THE CHARACTERIZATION OF PROPHENOLOXIDASE AND PHENOLOXIDASE FROM RED CLAW CRAYFISH, CHERAX QUADRICARINATUS
2015-11-05GUWeiCHENJingHUANGYanQingJINMingJianMENGQingGuoandWANGWen
GU Wei, CHEN Jing, HUANG Yan-Qing, JIN Ming-Jian, MENG Qing-Guo and WANG Wen
(1. Jiangsu Key Laboratory for Biodiversity & Biotechnology and Jiangsu Key Laboratory for Aquatic Crustacean Diseases, College of Life Sciences, Nanjing Normal University, Nanjing 210023, China; 2. Key and Open Laboratory of Marine and Estuary Fisheries,Ministry of Agriculture of China, East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Shanghai 200090, China; 3. Jiangsu Province Rudong County Fisheries Technology Extension Station, Nantong 226400, China)
Abstract: In this study, a prophenoloxidase (proPO) gene was cloned from hemocytes of red claw cray fi sh,Cherax quadricarinatus. The open reading frame (ORF) of proPO gene is 1995 bp and encodes a 665 amino acids (aa) protein. The predicted molecular mass of the protein was 75.7 kD with an estimated pI of 6.23. It contained two putative tyrosinase copper-binding motifs with six histidine residues and one thiol-ester-like motif, which showed similar structural features of proPOs from other crustaceans. The amino acid sequence of C. quadricarinatus proPO showed similarity of 68%, 63%, 63% and 59% with that of the proPO of Pacifastacus leniusculus, Homarus gammarus, Homarus americanus and Procambarus clarkii, respectively. The recombinant proPO were expressed in Escherichia coli BL21 with pET-28a expression vector. After protein purification and antibody prepared, the titer of rabbit anti-proPO serum was above 1︰12800. The mRNA transcription of proPO and PO activity in C. quadricarinatus were high in hemocytes, hepatopancreas and gills, lower values were seen in other tissues (nerve, heart, intestine and muscle). The immune characterization of proPO gene was studied after Spiroplasma eriocheiris and Aeromonas hydrophila stimulation. After S.eriocheiris or A. hydrophila challenging, the mRNA transcription of proPO gene and PO activity in hemocytes, hepatopancreas and gill of C. quadricarinatus were up-regulated at different times to resist different stimulation. These results indicated that it was potentially involved in the acute response against invading bacteria in C. quadricarinatus.
Key words: prophenoloxidase; Spiroplasma eriocheiris; Aeromonas hydrophila; Cherax quadricarinatus
The red claw crayfish, Cherax quadricarinatus belongs to the phylum Arthropoda, class Crustacea,order Decapoda, family Parastacidae and genus Cherax. It is native to Australia and Papua New Guinea, and is a tropical species[1]. It was introduced to China and cultured intensively and semi-intensively as an important species in aquaculture in Jiangsu,Zhejiang, Fujian and Guangdong Provinces[2,3]. Recently, the frequent outbreak of diseases has caused drastic decrease in its production and catastrophic economic losses in the industry. Understanding the immune defense mechanisms of crayfish may be conducive to the development of better disease control strategies in the crustacean farming.
It is known that invertebrates lack adaptive immune system, and rely instead on innate immune system against invading pathogens. Once pathogens like bacteria or viruses enter the host, they activate a complex system of innate defense mechanisms. First, it initiates the prophenoloxidase (proPO)-activating system is initiated in the presence of several microbial wall components like β-1,3-glucan, lipopolysaccharide(LPS) and peptidoglycan, which leads to the production of phenoloxidase (PO) by an prophenoloxidaseactivating enzyme (ppA), resulting in melanisation through a complex enzymatic cascade[4]. PO catalyses to formate the quinone by phenolic compounds. And quinine spontaneously becomes melanin. Quinone has antifungal functions and melanin is involved in wound healing and in encapsulation reactions[5].
The molecular biology study of C. quadricarinatus is little, there are few genes were reported including Heat shock protein 70, DDX5 gene and so on[2,3,6].The proPO gene has not reported and studied in C.quadricarinatus. The proPO gene was cloned and its characterizations were studied in this paper. The main objectives of the present study were (1) to clone and identify the full-length cDNA of proPO from C. quadricarinatus, (2) to investigate the proPO mRNA transcription and specific enzyme activity of PO in different tissues, (3) to examine the temporal transcription of proPO mRNA and the variation of specific PO activity in C. quadricarinatus post Spiroplasma eriocheiris and Aeromonas hydrophila challenge.
1 Materials and methods
1.1 Animal and RNA isolation
C. quadricarinatus [(100±5) g] were purchased from a market in Nanjing, China, and cultivated in 100 L tanks. The hemolymph from the base of crayfish last leg was obtained using a 2 mL syringe, quickly added into anticoagulant solution (glucose, 2.05 g; citrate,0.8 g; NaCl, 0.42 g; double distilled water was added to 100 mL). Samples were immediately centrifuged at 2000 g, 4 for 10min to collect the hemocytes. The
℃extraction of total RNAs from hemocytes was conducted by the Trizol, following the manufacturer’s instructions. The total RNA concentration was determined by measuring the absorbance at A260. Electrophoresis was used to check the RNA integrity.
1.2 Reverse transcription and gene cloning
Five μg of total RNA was reverse-transcribed into cDNA with an M-MLV RTase cDNA Synthesis Kit(Takara, Japan). The sequences of degenerate primer pairs, proPO-F and proPO-R (Tab. 1) were designed based on the highly conserved nucleotide of proPO gene by using the CLUSTAL program[7]to clone the sequence of proPO gene of C. quadricarinatus. PCR was performed using 1.5 mmol/L MgCl2, 0.2 mmol/L dNTP, 0.2 mmol/L of each of primers, 1 U TaqDNA polymerase (Takara, Japan) and 5 ng of cDNA. The ampli fi cation program consisted of 5min at 94℃ followed by 35 cycles of 94 ℃ for 30s, 52 ℃ for 40s,72 ℃ for 40s and a fi nal elongation step of 72℃ for 5min. PCR amplicons were size separated and visualized on an ethidium bromide stained 1% agarose gel.Amplicons of expected sizes were purified with an Agarose Gel DNA Purification Kit (Takara, Japan),and then subcloned into the pMD-19T cloning vector(Takara, Japan). Positive clones containing inserts of an expected size were sequenced using M13 primers,and sequenced at Invitrogen, Shanghai.
1.3 Rapid ampli fi cation of cDNA ends (RACE)
The proPO gene partial cDNA sequence from C.quadricarinatus was extended by using 5′ and 3′RACE (SMARTTMcDNA kit). A total of three genespeci fi c primers (Tab. 1) were designed based on the genes partial cDNA sequences. The 3′ RACE PCR reaction was carried out in a total volumeof 50 μL containing 2.5 μL (800 ng/μL) of the first-strand cDNA reaction as a template, 5 μL of 10 × Advantage 2 PCR buffer, 1 μL of 10 mmol/L dNTPs, 5 μL(10 mmol/L) gene-speci fi c primer, 1 mL ofUniversal Primer A Mix (UPM; Clonetech, USA), 34.5 μL of sterile deionized water, and 1 U 50 × Advantage 2 polymerase mix (Clonetech, USA). For the5′ RACE,UPM was used as forward primers in PCR reactions in conjunction with the reverse gene-speci fi c primers.PCR ampli fi cation conditions for both the3′ and 5′RACE were as follows: 5 cycles at 94 ℃ for 30s, 72℃for 3min; 5 cycles at 94 ℃ for 30s, 70 ℃ for 30s, and 72 ℃ for 3min; 20 cycles at 94 ℃ for 30s, 68℃ for 30s,and 72 ℃ for 3min. After linking into the vector, the samples were sequenced at Invitrogen.
1.4 Sequence analysis of proPO
The cDNA sequence and deduced amino acid sequence of proPO gene were analyzed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast).Translation and protein analyses were performed using ExPASy tools (http://us.expasy.org/tools/). The ClustalW Multiple Alignment program (http://www.ebi.ac.uk/clustalw/) was used to create the multiplesequence alignment. An unrooted phylogenetic treewas constructed based on the amino sequences alignment by the neighbor-joining (NJ) algorithm embedded inMEGA 4 program. The reliability of the branching was tested by bootstrap resampling (1000 pseudo-replicates).
1.5 Protein expression, purification andantibody preparation
ProPO-ORF-F and proPO-ORF-R were designed to amplify the ORF of C. quadricarinatus proPO gene.After digested with restriction enzymes (EcoRⅠand Xho Ⅰ ), theproduction was ligated into the pET-28a expression vector (Novagen, USA). The resulting recombinant plasmids pET-proPO transformedto E. coli BL21 (DE3) (Trans, China) for protein expression.The recombinant proteins were analyzedby SDSPAGE. High-Affinity Ni-NTA (nickel-nitrilotriacetic acid) Resin (Jinsite, China) was used to purify the recombinant proteins according to the instructions.The New Zealand White rabbits were immunized with 100 μg of purified protein that was homogenized in complete Freund’s adjuvant for three times at 2-week intervals. A boost injection in incomplete Freund’s adjuvant was given for another week. Rabbit serum was collected seven days after the last immunization.Antibody titer was determined from the immunized rabbits by ELISA. Briefly, the optimum concentration(0.5 μg per well) of recombinant protein diluted in 0.05 mol/L carbonate buffer, was coated onto a 96-well plate (Corning, USA) and then incubated 12h at 4 ℃ with blocking solution (5% skimmed milk powder in TBS). The plate was washed three times with TBST. Next, rabbit antiserum (primary antibody)of different dilutions was added to each well and incubated for 1h. After washing with TBST, the plates were incubated with 100μL of a 1︰5000 dilution of HRP-conjugated goat anti-rabbit IgG for 30min. The plates were washed again and 100 μL of TMB/H2O2substrate was added to each well. Optical density was measured at 450 nm with a Model 680 micro-plate reader (Bio-Rad, USA).
1.6 Tissue distributionof proPO mRNA transcription and PO activity
A sample of total RNA at 5 μg from C. quadriccarinatus tissues including nerve, heart, intestine,hemocytes, gill, and hepatopancreas were reversetranscribed respectively into cDNA with a PrimeScript RT reagent Kit (Takara, Japan), respectively. Realtime PCR was carried out with a Mastercycler ep realplex (Eppendorf, Germany) to study the transcription of proPO mRNA in tissues, respectively. The PCR reaction was performed in a 25 μLvolume with a SYBR Premix Ex TaqTMKit (Takara, Japan), 2 μmol/L of each specific primer and 1 μL of cDNA in Mastercycler ep realplex (Eppendorf,Germany). The primers pair using in real-time PCR was list in Tab. 1.The primers β-actin-F and β-actin-R were used to amplify β-actin fragment that was used as a housekeeping gene. The relative transcription level ofproPO was calculated according to the 2−ΔΔCTmethod[8]. Statistical analysis was performed using SPSS software (Ver11.0).Data represent the mean ± standard error (S.E.). Statistical significance was determined by one-way ANOVA and post-hoc Duncan multiple range tests. The signi fi cance level was set at P<0.05.

Tab. 1 Degenerate and speci fi c primers used for proPO cloning in the experiment
Three unchallenged C. quadricarinatus were dissected in cold PBS to isolate tissues, including hepatopancreas, gill, muscle, heart, nerve, intestine and hemocytes. The dissected tissues were added to nine volume of cold lysis buffer (415 mmol/L NaCl,100 mmol/L glucose, 10 mmol/L cacodylic acid, 5 mmol/L CaCl2, pH 7.0) and homogenized. Cell debris was removed by centrifuging at 3000 g for 10min to prepare crude PO samples after sonication at 40 W for 2min. PO activity was quantified by monitoring the rate of dopachrome formation by using a colorimetric assay[9]. 100 μL of HLS or crude PO sample was mixed with 250 μL 100 mmol/L PBS (pH 6.8) in a 1.5 mL Eppendorf tube. Then, 350 μL of the pre-cold 15 mmol/L DOPA dissolved in PBS was added to the Eppendorf tube. The tube was immediately capped,inverted, warmed at 30℃, blanked on its own absorbance and monitored for changes in optical density at 490 nm for over 10min. PO activity was recorded as the maximum change in absorbance over any one minute interval (ΔOD490nm/min) during the first 10min of the assay. The total protein of different samples was determined by the Bradford method using BSA as standards.
1.7 proPO mRNA transcription and PO activity under pathogen stimulation
The C. quadricarinatus were cultivated for 1 week before treatment. S. eriocheiris was maintained in pure culture in R2 liquid medium at 30 ℃ . The A. hydrophila was grown at 28 ℃ in TSB medium. The C. quadricarinatus in the experimental group I (30 individuals)received an injection of 100 μL S. eriocheiris (107spiroplasmas/mL) individually. The C. quadricarinatus in the experimental group II (30 individuals) received an injection of 100 μL A. hydrophila (105cells/mL)individually. The C. quadricarinatus (30 individuals)receiving an injection of 100μL PBS (pH=7.0) individually, were used as control groups. Every 3 individuals for one group were randomly collected at 0h, 1h,3h, 6h, 12h, 24h, 48h and 72h post-injection.
The total RNA from hemolymph, gill and hepatopancreas was extracted from different groups by the Trizol technique. After reverse-transcribed into cDNA,real-time RT-PCR was carried out to measure the transcription levels of proPO gene. The total crude PO from different tissues (hemolymph, gill and hepatopancreas) was prepared. ThePO activity at different time after pathogen challenge was measured. The methods of experiment and analysis were list in 1.6.
2 Results
2.1 Sequence of proPO cDNA
The full-length proPO gene cDNA of C. quadricarinatus consisted of 2951 bp, including 75 bp in the 5′-untranslated region, an open reading frame (ORF)of 1995, and 881 bp in the 3′-untranslated region including a stop codon (TAG), putative polyadenylation consensus signal (AATAAA)and a poly (A) tail. The ORF of proPO cDNA was found to be composed of 665 amino acids (aa). The calculated molecular mass of the proPO was 75.7 kD with an estimated pI of 6.23. The C. quadricarinatus proPO cDNA sequence and their deduced amino acid sequence were submitted to the NCBI GenBank under accession number JQ954858.
A thiol ester motif (GCGWPEHL) from 558 to 565 that is present in the complement components, C3,C4 and a2-macroglobulin, was also observed in C.quadricarinatus proPO. No hydrophobic signal was found in the proPO sequence. Three N-linked glycosylation sites and two putative tyrosinasecopper binding motifs with six histidine residues (177, 181, 203 in copper A, and 337, 341, and 377 in copper B) were conserved in what.
2.2 Phylogenetic analysis of proPO
The amino acid sequence of C. quadricarinatus proPO showed similarity of 68%, 63%, 63% and 59%with that of the proPO of Pacifastacus leniusculus,Homarus gammarus, Homarus americanus and Procambarus clarkii, respectively. The phylogenetic analysis indicated that the proPO of C. quadriccarinatus is homologous to that of P. leniusculus and P.clarkii. The proPO of C. quadricarinatusis is close to lobsters proPO and far away from crabsproPO (Fig. 1).
2.3 Protein expression, purification and antibody preparation
Express this as recombinant protein, proPO, with an apparent molecular weight of around 66 kD, was detected by SDS-PAGE (Fig. 2, lane 2), which could be purified by High-Affinity Ni-NTAResin (Fig. 2,lanes 3). The titer of anti-proPO antibody against rabbit was determined by ELISA (Fig. 3). The titer of anti-proPO serum was above 1︰12800 and below 1︰51200 as tested by ELISA.
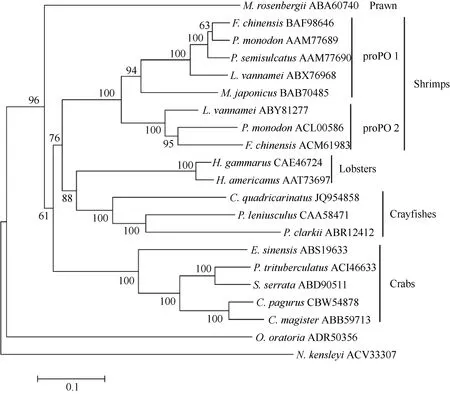
Fig. 1 Neighbor-joining phylogentic tree of proPO amino acid sequences from different species of animals

Fig. 2 Analysis of recombinant C. quadricarinatus proPO by SDS-PAGE

Fig. 3 Antibody titer was determined by ELISA
2.4 Tissue distribution of proPO mRNA transcription and PO activity
As determined by real-time RT-PCR, proPO transcription was widely observed in the nerve, hepatopancreas, hemocytes, gill, heart, intestine and muscle of C. quadricarinatus (Fig. 4A). The transcription of proPO was highest in hemocytes, followed by hepatopancreas and gills, lower levels were seen in other tissues (heart, intestine, muscle and nerve).
Similar to proPO mRNA transcription in tissues of C. quadricarinatus, the PO activities also could be detected in all examined tissues by the traditional colorimetric method. The highest activities were observed in hepatopancreas, followed by hemocytes and gill,and lower PO activities were seen in other tissues (intestine, heart, muscle and nerve) (Fig. 4B).
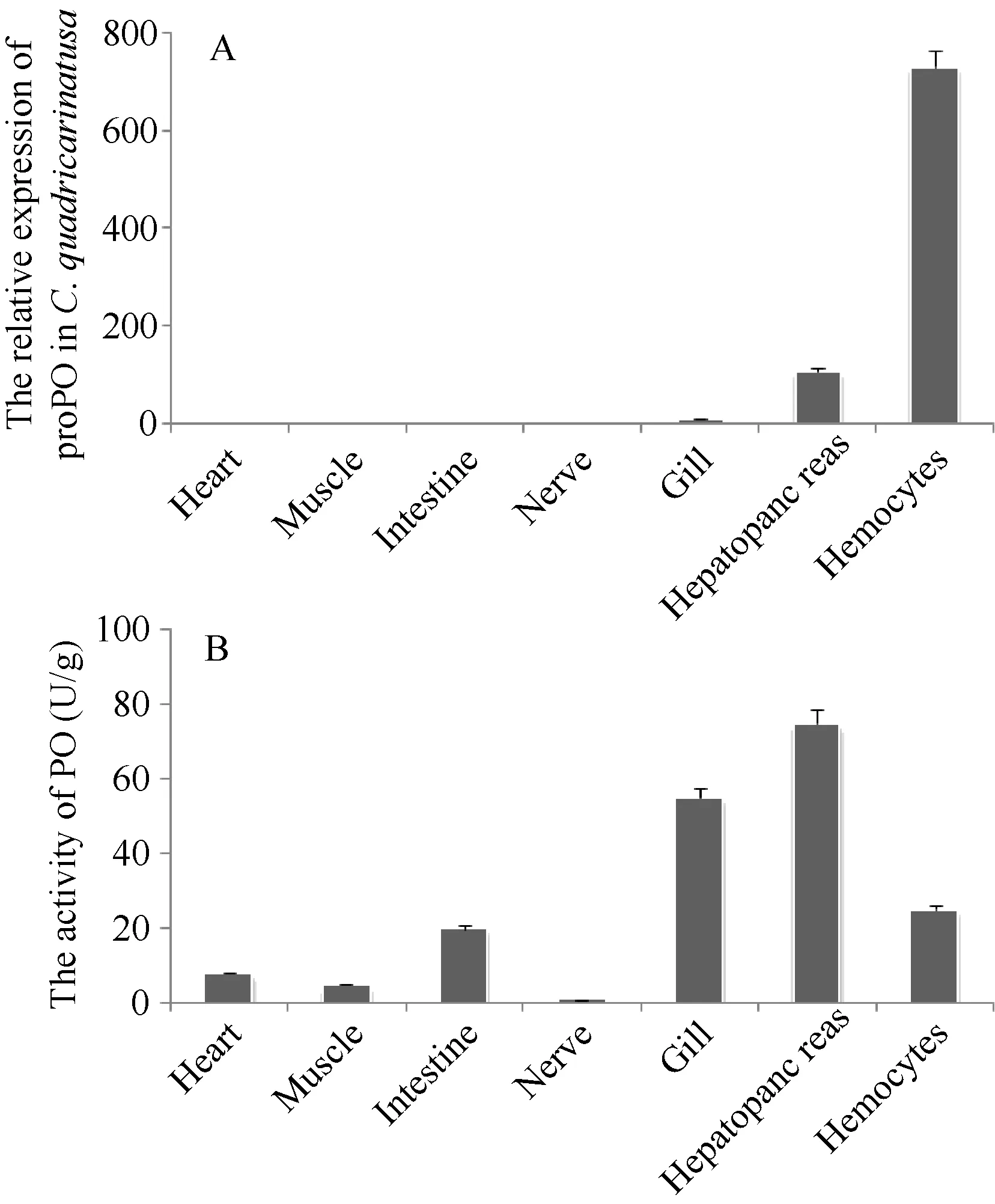
Fig. 4 Tissue-dependent proPO mRNA transcripts (A) and PO activity (B) of C. quadricarinatus
2.5 ProPO mRNA transcription pattern under pathogens stimulations
The proPO mRNA transcription pattern after pathogens challenge are shown in Fig. 5. The transcript of proPO mRNA in hemocytes was up-regulated at 48 and 72h after the S. eriocheiris or A. hydrophila challenge. The proPO mRNA transcription in hemocytes was also up-regulated at 6h after the A. hydrophila stimulation (Fig. 5A). The proPO mRNA in hepatopancreas was up-regulated at 1h after the S.eriocheiris or A. hydrophila challenge. And after S.eriocheiris challenge, proPO mRNA in hepatopancreas was also up-regulated at 3, 6 and 12h (Fig. 5B).The proPO mRNA transcription pattern in gills after A.hydrophila challenge is up-regulated at 24, 48 and 72h.The transcript of proPO mRNA in the gills was up-regulated at 1 and 72h after the S. eriocheiris challenge (Fig. 5C).
2.6 PO activities variation in C. quadricarinatus following pathogen stimulation

Fig. 5 The transcript levels of proPO in hemocytes (A), hepatopancreas (B) and gills (C) of C. quadricarinatus after S. eriocheiris or A. hydrophila challenge
Similar to proPO mRNA transcription, the PO activities were increased during the time course of hemocytes, hepatopancreas and gills post S. eriocheiris or A. hydrophila stimulation (Fig. 6). There were two peaks of PO activities in hemocytes post-stimulation. The first peak was observed at 1h post-injection, which was 1.88-fold to that in control group (P>0.05). Afterwards, the PO activities decreased to normal level at 3h. As time progressed, the PO activities was increased at 12, 24, 48 and 72h post pathogen stimulation (also at 6h post A. hydrophila stimulation) (Fig. 6A). The PO activities remained at normal level during the first 3h after pathogen stimulation, then increased from 6h (Fig. 6B). Similar to the PO activities in hemocytes, there were two peaks of the PO activities in gills post pathogen stimulation.The PO activities increased at 1 and 3h, and reached the first peak at 3h. After decrease to normal level at 6h (also at 12h post S. eriocheiris stimulation), the PO activities gradually increased after 12h post pathogenic stimulation (Fig. 6C).
3 Discussion

Fig. 6 The PO activity in hemocytes (A), hepatopancreas (B) and gills (C) of C. quadricarinatus after S. eriocheiris or A. hydrophila challenge.
The first proPO gene was cloned from the freshwater crayfish Pacifastacus leniusculus in 1995[10]. To date, more than 40 proPO genes have been obtained from about 30 species in crustaceans, such as P. leniusculus, Macrobarchium rosenbergii[11,12],Penaeus monodon[13], Litopenaeus vannamei[14—16],Fenneropenaeus chinensis[17,18], Marsupenaeus japonicus[19], P. semisculcatus (AAM77690), P. clarkii[14],H. americanus (AAT73697), H. gammarus[20], Eriocheir sinensis[21], Portunus trituberculatus[22], Scylla serrata[7], Cancer pagurus (CBW54878) and C.magister[23]. The C. quadricarinatus proPO has not cloned and studied its characteristic. Therefore the immune characteristic of C. quadricarinatus proPO was studied in this paper.
In the present study, the cDNA encoding prophenoloxidase was cloned from C. quadricarinatus.The 2951 bp full-length cDNA contained a 1995 bp open reading frame (ORF) encoding a putative proPO protein of 665 amino acids, a 5′-UTR of 75 bp and a long 3′-UTR (881 bp). The analysis of the amino acid sequence of this fragment showed that it contained six histidine residues which define two putative copper binding motifs for tyrosinase and also contained a thiol ester-like motif (GCGWPEHL) just like the structural features of other proPOs from other crustacean. This sequence has similarity of 69%, 63%, 63%and 59% to proPO-deduced amino acid from P.leniusculus, H. gammarus, H. americanus and P.clarkii. As a unique defense system of invertebrate,proPO system is of pivotal importance in crustacean immune response. The MW of proPO from crustacean is almost about (75—80) kD. The calculated MW of the proPO was 75.7 kD which is bigger than expression in E. coli (66 kD) (Fig. 2). SDS-PAGE is a common method to determinate the MW of protein. But sometimes MW analyzed using SDS-PAGE is inconsistent with the calculated MW. This may be caused by inappropriate electrophoresis conditions, changes of protein structural, irregular electric charge distribution and post-translational modification such as phosphorylation[24]. But the reasons for the deviation have not been fully elucidated now. Phylogenetic analysis revealed that C. quadricarinatus proPO is distinct far away from proPOs of Insecta, and is also distinct from proPO of shrimps, crabs and prawn. Multiple alignments indicated that the proPO of C. quadricarinatus forms monophyletic subgroup with proPO of P.Leniusculus and P. clarkii. The present study also indicated that the decapod crustacean proPOs can be probably classified into five distinct branches: prawn,crayfish, lobster, shrimp, and crab.
The distribution of proPO in different tissues has been investigated in a variety of organisms. In the present study, the proPO of C. quadricarinatus were highly transcription in hemocytes, hepatopancreas and gills, lowly transcript levels were seen in other tissues(muscle, heart, intestine and nerve). The proPO mRNA transcripts were strongly expressed in hemocytes of C. quadricarinatus. The same situation appears in mud crab S. serrata[7], crayfish P. Leniusculus[25], white shrimp L. vannamei[14—16], freshwater prawn M. rosenbergii[11], F. chinensis[17]and P. trituberculatus[22]. The proPO transcript was located in hemocytes, and released into the circulation, but not transcription in other tissue of crayfish P. leniusculus[25]. But the PO activities were detectable in all examined tissues, including muscle, heart, intestine and nerve with the highest level of both in hepatopancreas. The possibility of the widely distributed presence of PO activities could be the result of the proPO released into the circulation and other different tissues. The high PO activity in hepatopancreas might be caused of the cooperation of multiple POs or the cross interference of hemocyanin, which had been demonstrated to possess PO activity in crayfish[26—28].
Both proPO mRNA transcription and protein activity levels post stimulation were constructive to understand its roles in the immune mechanisms. In the present study, C. quadricarinatus proPO mRNA transcriptions in hemocytes, hepatopancreas and gills were up-regulated after S. eriocheiris and A. hydrophila challenges. The mRNA transcriptions of proPO in hemocytes were up-regulated at 48h and 72h post S.eriocheiris or A. hydrophila stimulation (also upregulated at 6h post A. hydrophila stimulation). The PO activity in hemocytes signi fi cantly increased and reached two peaks at 1h and 72h after pathogen stimulation. The up-regulation of proPO mRNA transcriptions and PO activity in response to different stimulations was reported in S. serrata[7], Astacus astacus[29], H. gammarus[20]and E. sinensis[21], which was in agreement with our results. The changing trends of proPO mRNA transcriptions and PO activity in hepatopancreas in response to stimulations were different. The mRNA transcriptions of proPO in hepatopancreas were up-regulated at the first 12h after pathogenic challenging. But the PO activity increased beginning at 6h. This case maybe result from that proPO system is an energy-consuming, multifunctional mechanism[12], it might function stepwise in different phases of the host-pathogen interaction. The mRNA transcriptions of proPO in gills were up-regulated at 1h and 72h post S. eriocheiris stimulation and at 24h, 48h and 72h post A. hydrophila stimulation. The PO activity in gills signi fi cantly increased and reached two peaks at 3h and 72h after pathogen stimulation. The increase of proPO transcription and PO activity at first several hours maybe cause by the stimulation of pathogen. And the proPO transcription and PO activity increased at 72h maybe cause by appeared of immune response. This result states the proPO may have relationship with the immune response of hosts to resist pathogen. But further investigations are required to better understand the regulation mechanism of proPO to resist pathogen stimulation.
In conclusion, proPO gene was cloned here for the first time from hemocytes of C. quadricarinatus.The proPO immune functions of C. quadricarinatus in hemocytes, hepatopancreas and gill were confirmed by real-time PCR, and PO activity was also analyzed in hemocytes, hepatopancreas and gill after S. eriocheiris and A. hydrophila challenges. The result will help us to understand the characteristic and function of proPO from C. quadricarinatus.
