.aerobicrespirationP30041Peroxiredoxin-613.40.670.332.03cytoplasmcellredoxhomeostasisQ04941Proteolipidprotein28.60.670.332.03MembraneChemotaxis;cytokine-me-diatedsignalingpathwayP34931Heatshock70kDaprotein12.40.670.322.09Cytoplasm;NucleusmolecularchaperonesP34931Carbonicanhydrase98.30.670.322.09Membranone-carbonmetabolicprocessP15311Ezrin17.40.670.322.09CytoplasmCytoskeletonP21796Voltage-dependentanion-selectivechannelprotein35.70.680.322.13MitochondrionapoptoticP07195L-lactatedehydrogenaseBchain16.80.680.322.13CytoplasmGlycolysis;L-lactatede-hydrogenaseactivityP6282960SribosomalproteinL2321.40.680.322.13RibosomeribosomalproteinimportintonucleusP31930Cytochromeb-c1com-plexsubunit1,mito-chondrial15.80.660.312.13Mitochondrioninnermem-braneElectrontransport;Re-spiratorychainP07737Profilin-127.10.680.312.19CytoplasmcytoskelectonP53597Succinyl-CoAligasesub-unitalpha,mitochondrial19.10.690.312.23MitochondriontricarboxylicacidcycleQ9VHD8Septin-92.60.690.312.23CytoplasmCellcycleQ03403Trefoilfactor236.40.690.312.23ExtracellulardigestionO75390Citratesynthase,mito-chondrial9.90.690.312.23MitochondriontricarboxylicacidcycleP3901940SribosomalproteinS1927.60.690.312.23Cytoplasm;NucleusRibonucleoprotein
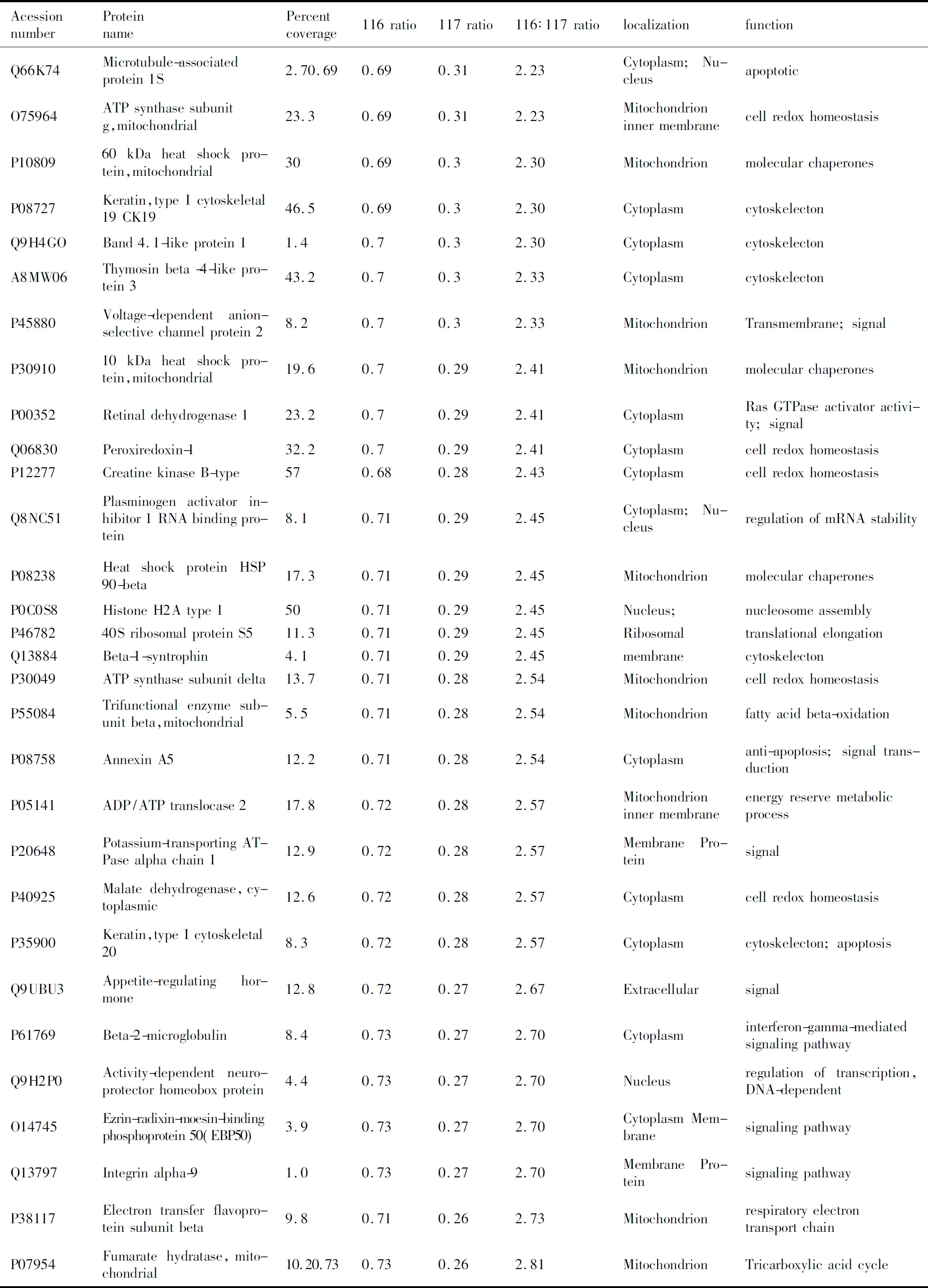
AcessionnumberProteinnamePercentcoverage116rati-o117rati-o116∶117rati-olocalizationfunctionQ66K74Microtubule-associatedprotein1S2.70.690.690.312.23Cytoplasm;NucleusapoptoticO75964ATPsynthasesubunitg,mitochondrial23.30.690.312.23Mitochondrioninnermem-branecellredoxhomeostasisP1080960kDaheatshockpro-tein,mitochondrial300.690.32.30MitochondrionmolecularchaperonesP08727Keratin,typeIcytoskele-tal19CK1946.50.690.32.30CytoplasmcytoskelectonQ9H4GOBand4.1-likeprotein11.40.70.32.30CytoplasmcytoskelectonA8MW06Thymosinbeta-4-likeprotein343.20.70.32.33CytoplasmcytoskelectonP45880Voltage-dependentanion-selectivechannelprotein28.20.70.32.33MitochondrionTransmembrane;signalP3091010kDaheatshockpro-tein,mitochondrial19.60.70.292.41MitochondrionmolecularchaperonesP00352Retinaldehydrogenase123.20.70.292.41CytoplasmRasGTPaseactivatorac-tivity;signalQ06830Peroxiredoxin-132.20.70.292.41CytoplasmcellredoxhomeostasisP12277CreatinekinaseB-type570.680.282.43CytoplasmcellredoxhomeostasisQ8NC51Plasminogenactivatorin-hibitor1RNAbindingprotein8.10.710.292.45Cytoplasm;NucleusregulationofmRNAsta-bilityP08238HeatshockproteinHSP90-beta17.30.710.292.45MitochondrionmolecularchaperonesP0C0S8HistoneH2Atype1500.710.292.45Nucleus;nucleosomeassemblyP4678240SribosomalproteinS511.30.710.292.45RibosomaltranslationalelongationQ13884Beta-1-syntrophin4.10.710.292.45membranecytoskelectonP30049ATPsynthasesubunitdelta13.70.710.282.54MitochondrioncellredoxhomeostasisP55084Trifunctionalenzymesubunitbeta,mitochon-drial5.50.710.282.54Mitochondrionfattyacidbeta-oxidationP08758AnnexinA512.20.710.282.54Cytoplasmanti-apoptosis;signaltransductionP05141ADP/ATPtranslocase217.80.720.282.57Mitochondrioninnermem-braneenergyreservemetabolicprocessP20648Potassium-transportingATPasealphachain112.90.720.282.57MembranePro-teinsignalP40925Malatedehydrogenase,cytoplasmic12.60.720.282.57CytoplasmcellredoxhomeostasisP35900Keratin,typeIcytoskele-tal208.30.720.282.57Cytoplasmcytoskelecton;apoptosisQ9UBU3Appetite-regulatinghor-mone12.80.720.272.67ExtracellularsignalP61769Beta-2-microglobulin8.40.730.272.70Cytoplasminterferon-gamma-media-tedsignalingpathwayQ9H2P0Activity-dependentneu-roprotectorhomeoboxprotein4.40.730.272.70Nucleusregulationoftranscrip-tion,DNA-dependentO14745Ezrin-radixin-moesin-bind-ingphosphoprotein50(EBP50)3.90.730.272.70CytoplasmMembranesignalingpathwayQ13797Integrinalpha-91.00.730.272.70MembranePro-teinsignalingpathwayP38117Electrontransferflavo-proteinsubunitbeta9.80.710.262.73MitochondrionrespiratoryelectrontransportchainP07954Fumaratehydratase,mi-tochondrial10.20.730.730.262.81MitochondrionTricarboxylicacidcycle

AcessionnumberProteinnamePercentcoverage116rati-o117rati-o116∶117rati-olocalizationfunctionP23528Cofilin-129.50.740.262.85CytoplasmRhoproteinsignaltrans-duction;cytoskelectonP14091CathepsinE3.70.740.252.96Endosomeaspartic-typeendopepti-daseactivityP30046D-dopachromedecarbox-ylase220.740.252.96CytoplasmmelaninbiosyntheticprocessQ13423NAD(P)transhydroge-nase7.10.740.252.96MitochondrionTricarboxylicacidcycleP09525AnnexinA48.20.740.252.96CytoplasmsignaltransductionP98088Mucin-5AC(Fragments)9.80.740.252.96SecretedcelladhesionP30044Peroxiredoxin-5,mito-chondrial42.10740.252.96MitochondrioncellredoxhomeostasisP48735Isocitratedehydrogenase[NADP],mitochondrial37.40.730.243.04Mitochondrion2-oxoglutaratemetabolicprocessP08263GlutathioneS-transferaseA17.20.740.243.08CytoplasmglutathionemetabolicprocessP06703ProteinS100-A68.90.760.243.17Cytoplasm;NucleusS100betabinding;signaltransductionP36957Dihydrolipoyllysine-resi-duesuccinyltransferasecomponentof2-oxoglut-aratedehydrogenasecomplex,mitochondrial4.60.750.233.26MitochondriontricarboxylicacidcycleP6291760SribosomalproteinL86.20.770.233.35CytoplasmTranslationalelongationP00403Cytochromecoxidasesubunit24.40.770.233.35Mitochondrioninnermem-branemitochondrialelectrontransport,cytochromectooxygenQ86XP6Gastrokine-219.60.770.223.50SecretedsignalP07098Gastrictriacylglycerolli-pase17.80.760.213.62SecretedtriglyceridemetabolicprocessP13073Cytochromecoxidasesubunit4isoform1,mi-tochondrial12.40.780.213.71Mitochondrioninnermem-branecytochrome-coxidaseac-tivity,proteinbindingP20142Gastricsin14.70.760.23.8Secreteddigestion,proteolysisP10606Cytochromecoxidasesubunit5B47.30.770.23.85Mitochondrioninnermem-branerespiratoryelectrontransportchainP00790PepsinA17.50.710.164.44Secreteddigestion,proteolysisO60218Aldo-ketoreductasefam-ily116.80.80.184.44Cytoplasmdigestion,oxidation-reduc-tionprocessP25815ProteinS100-P28.40.810.184.50Nucleus.Cyto-plasmendothelialcellmigrationQ92522HistoneH1x12.70.770.164.81Nucleus.Cyto-plasmnucleosomeassembly;DNAbindingP56856Claudin-183.80.840.165.25Cellmembranecalcium-independentcell-celladhesion,tightjunc-tionassemblyQ96NY7Chlorideintracellularchannelprotein618.00.850.146.07Cytoplasmmembranechloridechannelcomplex;IontransportQ9NS71Gastrokine-138.20.820.136.31SecretedDigestion;signalQ7Z673Ras-relatedproteinRab-441.90.90.0910.00CellmembranesmallGTPasemediatedsignaltransductionQ5D1E8ZincfingerCCCHdo-main-containingprotein12A2.50.890.0811.13Cytoplasm;Nu-cleusApoptosis;Differentiation
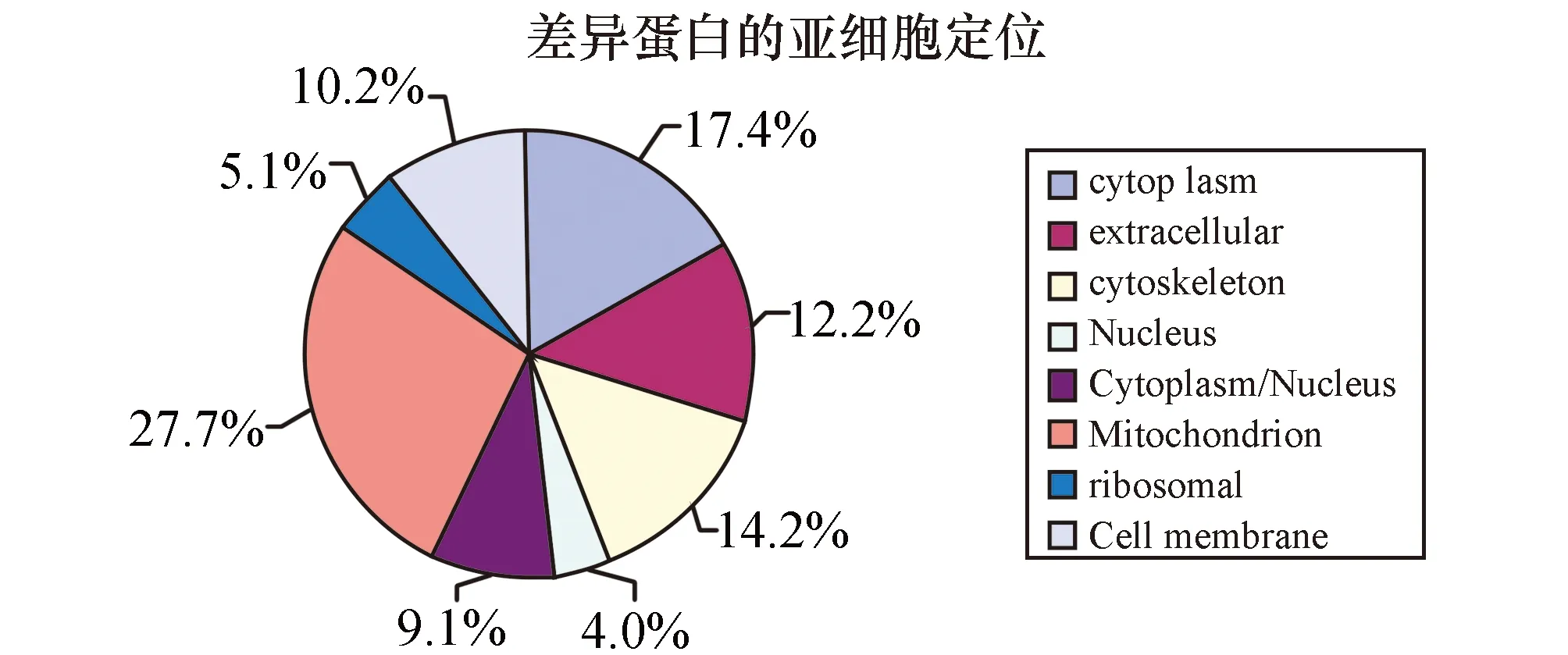
图1 被鉴定蛋白质的亚细胞定位
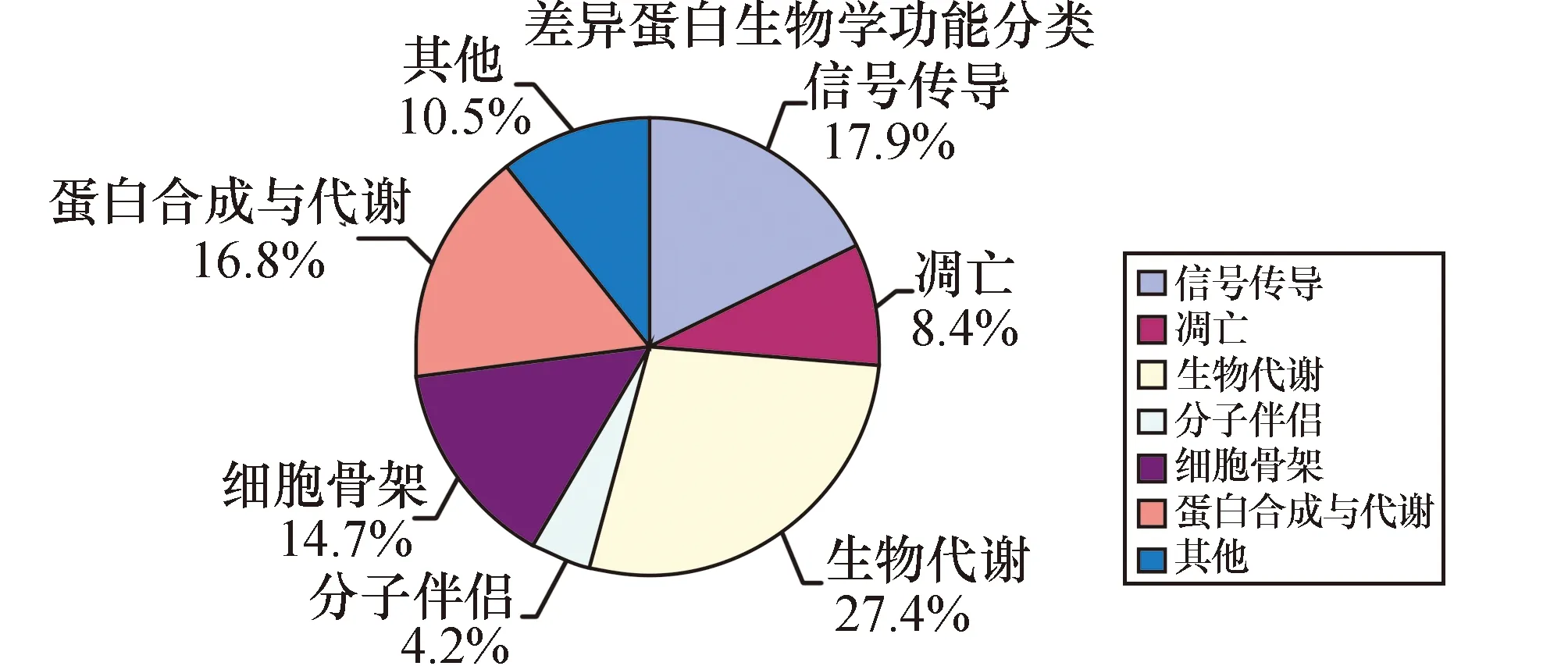
图2 Nano-RPLC-MS/MS鉴定的GAC和NGEC差异蛋白质功能分类
3 讨 论
蛋白质系基因功能的最终执行者,基因作用的发挥通过其表达产物蛋白质实现,所以蛋白质组学(Proteomics)的研究受到研究者们高度关注。比较蛋白质组学是蛋白质组学最重要的研究策略,有研究报道,采用比较蛋白质组学技术,通过比较肿瘤与其起源的正常组织,或肿瘤发展不同阶段组织中蛋白质在表达水平和修饰状态上的差异,发现了诸多癌变相关的特异性蛋白质[5-8]。
程爱兰等[9]人运用2-DE技术分别从鼻咽癌和正常的鼻咽上皮组织中纯化出鼻咽癌细胞和正常鼻咽上皮细胞,发现了29 个差异蛋白质,其中 15 个蛋白质仅在 NPC 中表达或者高表达,而14 个蛋白质在 NPC 中表达下调或者缺失。Neubauer等[10]应用2DE技术,发现了区别 (ER)+/(PR)+和ER+/PR-乳腺预后的相关蛋白质transgelin、cydophilinA及Neudesin,这些蛋白质的差异表达,并发现ER+/PR+乳腺癌预后好于ER+/PR-的乳腺癌。
蛋白质组技术,是后基因时代基因功能研究的重要技术,近年来在蛋白质组研究中新兴的多维液相色谱( MD-LC)技术,是一种非胶的分离方法,利用两种或多种分离机理不同的色谱模式组合对样品进行正交分离。因便于接口控制、易与质谱连接、自动化程度高等优势,已经作为与2DE互补的技术在大规模蛋白质组研究中发挥了重要作用[11]。
本实验中采用的是强阳离子交换色谱(SCX)和反相高效液相色谱(HP-RPLC)多分离模式来分离蛋白质,很大程度上克服了2DE的局限性,其与质谱的联用是目前蛋白质组学研究技术中发展较快的领域。采用基于非胶路线的液相色谱分离方法和iTRAQ标记定量蛋白质组学技术,所需样本量少,可以很好与LCM技术结合运用。
本研究样本经过SCX的预处理后,复杂程度大大降低,同时iTRAQ标记的引入,保证了分离过程中化学行为的一致性,可直接对差异蛋白进行对比,提高了实验结果的准确性。这种实验技术的联用可替代传统的2-DE、DIGE技术,是对胃癌等实体瘤进行差异分析的有效方法[12]。
采用iTRAQ同位素4标试剂盒进行2标116、117标记,116标记正常胃黏膜上皮的总蛋白,117标记胃低分化腺癌的总蛋白,通过比较标记同一蛋白的116、117的相对丰强度来进行相对定量。肽段使用强阳离子交换/反相纳升级液相色谱(SCX/nanoHPLC)进行电喷雾串联质谱分析(ESI-MS/MS),使用Trans Proteomic Pipeline 软件通过Peptide Prophet(P≥0.90)和Protein Prophet(P≥0.95)来过滤肽段和蛋白结果,可获得置信度在0.95以上的蛋白结果,鉴定出胃癌差异表达蛋白质分子,包括高表达和低表达蛋白质分子。高表达的蛋白主要是细胞骨架蛋白和分泌蛋白比如Vimentin、Myosin-13、Decorin、Lumican,低表达的蛋白集中在信号传导、生物代谢、蛋白合成与代谢等生物学功能上如Ras-related protein Rab-44、NAD(P) transhydrogenase、60S ribosomal protein L8等。这些蛋白的差异表达都与肿瘤的发生发展相关,表明胃癌发病机制的复杂性。
胃癌的发生发展的过程是一个多因素、多步骤、多分子参与的复杂事件,可能包括相关癌基因激活和(或)抑癌基因失活,上调和(或)下调相关蛋白质的表达,经其相应信号传导途径,导致胃癌的发生与发展[13-15]。
综上所述,人低分化胃癌组织与正常胃黏膜上皮组织的蛋白质表达存在差异。但胃癌发生发展的确切分子机制、蛋白质间相互作用、及其蛋白质表达调控网络,有待今后进一步深入研究。
[1] Alberts SR,Cervantes A,Van VC.Gastric cancer:epidemiology,pathology and treatment[J].Ann Onco1,2003,14 Suppl 2:31-36.
[2] Ishigami S,Natsugoe S,Nakajo A,et al.Prognostic value of CCR7 expression in Gastric cance[J].Hepatogastroenterology,2007,54(76):1025-1028.
[3] Binninger-Schinzel D,Muller D,Wolf T,et al.Characterization of a chemokine Receptor CCRS-negative T cell line and its use in determining human immunodeficiency virus type1 phenotype[J].J Med Virol,2008,80(2):192-200.
[4] Qiu MZ,Lin JZ,Wang ZQ,et al.Cut off value of carcinoembryonic antigen and carbohydrate antigen 19-9 elevation levels for monitoring recurrence in patients with resectable gastric adenocarcinoma[J].Int J Biol Markers,2009,24(4):258-264.
[5] Yu Z,Yu JK,Fang XF,et al.Proteomic biomarker predicts therapeutical effects of oxaliplatin combining with fluoropyrimidine in metastatic gastric cancer patients by the SELDI-proteinchip platform[J].Hepatogastroenterology,2013,60(127):1800-1804.
[6] Humphries JM,Penno MA,Weiland F,et al.Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer[J].Biochim Biophys Acta,2014,1844(5):1051-1058.
[7] Ge X,Lyu ZX,Liu YI,et al.dentification of FkpA as a key quality control factor for the biogenesis of outer membrane proteins under heat shock conditions[J].J Bacteriol,2014,196(3):672-680.
[8] Zhang ZQ,Li XJ,Liu GT,et al.Identification of Annexin A1 protein expression in human gastric adenocarcinoma using proteomics and tissue microarray[J].World J Gastroenterol,2013,19(43):7795-7803.
[9] 程爱兰,黄卫国,张鹏飞,等,激光捕获显微切割技术纯化的鼻咽癌和正常鼻咽上皮细胞的比较蛋白质组学研究[J].生物化学与生物物理进展,2008,35(5):521-530.
[10] Neubauer H,Clare SE,Kurek Retal.Breast cancer proteomics by laser capture microdissection,sample pooling,54-cm IPG IEF,and differential iodine radioisotope detection [J].Electrophoresis,2006,27(9):1840-1852.
[11] Kocˇevar N1,Grazio SF,Komel R.Two-dimensional gel electrophoresis of gastric tissue in an alkaline pH range[J].Proteomics,2014,14(2-3):311-321.
[12] Hu W,Wang J,Luo G,et al.Proteomics-based analysis of differentially expressed proteins in the CXCR1-knockdown gastric carcinoma MKN45 cell line and its parental cell [J].Acta Biochim Biophys Sin (Shanghai),2013,45(10):857-866.
[13] 黄昊,韩勇,吴健,等.胃癌细胞MGC-803耐药细胞株的建立及分泌蛋白差异分析[J].北京大学学报:医学版,2014,46(2) :183-189.
[14] Biff,Yang Q,Wang Y,et al.Expression and significance of pro-apoptotic protein Bid in endometrial carcinoma[J].Modern Oncology,2012,20(3):589-592.
[15] 陈思颖,蔡江霞,张蔚鹏,等.紫杉醇诱导的乳腺癌耐药细胞的蛋白质组学研究[J].中国药学杂志,2014,49(10) :825-832.
ProteomicsAnalysisofDifferentialExpressionProteinsinGastricCarcinoma
LI Suyun,CHEN Suqiong,XIE Yuanjie,et al
(CancerResearchInstitute,UniversityofSouthChina,Hengyang,Hunan421001,China)
ObjectiveTo identify the differential expression proteins between gastric adenocarcinoma cells (GAC) and normal gastric adenoepithelial cells (NGAC),and to look for early diagnosis of Gastric carcinoma (GC )specific proteins.MethodsThe total proteins were extracted from the tissues.The total proteins labeled with different iTRAQ stable isotope were segregated with Nano-RPLC-MS/MS,to obtain relative abundance of peptides information.The differential expression proteins were identified through SWISSPORT database.ResultsThrough Quantitative proteomic analysis of GAC and NGAC,319 proteins were identified with MS,88 of which were differential expression proteins.There were 12 Up-regulation and 76 Down-regulation proteins in GAC.The identified proteins can be divided into seven groups according to protein function:biological oxidation,signal transduction,protein composition and metabolism,molecular chaperones,cytoskeleton,apoptotic and others.ConclusionThe differentially expressed protein molecules were successfully screened and identified in gastric carcinoma,and the expression changing of these proteins may be involved in the genesis and development of gastric carcinoma.
gastric carcinoma; iTRAQTM; multidimensional liquid chromatography; tandem Mass spectrometry
10.15972/j.cnki.43-1509/r.2015.01.004
2014-05-30;
2014-10-20
湖南省自然科学基金资助项目(12JJ3102),湖南省卫生厅科研基金资助项目(B2012-046),衡阳市科技局资助项目(2013HKJ19),湖南省高校创新平台开放基金资助项目(13K082),地方高校国家级大学生创新创业训练计划资助项目(2012105555017).
*通讯作者,E-mail:hexiusheng@hotmail.com.
R735.2
A
(此文编辑:蒋湘莲)
