Change of MicroRNA-134, CREB and p-CREB expression in epileptic rat
2015-12-08YanZhuChengShanLiYuanYeWangShengNianZhou
Yan Zhu, Cheng-Shan Li, Yuan-Ye Wang, Sheng-Nian Zhou
1Department of ICU, Zaozhuang Municipal Hospital (Jining medical college affiliated hospital), Zaozhuang, Shandong, P. R. China
2Department of Neurology, Qilu Hospital of Shandong University and Brain Science Research Institute, Shandong University, Jinan, Shandong, P. R. China
Change of MicroRNA-134, CREB and p-CREB expression in epileptic rat
Yan Zhu1*, Cheng-Shan Li1, Yuan-Ye Wang1, Sheng-Nian Zhou2
1Department of ICU, Zaozhuang Municipal Hospital (Jining medical college affiliated hospital), Zaozhuang, Shandong, P. R. China
2Department of Neurology, Qilu Hospital of Shandong University and Brain Science Research Institute, Shandong University, Jinan, Shandong, P. R. China
ARTICLE INFO
Article history:
Received15 January 2015
Received in revised form 20 February 2015
Accepted 15 March 2015
Available online 20 April 2015
MicroRNA-134
CREB
p-CREB
Quantitative PCR
Western blot
Immunohistochemistry
Objective: To To investigate the changes of MicroRNA-134, CREB and p-CREB expression in epileptic rat brains in order to elucidate the molecular mechanisms of epilepsy, providing new ideas for clinical treatment. Methods: Sixty-four Spraque-Dawley (SD) rats were divided into groups randomly, including control group, six hours after seizure group, 24-hour group, threeday group, one-week group, two-week group, four-week group, and eight-week group. All groups were placed under a pilocarpine-induced epilepsy model except the control group, and all rats were decapitated in different points of time. Brain specimens were taken for quantitative PCR experiments, immunohistochemistry and Western blot experiments. The results of the epilepsy model groups and the control group were compared. Results: There were no significant differences between the six hours after seizure group, the 24-hour group and the control group about the MicroRNA-134 levels. MicroRNA-134 in the hippocampus tissue of the three-day group significantly reduced compared with the control group; same result was observed with the one-week, two-week, four-week and eight-week groups. The CREB and p-CREB levels in the three-day group's rat hippocampus significantly increased compared with the control group; and the high levels of CREB and p-CREB were constantly maintained in the one-week, two-week, four-week and eight-week groups. Conclusions: The MicroRNA-134 level of the epileptic rat hippocampus is significantly lower than normal after three days, and continues to maintain a low level; while CREB and p-CREB levels are rsignificantly increased after three days, and continue to remain at a high level. MicroRNA-134 plays a role in inhibiting synaptic plasticity by inhibiting CREB and p-CREB expressions.
1. Introduction
Intractable epilepsy[1-4] (IE) is a disease that does not involve organic diseases in the central nervous system; healing is delayed after more than two years of formal antiepileptic treatment that a variety of anti-epileptic drugs are combined. The disease still cannot be controlled, even by achieving an effective blood concentration. Patients cannot live a normal life[5-9]. There may still be possible varieties of pathogenesis, such as the multi-drug resistance mechanism and the neural network restructuring mechanism[10-14]. According to the research of Du et al, mossy fiber sprouting is a common pathological basis for the pathogenesis of epilepsy with the basis of synaptic plasticity[15]. Mossy fiber sprouting is the basis of seizures, which may also be the result of repeated long-term epileptic seizures. Therefore, epilepsy can be treated fundamentally by clearly investigating the basic molecular biology of the mossy fiber sprouting only. Studies by Cohen[16] and Bassell[17] show that certain types of mammalian MicroRNA are associated with synaptic plasticity. MicroRNA-134 in rat hippocampus inhibits the formation
of synapses that exists only in brain tissue[18]. Studies by Gao[19] found that MicroRNA-134 regulates synaptic plasticity by regulating the phosphorylated cAMP-response element binding protein (CREB) and CREB. Various findings relevant to MicroRNA-134, CREB and phosphorylated CREB variations in rat hippocampus need to undergo further studies. MicroRNA-134, CREB and phosphorylated CREB variations in rat hippocampus may provide new insights for treating epilepsy by molecular targeted therapy, which could also be used as a foundation for further studying the mechanisms of epilepsy. Therefore, in this study, changes of MicroRNA-134, CREB, and phosphorylated CREB in the hippocampus of epileptic rats needs further analysis, as reported below.
2. Materials and methods
2.1. Experimental animals
Sixty-four clean Spraque-Dawley (SD) male rats were selected for the study; body weight was approximately 200g; and the rats were reared on free diet in the laboratory by animal laboratory professionals.
2.2. Method
2.2.1. Animal groups
Eight rats were randomly selected as the control group and the remaining rats were introduced to an epilepsy model. The epilepsy model rats were randomly divided into 7 groups; six hours after seizure group, 24-hour group, three-day group, one-week group, two-week group, four-week group and eight-week group; each group have at least five animals.
2.2.2. Establish an epileptic rat model
Lithium chloride was intraperitoneally-administered to the rats, based on the 127 mg/kg dosage. After eighteen hours, the rats were treated with a 40 mg/kg dose of pilocarpine, administered intraperitoneally. Twenty minutes before administering pilocarpine, the rats were intraperitoneally-administered with 1 mg/kg of atropine in order to prevent serious cholinergic reactions. Saline was administered to the control group with the same dosage. When seizures reached stage 4 or 5, the rats were given 10mg/kg of diazepam to terminate the seizures after one hour. According to Racine Stages, “0” no abnormal reactions; “1” blinking, rhythmic chewing-clonic movements and other facial expression appear;“2” paroxysmal nods appear; “3” bilateral forelimb clonus occurs;“4” hind limb standing; and “5” all symptoms above with falling movements occurs.
2.2.3. Preparation and production of tissue specimen for biopsy
Each group was anesthetized by 3.5% chloral hydrate; intraperitoneally administering a 0.1 mL/kg dosage after 6 hours, 24 hours, three days, one week, two weeks, four weeks and eight weeks, respectively. The hippocampus was peeled and placed into EP tubes without RNA enzymes; and stored in a refrigerator at -80 ℃ to be used as materials for quantitative PCR and western blot experiments. After anesthetization, the rats were perfusion-fixed with 4% paraformaldehyde (250 mL); the hippocampus was removed, separated from brain, and fixed with a 4% paraformaldehyde for approximately 2 hours. The specimens were embedded in paraffin and cut into slices (5 μm) to be used as materials of immunohistochemistry.
2.2.4. Quantitative PCR experiments
Approximately 50-100 mg of hippocampus tissue was placed in the tube and 1 mL of RNAiso Plus was added with a dropper. The specimen was homogenized on ice until the polishing fluid became clear, and moved to a tube without RNase. 1.2 mL of chloroform was added, and the centrifuge tube was stamped for 30 seconds after the turbulence. The specimen was placed on ice for 5 minutes, and placed under a centrifuge speed of 12 000 r/minute for fifteen minutes. The centrifugal supernatant was transferred to a fresh tube without RNase, added 0.5 mL of chloroform, and placed on ice for ten minutes after turbulence; the specimen was centrifuged for fifteen minutes with the same speed. Afterwards, the supernatant was discarded, added with 75% ethanol, and was centrifuged for seven minutes with a speed of 7 500 r/minute. The alcohol was discarded; and after drying the tube, RNase-free water was added to dissolve the RNA pellet.
After determining the RNA concentration, the RNA was retained at 1.8-2.0 during 260 nm/280 nm, as a material for cDNA reverse
transcription; reverse transcription procedures were all done on ice. During the experiment, the centrifuge tube and Pipette tips were all RNase free. After reverse transcription, the specimen was cooled on ice. Spare cDNA results were stored at -20 ℃.
Fluorescence quantitative PCR; ① each cDNA tissue sample was pipetted three times; ② Fluorescence quantitative PCR reaction steps (95 ℃ for 30 s, 95 ℃ for 5 s, 60 ℃ for 20 s, 72 ℃ for 10 s, 35 cycles ); ③ After PCR amplification reactions were completed, the temperature was gradually increased from 55 ℃ to 95 ℃ at a speed 0.5 ℃ every 5 seconds to draw a melting curve. The RNA reverse transcription and PCR fluorescence quantitative reaction systems are shown in Table 1.
2.2.5. Western blot experiments
One μL of phosphatase inhibitors (sodium pyrophosphate), 1 mL Lysis buffer and 5 μL PMSF were added in a uniform shockresistant centrifuge tube. The contents were made into protein extracts and placed on ice. As for the 100 mg rat hippocampus, 600 μL protein extracts were added; tissue homogenates were homogenized with a glass homogenizer until it become clear; and was transferred into a new centrifuge tube and centrifuged at 10 000 r/min for five minutes. The supernatant was dispensed into two centrifuge tubes. After determining the protein concentration in the supernatant, 50 μg of protein and a volume of 5×SDS solution were added in a uniform shock-resistant centrifuge tube. The tube was sealed, submersed in boiling water for ten minutes, and stored in the refrigerator at 4 ℃.
Protein concentration assay solution and standard protein solution were prepared according to the manual of BCA protein kit. Three μL of brain homogenate and 57 μL of buffer were thoroughly mixed, which were added to 3 wells of the 96-well plates. Each well was filled up to 20 μL; the protein concentration assay solution was added into each well, placed in the incubator for half an hour, and the A562 wavelength was measured and plotted. The protein concentration's standard curve was drawn, and the protein concentration was calculated based on the standard protein concentration curve. According to the molecular weight of the protein, a 10% separating gel was used; which was solidified to 5% with a stacking gel comb. After the stacking gel was solidified, the comb was pulled out, and a liquid sample was poured to begin the electrophoresis.
A 1×SDS buffer was initially poured for electrophoresis, the marker protein and the hippocampal protein of each group were added; β -mercaptoethanol was added afterwards, and boiled with an initial voltage of 60 volts. After the gel sample started to separate, the voltage level was raised to 120 volts, and the current voltage was maintained until the bromine finland indicator gel electrophoresis reached the next downstream. A stack was placed together into an electrophoresis tank in the following order: PVDF membrane, gel electrophoresis, a three-sheet filter; and was fixed on ice with an electrical current of 250 mA for 60 minutes. The PVDF membrane was transferred to a 5% skimmed powder box for an hour with a temperature of 37 ℃ combined with tissues from non-specific antibody binding sites. The CREB, p-CREB and β-actin antibody were refrigerated at 4 ℃ for spare, after diluting 100, 500, and 1 000 folds. According to the displayed electrophoretic bands, the PVDF CREB, p-CREB and β-actin proteins reside were cut and immersed overnight into its corresponding primary diluted antibody liquid, at a temperature of 4 ℃. The PVDF strips were washed four times with Tris-Buffered Saline Tween-20 (TBST). The Goat anti-rabbit secondary antibody was diluted 4 000 times with TBST. The PDVF membranes were incubated for an hour in the secondary diluted antibodies at 37 ℃; and were immersed in TBST to be shakewashed for four times, at about ten minutes each time. Luminescentliquid-one and luminescent-liquid-two were mixed at a 1:1 ratio, and was added to the membrane. Gel strip images were obtained when light was emitted from the UV gel imager. Quantity one was the image analysis software used to calculate the optical density of each band in order to obtain the expression of the tissue's protein.
2.2.6. Immunohistochemistry
Tissue slices were immersed in a xylene solution Ⅰ and Ⅱ, and incubated for 20 minutes at 60 ℃. These were then immersed in a 100%, 95%, 80%, and 70% alcohol solution for five minutes, one at a time; and tissue slices were soaked in a PBS solution for 5 minutes. The slices were immersed in a citric acid solution and placed in a microwave for antigen retrieval; the temperature was raised to the maximum level for 3 or 4 minutes until the solution boiled; then, the temperature was adjusted and maintained at a low level for 20 minutes. The slices were added in the PBS solution for three minutes after cool down to room temperature for cleaning, which was washed for a total of three times. 3% hydrogen peroxide solution was added in the slice, and was kept at room temperature for 15 minutes; then the slice was washed three times with a PBS solution, three minutes per time.
Dropwise Goat serum was added and incubated for half an hour at 37 ℃. Diluted CREB and p-CREB antibody solutions were added. The slices were incubated at 37 ℃ for two hours. After incubation, the slices were washed three times with a PBS solution, five minutes each time. Secondary antibody was added to the slice, incubated for 20 minutes at 37 ℃, and was washed three times again with a PBS solution, five minutes each time. SABC solution was added in the slices, incubated for 20 minutes, and washed three times with a PBS solution at 37 ℃, five minutes each time.
A drop of color-liquid from the DAB chromogenic kit and 1ml double distilled water were mixed without light; the color-liquid was
added on the slices. When the color darkened from the background, the reaction was stopped by adding water. The slices were washed three times with tap water, placed in a hematoxylin stain for four minutes, washed with tap water three times again, and dipped in a 1% hydrochloric acid for one second for differentiation. After washing three times with tap water, the slices were soaked in a lithium saturated solution for twenty seconds; then, the slices were observed under the microscope after staining the nuclei of neurons examining under the microscope, after the slices were dried.
2.3. Experimental equipment
Ultra-pure water system (MILLI-Q)(Millipore USA); Optical microscope (BX 51) (Olympus, Japan); CFX96 Real time PCR instrument, electrophoresis and electroporation instrument(Bio-Rad America); gel/luminescent image analysis system (ChemiDoc XRS); Visible / UV gel scanning analysis system UVP (Britain); slicers (RM2135)(Leica German); thermostatic water bath(Jiangsu instrument).
2.4. Reagents
Total protein extraction kit (Nanjing KGI); BCA Protein Assay Kit (Enhanced)(Haibi sky); β-actin, P-CREB polyclonal antibody (Santa America);paraformaldehyde PVDF membrane (Millipore USA);pilocarpine, lithium chloride, glycine, Tris base (Sigma USA);Tween-20, Triton-X100, SDS, Protein Marker (Fermentas); SABC kit, DAB chromogenic kit (Beijing Zhongshan Golden Bridge); IgG, CREB polyclonal antibody (Cell signaling technology USA); 0.01 M PBS buffer, polylysine on SDS-PAGE sample buffer (5×) (Wuhan Boster China); kim milk, citrate buffer, and miR-134-specific reverse transcription and PCR primers (Guangzhou RiboBio Ltd.); DEPC (Invitrogen America); SYBR Premix Ex Taq™ Ⅱ, RNAiso Plus (Takara China); PrimeScript RT reagent Kit (Takara China).
2.5. Statistical analysis
IBM SPSS19.0 software was applied for statistical analysis. Normally distributed data were expressed in, and the measured data was compared using ANOVA analysis. Independent t-test sample was used for comparing the groups. P<0.05 represents statistical significance.
3. Result
3.1. MicroRNA-134 expression in each group of rats
After 10-15 cycles of PCR amplification, the MicroRNA-134 CT values of the six hours after seizure group and the 24-hour group had no significant difference compared with the control group, the result was not statistically significant (P>0.05). The MicroRNA-134 expression levels of the three-day-group began to decrease, and MicroRNA-134 levels in the hippocampus of the three-day, one-week, two-week, four-week and eight-week groups decreased significantly, compared with the control group. The difference had statistical significance (P<0.05), as shown in Table 2.
3.2. Immunohistochemistry
3.2.1. CREB expression of a rat model's brain tissue
CREB was expressed in all cells of brain CA3, CA1 neurons of the three-day group's hippocampal nucleus; the CREB expression was significantly more than the control group, shown in Figure 1.
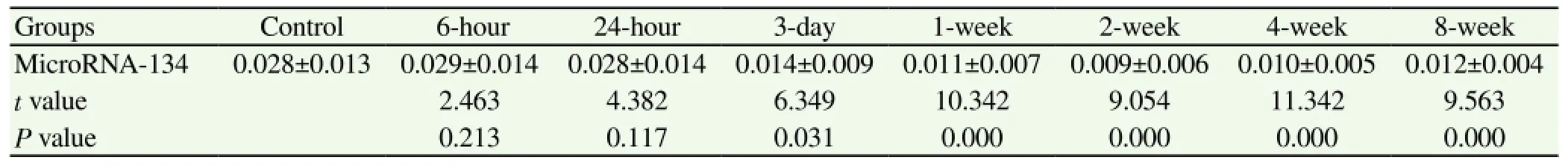
Table 2 Comparison of MicroRNA-134 values in the rat hippocampus of each group.
Figure1-a, CREB expression figure in CA3, neurons of the control group's nucleus, as shown by the arrow; Figure1-b, CREB expression in CA3, neurons in the three-day group's nucleus, as shown by the arrow. Color bands in Figure1-b darkened, indicating that CREB neurons are significantly more than the control group. Figure1-c, CREB expression in CA1, neurons in the control group's nucleus, as shown by the arrow; Figure1-d, CREB expression in CA1, neurons in the three-day group's nucleus, as shown by the
arrow; color bands in figure-d darkened, indicating that CREB neurons are significantly more than the control group.
3.2.2. p-CREB expression of the rat model's brain tissue
In the CA3, CA1 region of the hippocampal, the p-CREB expression levels of the three-day group were significantly more than the control group (Figure 2).
Figure-1a, p-CREB expression in CA3, neurons in the control group's nucleus, as shown by the arrow; and Figure1-b, p-CREB expression in CA3, neurons in the three-day group's nucleus, as shown by the arrow; color bands in Figure1-b are darker, indicating that p-CREB neurons are significantly more than the control group. Figure1-c, p-CREB expression in CA1, neurons in the control group's nucleus, as shown by the arrow; and Figure1-d, p-CREB expression in CA1, neurons in the three-day group's nucleus, as shown by the arrow; color bands of figure-d are darker, indicating that p-CREB neurons are significantly more than the control group.
3.3. Western blot experiment
The CREB expression of the six hours after seizure group and the 24-hour group showed no significant difference compared with the control group, the result was not statistically significant (P>0.05). Three days after seizures, the expression of CREB began to increase; and the expression of the three-day, one-week, two-week, four-week and eight-week groups significantly increased compared with the control group, the results had statistical significance (P<0.05), shown in Table 3. The expression of p-CREB of the six hours after seizure group increased significantly compared with the control group. The expression levels continued to remain high for eight weeks -- the results were statistically significant (P<0.05), shown in Table 4. The PVDF membrane strips and the CREB levels of the three-day group were both more than the control group compared with β-actin; and p-CREB began to increase in the six hours after seizure group for eight weeks, shown in Figure 3.
With β-actin as a reference and CREB in the normal control group, the six hour after seizure group and the 24-hour group had no obvious PVDF membrane strips. The PVDF membrane strips of three-day group was obvious for eight weeks. The p-CREB of the six hours after seizure group had significant number of PVDF membrane strips until the end of the experiment; indicating that the CREB and p-CREB expressions increased significantly.

Table 3 Comparison of CREB expression levels in the hippocampus between the epilepsy groups and the control group.

Table 4 Comparison of p-CREB expression levels in the hippocampus between the epilepsy groups and the control group
4. Discussion
This study aims to investigate the expression level changes of MicroRNA-134, CREB and phosphorylated CREB; in different time periods after seizures of the rat's hippocampus; exploring variations of the hippocampus substances mentioned above; which may give us a better understanding of the pathogenesis of epilepsy. This
study can also help us to learn more about the signaling pathway of MicroRNA-134, and its interaction with CREB and phosphorylated CREB. Grasping the significance of the pathogenesis of epilepsy may contribute to its further in-depth study, at a genetic-level. All of these may provide new ideas for the gene therapy of epilepsy.
4.1. Pathogenesis of epilepsy
There are a variety of concepts in the pathogenesis of epilepsy; but the most common pathological mechanism is the mossy fiber sprouting. Mossy fiber sprouting is based on synaptic plasticity. A rat's newborn synaptic epileptic brain tissue is considered as a pathological basis, due to the abnormal discharge activities of the rat hippocampus. In normal circumstances, normal MicroRNA-134 concentrations can effectively inhibit abnormal formations of synapses. Studies have shown that MicroRNA-134 affects the development of synapses, and that large amounts of MicroRNA-134 can induce the dendritic spine volume to reduce significantly. Currently, the common causes of the pathogenesis of epilepsy[21-23] include: MicroRNA-134, CREB and phosphorylated CREB variations; these are still in the experimental stage and are being researched in epileptic rats findings still needs to be further confirmed. Studies of Nakagawa[24], Lubin[25] and Eacker[26] have shown: the signal transduction pathway mediated by MicroRNA-134 can express by inhibiting specific genes, and that the amount of CREB and phosphorylated CREB inhibits the development of synapses preventing mossy fiber sprouting.
4.2. Expression of CREB and p-CREB inhibited by MicroRNA-134 (mi-R134)
The study showed that after the PCR amplification of the rat hippocampus, the MicroRNA-134 CT values of the six hours after seizure group and the 24-hour group had no significant difference compared with the control group. However, MicroRNA-134 expression of the rat hippocampus of the three-day group was significantly lower than the control group; and in one, two, four, and eight weeks, MicroRNA-134 expression levels were still significantly lower than the control group. All these findings showed that, after the seizures, the epileptic content pertaining to the MicroRNA-134 of rat hippocampus-reduced in 3 days, and continuously maintained a low level. These results are similar to Yang Xiaolan's findings; which may also be related to the effects of pilocarpine-inhibiting the MicroRNA-134 expression and reducing the inhibition of synaptic plasticity related issues.
Immunohistochemistry and Western blot experiments have shown that the CREB expression of rat hippocampus levels of the six hours after seizure group and the 24-hour group had no obvious differences compared with the control group; on the other hand, the levels of the three-day, one-week, two-week, four- week, and eightweek groups significantly increased. The three-day group's p-CREB was also significantly higher, and this level was maintained for eight weeks. Therefore, the CREB and p-CREB expressions both increased; which may be related to the stimulation of the mossy fiber sprouting. In addition, both levels had the same trend; suggesting that CREB stimulates mossy fiber sprouting through p-CREB. Due to this observation, CREB and p-CREB had opposite trends verifying that CREB can be regulated by p-CREB. When the amount of MicroRNA-134 decreased, the CREB and p-CREB expressions would increase. This promotes a new abnormal synaptic formation, which leads to high levels of mossy fiber sprouting - forming an abnormal nerve grid loop in the brain tissue. Seizures occur through constant abnormal high-frequency discharge; but regulating the correlation of these three substances needs to be further confirmed. This study also provides us with new ideas for epileptic gene therapy applications, and developing new drugs at the genetic level - making it eventually possible to treat or prevent seizures.
4.3. Prospects
Although this study found that the MicroRNA-134 expression of the three-day group decreased and maintained a low level for eight weeks, this case still does not provide a clear answer after eight weeks. In addition, 24 hours after the seizure, the 24-hour group did not show any significant difference in the MicroRNA-134 levels, compared with the control group; but there were significant differences with the three-day group. Thus, these findings should still be refined, as well as a more precise timing including the findings on CREB and p-CREB. The detailed steps in regulating the MicroRNA-134, CREB and p-CREB levels are still not clear; which requires further experiments and new concepts exploring the signaling pathways. This also requires multi-disciplinary researchers to explore further and search for strong evidence that could be used as a basis for treating epilepsy.
In a word, the cause of epilepsy is still complex and vivid; and until now, the study of its pathogenesis is still in the animal experimental stage. The diversity of its pathogenesis further increased, which made it more difficult for clinicians and researchers. The mechanism of epilepsy still needs to be confirmed through more extensive research.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
This study was supported by Natural Science Foundation of China (No: 81371439).
[1] Chin JH. The global fund for epilepsy: a proposal. Neurology 2013; 80(8): 754-755.
[2] Newton CR, Garcia HH. Epilepsy in poor regions of the world. Lancet 2012; 380(9848): 1193-1201.
[3] Hui YY, Ahmad N, Makmor-Bakry M. Pathogenesis of epilepsy: challenges in animal models. Iran J Basic Med Sci 2013; 16(11): 1119-1132.
[4] Laxer KD, Trinka E, Hirsch LJ, Cendes F, Langfitt J. The consequences of refractory epilepsy and its treatment. Epilepsy Behav 2014; 37: 59-70.
[5] Griessenauer CJ, Bilal M, Kankirawatana P, Kulbersh B, Tubbs RS, Rozzelle C. Lymphatic malformation of the tongue with coexisting intractable epilepsy treated with corpus callosotomy: a case report. Clin Neurol Neurosurg 2014; 117: 68-70.
[6] Gross RE, Drane DL. Caveat E. Bitemporal intractable epilepsy: could it be surgically treated? Stereotact Funct Neuros 2013; 91(2): 134-137.
[7] Novy J, Logroscino G, Rossetti AO. Refractory status epilepticus: a prospective observational study. Epilepsia 2010; 51(2): 251-256.
[8] Hodaie M, Wennberg RA, Dostrovsky JO, Lozano AM. Chronic anterior thalamus stimulation for intractable epilepsy. Epilepsia 2002; 43(6): 603-608.
[9] Kerrigan JF, Litt B, Fisher RS, Cranstoun S, French JA, Blum DE, et al. Electrical stimulation of the anterior nucleus of the thalamus for the treatment of intractable epilepsy. Epilepsia 2004; 45(4): 346-354.
[10] Rossetti AO, Lowenstein DH. Management of refractory status epilepticus in adults: still more questions than answers. Lancet Neurology 2011; 10(10): 922-930.
[11] Rangaprakash D. Connectivity analysis of multichannel EEG signals using recurrence based phase synchronization technique. Comput Biol Med 2014; 46: 11-21.
[12] Ghareeb F, Duffau H. Intractable epilepsy in paralimbic Word Health Organization Grade II gliomas: should the hippocampus be resected when not invaded by the tumor? J Neurosurg 2012; 116(6): 1226-1234.
[13] Kwan P, Brodie MJ. Potential role of drug transporters in the pathogenesis of medically intractable epilepsy. Epilepsia 2005; 46(2): 224-235.
[14] Hodaie M, Wennberg RA, Dostrovsky JO, Lozano AM. Chronic anterior thalamus stimulation for intractable epilepsy. Epilepsia 2002; 43(6): 603-608.
[15] Du CL, Deng XJ, Wang F. Change and Significance of Semaphorin-3A and Mossy Fiber Sprouting in Epileptic Rats Induced by Pilocarpine. Chin J Neuroimmunol Neurol 2008; 15(6): 434-437.
[16] Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci USA 2011; 108(28): 11650-11655.
[17] Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 2008; 60(2): 201-214.
[18] Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med 2012; 18(7): 1087-1094.
[19] Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 2010; 466(7310): 1105-1109.
[20] Jin XF, Wu N, Li J. Roles of microRNA in drug addiction: research advances. J Int Pharm Res 2013; 40(5): 519-524.
[21] Huo ZH, Lin ZG, Shen H. Effect of simvastatin on chronic temporal epilepsy in rats. Chin J Neuromed 2012; 11(3): 250-253.
[22] Liu HQ, Meng L, Feng RF. The protective effect of edaravone on hippocampal neurons in kainic acid-induced epileptic rats. J Brain Nervous Dis 2012; 20(6): 413-416.
[23] Wei L. Progression of the pathogenesis of epilepsy. J Neurosci Mental Health 2012; 12(3): 313-315.
[24] Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Science 2011; 124(Pt 6): 833-838.
[25] Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL. Epigenetic mechanisms: critical contributors to long-term memory formation. Neuroscientist 2011; 17(6): 616-632.
[26] Eacker SM, Dawson TM, Dawson VL. The interplay of microRNA and neuronal activity in health and disease. Front Cell Neurosci 2013; 7: 136.
ment heading
10.1016/S1995-7645(14)60333-3
*Corresponding author: Yan Zhu, MD, Department of ICU, Zaozhuang Municipal Hospital, NO. 47, Longtou Road, Zaozhuang, Shandong, P.R. China.
Tel: 0632-3288776
E-mail: zhuyan3486@126.com
Foundation project: It is supported by National Natural Science Foundation (No: 81371439).
杂志排行
Asian Pacific Journal of Tropical Medicine的其它文章
- A brief review on biomarkers and proteomic approach for malaria research
- Trigonelline protects the cardiocyte from hydrogen peroxide induced apoptosis in H9c2 cells
- In vitro cholinesterase inhibitory and antioxidant effect of selected coniferous tree species
- Monascus pilosus-fermented black soybean inhibits lipid accumulation in adipocytes and in high-fat diet-induced obese mice
- Antiprotozoal assessment and phenolic acid profiling of five Fumaria (fumitory) species
- Profile and geographical distribution of reported cutaneous leishmaniasis cases in Northwestern Saudi Arabia, from 2010 to 2013
